Abstract
Group B Streptococcus agalactiae bacteria (group B streptococci [GBS]) are the most common cause of serious bacterial infection in newborn infants. The majority of serotype III-related cases of neonatal disease are caused by a genetically related subgroup of bacteria, restriction fragment digest pattern (RDP) type III-3, suggesting that these strains possess unique genes contributing to virulence. We used genomic subtractive hybridization to identify regions of genomic DNA unique to virulent RDP type III-3 GBS strains. Within one of these III-3-specific regions is a 1,506-bp open reading frame, spb1 (surface protein of group B streptococcus 1). A mutant type III GBS strain lacking Spb1 was constructed in virulent RDP type III-3 strain 874391, and the interactions of the wild-type and spb1 isogenic mutant with a variety of epithelial cells important to GBS colonization and infection were compared. While adherence of the spb1 isogenic mutant to A549 respiratory, C2Bbe1 colonic, and HeLa cervical epithelial cells was slightly lower than that of the 874391 strain, invasion of the Spb1− mutant was significantly reduced with these cell lines compared to what was seen with 874391. The defect in epithelial invasion was corrected by supplying spb1 in trans. These observations suggest that Spb1 contributes to the pathogenesis of neonatal GBS infection by mediating internalization of virulent serotype III GBS and confirm that understanding of the population structure of bacteria may lead to insights into the pathogenesis of human infections.
For most of the last century, group B Streptococcus agalactiae bacteria (group B streptococci [GBS]) were best known as an important cause of bovine mastitis. For unknown reasons, these bacteria emerged in the mid-1970s as the most common cause of serious bacterial infection in newborn human infants and an important pathogen in parturient women (3, 4). GBS are subdivided into nine serotypes, based on the structure of the type-specific polysaccharide capsule. Type III GBS are of particular interest since this is the most common serotype identified in neonatal and maternal infections and the most common serotype causing neonatal meningitis (17, 19).
Our understanding of the pathogenesis of GBS infections is incomplete. Up to 40% of pregnant women carry GBS in their genitourinary or gastrointestinal tracts (3). The majority of neonatal infection cases presenting in the first week of life result from vertical transmission of bacteria from a colonized mother (4). It is postulated that early-onset GBS infection results from aspiration of infected vaginal secretions or amniotic fluid by the infant, followed by bacterial adhesion to and invasion of respiratory epithelium and endothelium (26, 27). Pneumonia and pulmonary hypertension occur in up to 80% of early-onset infections, thus supporting this hypothesis (3).
The specific molecular interactions that are responsible for the colonization of pregnant women and the subsequent colonization and infection of infants are not known. GBS bind to a variety of extra- and intracellular matrix proteins, including laminin, fibronectin, and cytokeratin 8 (29, 34, 35). Pretreatment of GBS with proteases decreases adhesion to epithelial cells, suggesting that GBS cell surface proteins are important to this process (33). Preincubation of epithelial cells with hydrophobic GBS proteins or preincubation of GBS with antibody directed against these proteins diminishes bacterial binding, findings that also support the existence of specific GBS epithelial cell adhesins (37). GBS expressing cell-associated β-hemolysin invade A549 epithelial cells more efficiently than do β-hemolysin-deficient mutant strains (14). Otherwise, the molecular mechanisms that permit adherent GBS to invade host epithelium and endothelium have not been clarified.
Restriction fragment digest patterns (RDP) have been used to divide serotype III GBS into three genetically related subgroups (22). RDP type III-3 GBS are responsible for over 90% of invasive neonatal infections in both Salt Lake City, Utah, and Tokyo, Japan, suggesting that these strains may cause most neonatal disease worldwide (32). RDP type III-2 strains, in contrast, are significantly more likely to be colonizing strains isolated from healthy pregnant women who subsequently give birth to healthy infants. These data suggested that invasive RDP type III-3 strains possess virulence factors not present in less virulent RDP type III-2 strains and that these virulence genes might be identified by genomic subtractive hybridization. This approach led to the identification of a novel protein of serotype III GBS, Spb1 (surface protein of group B streptococcus 1), that mediates invasion of epithelial cells, an important process in early-onset neonatal infection.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The isolation and characterization of strains 874391 and 865043 used in this study has been described previously (32). Strain 874391 is a virulent RDP type III-3 GBS and 865043 is a less virulent RDP type III-2 strain. Serotype III strain COH-1 was kindly provided by Craig Rubens, University of Washington. GBS were grown on Columbia Blood agar or Todd-Hewitt (TH) agar and in TH broth, with 10 μg of erythromycin/ml for antibiotic selection as indicated. Escherichia coli XL1-Blue (Stratagene, La Jolla, Calif.) was grown on Luria-Bertani agar or in LB broth, with 350 μg of erythromycin/ml or 100 μg of ampicillin/ml for antibiotic selection. The pBS KSII and pBC (Stratagene) and pCR II plasmids and λFIX II bacteriophage (Invitrogen, Carlsbad, Calif.) were obtained commercially. The conditionally replicating shuttle vector pHY304 was generously provided by C. Rubens, University of Washington. This plasmid contains a temperature sensitive ori which is active at 30°C but not at 37°C, an ermr erythromycin resistance gene, and a multiple cloning site derived from pUC19. The low-copy-number shuttle vector pMS3545 that also contains ermr was provided by G. Dunny, University of Minnesota (11).
Epithelial cell culture.
A549 respiratory epithelial cells (no. CCL-185; American Type Culture Collection [ATCC], Manassas, Va.) were propagated in Ham's F12K medium with 2 mM l-glutamine, 1.5 g of NaHCO3/liter, and 10% fetal bovine serum (FBS). C2BBe1 (no. CRL-2102; ATCC) cells were propagated in Dulbecco's modified Eagle's medium with 4 mM l-glutamine, 1.5 g of sodium bicarbonate/liter, 4.5 g of glucose/liter, 1.0 mM sodium pyruvate, 0.01 mg of human transferrin/ml, and 10% FBS. HeLa cervical epithelial cells (no. CCL-2; ATCC) were propagated in Eagle's minimum essential medium with 2 mM l-glutamine and Earle's balanced salt solution, 1.5 g of sodium bicarbonate/liter, 0.1 mM nonessential amino acids, 1.0 mM sodium pyruvate, and 10% FBS.
Genomic subtractive hybridization.
Genomic subtractive hybridization was performed as previously described (9). Briefly, DNA was prepared from virulent RDP type III-3 strain 874391 and avirulent RDP type III-2 strain 865043 by mutanolysin and proteinase K digestion (22). Genomic DNA from each strain was digested with TaqI restriction endonuclease (Gibco Invitrogen). Two pairs of oligonucleotide adapters were alternated for each round of subtractive hybridization: taqA (5′ CTAGGTGGATCCTTCGGCAAT 3′) and taqB (5′ CGATTGCCGA 3′); taqE (5′ AGGCAACTGTGCTAACCGAGGGAAT 3′) and taqF (5′ CGATTCCCTCG 3′). Three rounds of subtraction and amplification were performed using 0.5 μg of adapter-ligated virulent 874391 DNA for the first cycle, 200 ng for the second cycle, and 5 pg for the third cycle. Forty micrograms of TaqI-digested avirulent RDP strain 865043 was used in each subtraction. Following the final subtraction-amplification reaction, amplification products were cloned into pCR II vectors.
Southern blot and dot blot hybridization.
Genomic DNA was prepared from GBS as previously described (32). Nine DNA sequences that are present in all isolates of the RDP III-3 phylogenetic lineage but not in the RDP type III-2 lineage were identified by genomic subtractive hybridization (10). For dot blots, genomic DNA was applied to membranes and hybridized with radiolabeled DY-1 and spb1 probes. For Southern blots, genomic DNA was digested with EcoRI and electrophoresed in a 0.8% Tris-borate-EDTA agarose gel. Restriction fragments were transferred to nylon membranes and hybridized with fluorescein-labeled probes (Gene Images; Amersham Biosciences, Piscataway, N.J.) following the manufacturer's protocol.
Genomic library construction and screening.
A GBS RDP type III-3 874391 genomic DNA library was constructed in λFIX II (Stratagene) by partial digestion of genomic DNA with BglII restriction endonuclease and partial fill-in with dGTP and dATP according to the manufacturer's protocol. DNA sequencing was performed using ABI Prism 3700 DNA analyzers (Applied Biosystems, Foster City, Calif.).
spb1 deletion mutant.
The spb1 open reading frame (ORF) and approximately 200 bp of 5′- and 3′-flanking genomic DNA were amplified by PCR and subcloned into pBS KSII, and an 800-bp intragenic HindIII fragment was excised to create pBS/spb1−. The truncated spb1 gene was cloned into the targeting vector pHY304, transformed into competent 874391 strain GBS, and incubated at 30°C (15). An erythromycin-resistant colony was grown in TH broth with erythromycin at 30°C for 3 h, and the presence of the pHY304/spb1− plasmid was confirmed by plasmid purification and restriction digestion. Bacteria were then diluted 1:100 in fresh medium, incubated at 37°C for 3 h, and plated onto TH-erythromycin plates at 37°C. An erythromycin-resistant colony, containing an integrated copy of the targeting vector, was inoculated in 5 ml of TH broth without erythromycin, grown for 3 h at the permissive temperature of 30°C, and plated on TH plates at 37°C. Genomic DNA was prepared from erythromycin-susceptible colonies, and PCR amplification using spb1-specific primers Southern blotting with an spb1 probe from pBS/spb1− was performed to confirm the presence of a truncated spb1 gene (1).
To complement the spb1 mutation in trans, the complete spb1 coding region and 500 bp of upstream flanking sequence were cloned into pMS3545 and transformed into competent Spb1− strain GBS, and an erythromycin-susceptible colony was selected. The presence of the pMA3545/spb1 expression vector in this strain (Spb1c) was confirmed by plasmid purification and restriction digestion. To examine the effect of providing spb1 in trans on an RDP type III-2 strain, pMA3454/spb1 was also used to transform competent 865043 strain GBS.
Hemolytic activity of GBS strains.
The hemolytic activities of the GBS strains 874391 and Spb1 were determined by their ability to lyse sheep red blood cells as previously described (25). The hemolytic activities of these strains were also compared with that of the previously characterized type III GBS strain COH-1. The hemolytic activity of each strain was expressed as a hemolytic index, with the activity of strain COH-1 designated as 1.
Adherence to and invasion of epithelial cells.
Three epithelial cell lines were used in in vitro studies of GBS adherence and invasion. A549 pulmonary epithelial cells served as a model for neonatal respiratory colonization and infection, and HeLa cervical epithelial cells served as a model for maternal colonization and maternal infection. It has been hypothesized that neonatal infections after the first week of life may originate from a gastrointestinal rather than pulmonary focus (5). C2BBe1 colonic epithelial cells, derived from the Caco-2 cell line, were used as a model of intestinal colonization and possible late-onset neonatal infection. These cells form a polarized monolayer that is more homogeneous than Caco-2 cells and morphologically more comparable to the human colon since villi are located exclusively on apical surfaces (24).
An overnight culture of each bacterial strain was diluted 1:10 in TH broth and grown to an optical density at 600 nm of 0.900 (5 × 108 bacteria/ml). Bacteria were washed twice in phosphate-buffered saline (PBS) and diluted to the desired inoculum in 100 μl of tissue culture medium with a 1/100 dilution of HEPES. The ability of GBS to adhere to epithelial cell layers was compared by using the assay described by Tamura and Rubens (35), except that cultures of serial dilutions of bacteria were performed to quantify adherent cells instead of using radiolabeled bacteria. Adherence assays were performed at 4°C to limit the numbers of GBS invading epithelial cells during the assay. Adherent bacteria were removed from wells by scraping and sonicated briefly to disrupt chains, and quantitative cultures were plated on TH agar plates with antibiotic selection as indicated. In each case, several different inocula were tested over at least a fivefold range of multiplicity of infection (MOI; expressed as the number of bacteria per eukaryotic cell). Bacteria were tested in triplicate for each assay in at least three separate experiments.
The ability of GBS to invade epithelial cell layers was determined as described by Rubens et al. (27). Briefly, epithelial cells were grown to confluence in 24-well tissue culture plates (Corning). Following infection with GBS, cells were incubated for 2 h at 37°C in an atmosphere of 5% CO2. The medium was aspirated, and wells were gently washed three times with 500 μl of PBS. Adherent extracellular bacteria were killed by the addition of 1 ml of tissue culture medium with 5 μg of penicillin/ml and 100 μg of gentamicin/ml, followed by incubation for 2 h at 37°C in 5% CO2. Plates were washed twice with 500 μl of PBS, cells were lysed, and serial dilutions were plated as described above. Again, several different inocula were tested over a 10-fold range of MOI (expressed as the number of bacteria per eukaryotic cell). Bacteria were tested in triplicate for each assay in at least three separate experiments.
Statistical analysis.
Continuous variables were analyzed by Student's t or Mann-Whitney test.
RESULTS
Identification of spb1.
The genomic subtraction procedure of Lisitsyn et al. was modified to identify candidate virulence genes unique to RDP type III-3 GBS strains (20). TaqI, a restriction endonuclease resulting in relatively small restriction fragments (mean size, 1,200 bp), was selected for restriction digestions because of the tendency of PCR to preferentially amplify short DNA segments, resulting in a bias against the identification of genomic sequences present on large restriction fragments. DNA from TaqI-digested virulent RDP type III-3 strain was marked by ligation with oligonucleotide adapters prior to denaturation and mixing with TaqI-digested DNA from an avirulent RDP type III-2 strain. Following annealing, this mixture was amplified with primers corresponding to adapter sequences. Genomic DNA unique to virulent RDP type III-3 strains is amplified logarithmically by this strategy, whereas hybrid DNA molecules common to both RDP types are amplified arithmetically, and DNA unique to avirulent RDP type III-2 strains is not amplified. Following three cycles of subtraction and amplification, subtracted amplicons were cloned into plasmid vectors. The specificity of each probe for virulent RDP type III-3 strains was confirmed by hybridization to genomic DNA from a large panel of type III strains of GBS (10).
The RDP type III-3-specific sequence tag DY-1 was selected for further analysis. The 166-bp DY-1 nucleic acid and translated amino acid sequences are not homologous to known bacterial genes entered in the GenBank database. Additional sequence information from the flanking regions of the DY-1 sequence was obtained from genomic DNA clones from an RDP type III-3 library. The library was constructed using λFIX II phage vector, and the resulting library of 1.7 × 105 recombinant phage was amplified once. Multiple plaques hybridizing with the DY-1 sequence tag were purified and three overlapping genomic clones, with approximate sizes of 9, 22, and 23 kb, were identified. Since the boundaries of GBS III-3-specific segments of the chromosome were not known, a 6.4-kb SalI-BglII restriction fragment that was common to all phage clones and that hybridized with the DY-1 probe was subcloned and both strands were sequenced.
A 1,509-bp ORF, spb1 (the designation derives from surface protein of GBS 1), is the second of two relatively homologous ORFs (43% nucleic acid and 22% predicted amino acid identity) that are located adjacent to one another in the same transcription orientation 200 bp upstream of the DY1-1 probe sequence. Sequencing of >1,200 nucleotides of the genomic clone 3′ of spb1 failed to identify additional significant ORFs 3′ of spb1. Like the DY-1 probe, dot blot hybridization studies of 62 type III clinical isolates demonstrated that spb1 was present only in RDP type III-3 GBS strains and not in representative isolates from serotype III RDP type III-2 or III-1 strains (data not shown).
The spb1 coding region is preceded by a potential ribosomal binding site 10 bases upstream from the ATG start codon (Fig. 1). The predicted Spb1 protein (502 amino acids [aa]; Mr, 53,446) has the structural characteristics of a gram-positive cell wall-bound protein (Fig. 1). The N terminus of the predicted protein is comprised of a hydrophilic, basic stretch of 6 aa followed by a 23-aa hydrophobic, proline-rich core, consistent with a signal peptide. The hydrophilic mature protein terminates in an LPXTG domain that immediately precedes a hydrophobic 20-aa core and a short, basic hydrophilic terminus. The spb1 nucleotide sequence is not homologous to other known bacterial genes, and this ORF is not present in the recently reported type III or type V GBS genome sequences (16, 36). The translated amino acid sequence, however, shares segmental homology with a number of characterized proteins, including the fimbrial type 1 and 2 proteins of Actinomyces naeslundii (25 to 27% identity over 350 to 420 aa), the T6 surface protein of Streptococcus pyogenes (23% identity over 359 aa), and Hsf (27% identity over 260 aa) and High Molecular Weight protein 1 (HMW1) (25% identity over 285 aa) of Haemophilus influenzae (6, 18, 28, 31, 38). The function of the S. pyogenes T6 protein is unknown. Each of the other homologues plays a role in bacterial adhesion or invasion, suggesting that Spb1 may have a similar function in the pathogenesis of GBS infections. The type I and II fimbrial structural proteins of A. naeslundii (formerly A. viscosus), for example, bind to proline-rich salivary proteins, facilitating adhesion to tooth surfaces (13, 23). The HMW1 adhesin of H. influenzae binds glycoprotein receptors containing N-linked oligosaccharide chains with sialic acid at an α2-3 configuration on cultured human epithelial cells (30). Hsf (Haemophilus surface fibril) is a second protein associated with binding of H. influenzae to epithelial cells (7). The spb1 nucleic acid sequence is available from the GenBank database under accession number AF485279.
FIG. 1.
Nucleic acid and translated amino acid sequences of spb1. Shown are the nucleic acid sequence of the spb1 coding and 5′ noncoding regions and the translated amino acid sequence. A potential ribosomal binding site is underlined with dashes, putative transmembrane domains are underlined with solid lines, an arrow indicates the predicted signal peptide cleavage site, and the LPXTG cell wall-binding domain is shaded. The spb1 nucleotide sequence is available from the GenBank database under accession number AF485279.
Construction of an spb1 isogenic mutant strain of GBS 874391.
Southern blotting of genomic DNA with an spb1− probe was performed to identify bacterial strains with insertion-deletion mutations of the spb1 gene. A colony containing a single truncated copy of spb1 was selected for further study and designated strain Spb1− (data not shown). The colony morphology and microscopic appearance of Spb1− were indistinguishable from that of the parental strain, and both strains had identical growth curves in TH broth.
Hemolytic activities of 874391 and Spb1.
The abilities of strains 874391 and Spb1− to lyse sheep red blood cells were identical. Compared with that of strain COH-1 (hemolytic index of 1), both Spb1− and its parental strain 874391 had slightly more hemolytic activity (hemolytic index of 2).
Reduced epithelial cell adherence and invasion by the spb1 deletion mutant.
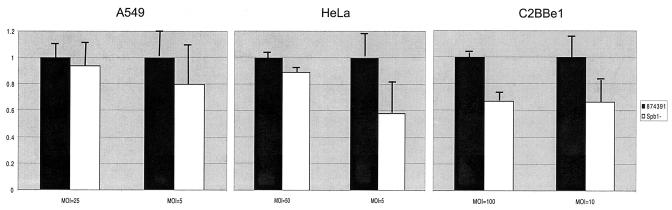
Overall, fewer Spb1− GBS adhered to all epithelial cell lines tested compared with the wild-type strain 874391. Adherence of Spb1− to A549 respiratory epithelial cells was reduced from 10.87% ± 1.39% to 10.191% ± 1.68% compared to that seen with GBS 874391 at a MOI of 25 and from 3.11% ± 0.62% to 2.49% ± 0.81% at a MOI of 5 (P value not significant) (Fig. 2). Adherence to HeLa cervical epithelial cells was reduced from 6.308% ± 0.60% to 5.62% ± 0.92% at a MOI of 50 and from 3.78% ± 0.73% to 2.2% ± 0.51% at a MOI of 5 (P value not significant). Adhesion of the Spb1− strain to C2Bbel colonic epithelial cells was reduced from 2.56% ± 0.06% to 1.72% ± 0.17% compared to the wild-type strain at a MOI of 100 (P < 0.001) and from 2.88% ± 0.53% to 1.92% ± 0.38% at a MOI of 10 (P value not significant).
FIG. 2.
Adherence of Spb1− GBS to epithelial cells. Graphs illustrate the relative adherence of strain Spb1− to A549 respiratory epithelial cells, HeLa cervical epithelial cells, and C2Bbe1 colonic epithelial cells compared to that of RDP type III-3 wild-type strain 874391. Bars represent the mean numbers of adherent bacteria plus standard errors of the mean (SEM) for each inoculum tested and also represent the reduction in invasion of recombinant strains relative to the wild-type strain 874391 for each inoculum tested. Spb1− had lower levels of adhesion than the wild-type strain, but only adherence to C2BBel cells was statistically significant at a MOI of 100 (P < 0.001). Data represent means ± SEM from three to seven separate experiments.
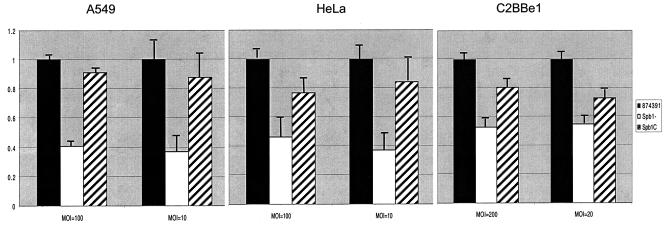
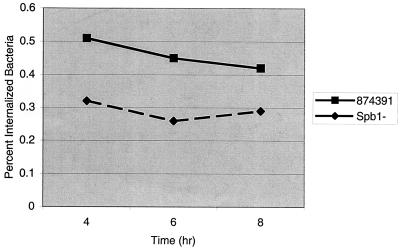
As previously observed, GBS invade epithelial cells inefficiently, with 1 to 2.5% of adherent cells internalized (14, 27). Invasion of each epithelial cell line by the Spb1− strain was significantly reduced compared with that of the wild-type strain 874391 (Fig. 3). Compared to wild-type 874391, Spb1− GBS invasion of A549 respiratory epithelial cells was reduced from 1.93% ± 0.01% to 0.079% ± 0.01% at a MOI of 100 (P < 0.001) and was reduced from 0.59% ± 0.10% to 0.22% ± 0.10% at a MOI of 10 (P < 0.001). At a MOI of 100, invasion of HeLa cervical epithelial cells by Spb1− was reduced from 0.121% ± 0.001% to 0.056% ± 0.01% (P < 0.001) compared to the wild-type strain, and at a MOI of 10, invasion was reduced from 0.173% ± 0.02% to 0.065% ± 0.01% (P < 0.001). Compared with that of wild-type 874391, Spb1− GBS invasion of C2Bbe1 colonic epithelial cells was reduced from 0.07% ± 0.01% to 0.04% ± 0.00% at a MOI of 200 (P < 0.001) and was reduced from 0.12% ± 0.01% to 0.07% ± 0.00% at a MOI of 20 (P < 0.001). The numbers of intracellular bacteria remained stable for at least 10 h after invasion (Fig. 4). Therefore, the reduced number of intracellular Spb1− GBS represents a reduction in invasion, rather than a reduction in intracellular survival, compared to the wild-type strain.
FIG. 3.
Invasion of epithelial cells is reduced in Spb1− GBS. Graphs illustrate the relative invasion of A549 respiratory epithelial cells, HeLa cervical epithelial cells, and C2Bbe1 colonic epithelial cells by RDP type III-3 wild-type strain 874391 (black bars), the spb1-deficient isogenic mutant strain Spb1− (white bars), and the Spb1− strain complemented in trans, strain Spb1c (stripped bars). Bars represent the reduction in invasion of recombinant strains relative to that of the wild-type strain 874391 for each inoculum tested. Spb1− invades all epithelial cells significantly less well than the wild-type strain 874391. Complementation of the Spb1− mutant in trans restores invasion to a level comparable to that of the parental strain. Data represent means ± SEM from three or four separate experiments.
FIG. 4.
Intracellular persistence of GBS in A549 respiratory epithelial cells. A549 cells were grown to confluence in 24-well tissue culture plates and infected with strain 874391 or Spb1 GBS at a MOI of 5 as described previously. Adherent extracellular bacteria were killed by the addition of tissue culture medium with 5 μg of penicillin/ml and 100 μg of gentamicin/ml followed by incubation at 37°C in 5% CO2. Epithelial cells were lysed at 4, 6, and 8 h after infection, and the percentage of the initial inoculum surviving within cells was quantified. Numbers of intracellular bacteria did not diminish significantly over 8 h. Data represent means from five or six separate experiments.
Complementation of the spb1 deletion restores the ability of GBS to invade epithelial cells.
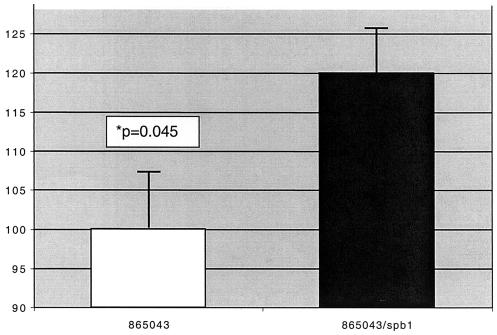
The Spb1− mutant strain was transformed with a plasmid-encoded copy of spb1 to create strain Spb1c. Spb1c invaded each type of epithelial cell in significantly greater numbers than did the Spb1− mutant. In the case of A549 and HeLa cells, complemented bacteria had a percentage of invasion that was lower than but not significantly different from that of wild-type 874391 cells. Invasion of C2Bbe1 cells by Spb1c was significantly greater than that by Spb1− at a MOI of 100 (P = 0.016) but not at a MOI of 10. Transforming the RDP type III-2 strain 865043 with a plasmid expressing spb1 in trans (creating strain 865043/spb1) significantly increased the numbers of bacteria invading A549 cells compared to the wild-type strain (P = 0.045). This result confirms the ability of the spb1 gene product to promote epithelial invasion (Fig. 5).
FIG. 5.
Invasion of A549 epithelial cells by GBS strains 865043 and 865043/spb1. The RDP type III-2 strain 865043 was transformed with pMS3545/spb1 to create strain 865043/spb1. Graph illustrates the relative invasion of A549 respiratory epithelial cells by each strain at a MOI of 100. Bars represent the invasion of 865043/spb1 (120.0% ± 7.4%) relative to that of the wild-type strain 865043 (100.0% ± 5.6%). Data represent means ± SEM from five separate experiments.
DISCUSSION
Our understanding of the pathogenesis of GBS infections is incomplete and has, to a great extent, focused on the role of host factors. The incidence of invasive GBS infections is strikingly correlated with age, with the vast majority of neonatal infections occurring in the first 6 weeks of life, and most within the first week (3). This age-related susceptibility has been attributed to the immaturity of innate host defenses in newborn infants, a hypothesis that might also help explain the increased frequency of these infections in premature infants. Serum antibody directed against the capsular polysaccharide facilitates opsonophagocytic killing of GBS, and the concentration of transplacentally acquired maternal anti-GBS capsular antibody in newborns is inversely correlated with their risk of invasive infection (2). Unfortunately, the majority of pregnant women, even those heavily colonized with GBS, do not develop antibody concentrations sufficient to provide passive immunity to their infants. The complex interrelationships between maternal GBS colonization and maternal and infant immune responses complicate the investigation of the role of bacterial factors in GBS infections. This study, however, confirms that bacterial genetic factors may contribute to GBS pathogenicity.
The development of sensitive genetic classification systems has provided compelling evidence that virulence is correlated with phylogeny in many bacterial species. In one study, over 90% of serotype III GBS that caused invasive neonatal disease in neonates in both Tokyo and Salt Lake City were RDP type III-3 (32). This finding is consistent with the results of Musser and colleagues, who used multilocus enzyme electrophoresis to classify GBS isolates from North America into two major divisions (21). Division I isolates consisted of a single electrophoretic type of serotype III GBS that were more likely to be isolated from ill neonates than from healthy carriers and that were genetically distinct from other type III isolates. These Division I isolates shared distinctive phenotypic characteristics that are also found in RDP type III-3 strains, such as growth lag in chemically defined medium containing 200 mM phosphate, suggesting that both of these classification systems identify the same group of hypervirulent type III GBS strains (22).
The positive correlation of virulence with chromosomal organization and the genetic disequilibrium of virulence with electrophoretic typing in previous studies led to the hypothesis that RDP III-3 strains possessed novel noncapsular virulence factors. Furthermore, these data suggest that differences in virulence are more likely to be related to the presence or absence (or activation or inactivation) of virulence genes than to the differential regulation of a factor common to both groups of bacteria. In this setting, subtractive genomic hybridization is likely to demonstrate differences in the genomes of virulent and less virulent type III GBS strains that contribute to pathogenicity. The presence or absence of these novel virulence factors may not be strictly linked to serotype. For example, an individual strain may have acquired these genes by horizontal transmission after the acquisition of type III capsulation genes and evolved in parallel with other type III lineages. Alternatively, a non-type III GBS strain with an enhanced ability to colonize or invade might acquire serotype III capsule genes (capsule switching) by the same process. These genetic events might explain the abrupt emergence of GBS as a major human pathogen in the last quarter century.
The more pronounced effect of the spb1 deletion on invasion of respiratory and cervical epithelial cells suggests that Spb1 might particularly contribute to the pathogenesis of early-onset neonatal infections and maternal infections. Adhesion to and invasion of respiratory epithelium and endothelium appear to be critical factors in neonatal pneumonia and systemic infection, and it is at this early stage that Spb1 may confer an advantage to RDP type III-3 GBS (27, 33). It was recently demonstrated that GBS C5a peptidase binds fibronectin and increases the invasive potential of GBS (8, 12). The cell-associated β-hemolysin also promotes GBS invasion of epithelial cells (14). Although the precise mechanism has not been described, it has been hypothesized that this hemolysin/cytotoxin may activate respiratory epithelial cells, increasing bacterial uptake (14). Spb1 differs from these ubiquitous virulence factors in that it is present only in more virulent type III GBS strains. It may provide RDP type III-3 strains with a numerical advantage over less pathogenic strains by increasing the number of bacteria that successfully invade the epithelial barrier.
The reduction in invasion of epithelial cells by the Spb1− mutant strain is considerably greater than the reduction in adherence of these bacteria. This observation, and the inefficiency of invasion of epithelial cells by GBS, suggests that not all adherent GBS have the potential to be internalized. Spb1 may mediate a novel pathway of bacterial invasion. Alternatively, Spb1 engagement by epithelial cells might trigger an inflammatory process that more globally enhances bacterial uptake.
Understanding of the population structure and molecular epidemiology of infectious agents may provide insights into the pathogenesis of diseases that are precluded by examination of genetically heterogeneous organisms. This study provides further evidence for the clonal organization of GBS strains and demonstrates that the increased virulence of certain type III GBS is attributable to unique genetic factors.
Acknowledgments
We thank E. Tuomanen for many helpful discussions.
This work was supported by NIAID grant R01 AI40918 (E.E.A. and J.F.B.), NCI grant P30 CA21765 (E.E.A.), the American Lebanese Syrian Associated Charities, and the University of Utah Undergraduate Education Program (D.V.M.). E.E.A. is an Established Investigator of the American Heart Association.
Editor: V. J. DiRita
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, K. Struhl, L. M. Albright, D. M. Coen, and A. Varki. 1994. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 2.Baker, C. J. 1976. Correlation of maternal antibody deficiency with susceptibility to neonatal group B streptococcal infection. N. Engl. J. Med. 294:753-756. [DOI] [PubMed] [Google Scholar]
- 3.Baker, C. J. 2000. Group B streptococcal infections, p. 222-237. In D. L. Stevens, and E. L. Kaplan (ed.), Streptococcal infections: clinical aspects, microbiology, and molecular pathogenesis. Oxford University Press, New York, N.Y.
- 4.Baker, C. J., and F. F. Barrett. 1973. Transmission of group B streptococci among parturient women and their neonates. J. Pediatr. 83:919-925. [DOI] [PubMed] [Google Scholar]
- 5.Baker, C. J., and M. S. Edwards. 1995. Group B streptococcal infections, p. 980-1054. In J. Remington, and J. O. Klein (ed.), Infectious diseases of the fetus and newborn infant, 4th ed. W. B. Saunders, Philadelphia, Pa.
- 6.Barenkamp, S. J., and E. Leininger. 1992. Cloning, expression, and DNA sequence analysis of genes encoding nontypeable Haemophilus influenzae high-molecular-weight surface-exposed proteins related to filamentous hemagglutinin of Bordetella pertussis. Infect. Immun. 60:1302-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barenkamp, S. J., and J. W. St. Geme III. 1996. Identification of a second family of high-molecular adhesion proteins expressed by non-typeable H. influenzae. Mol. Microbiol. 19:1215-1223. [DOI] [PubMed] [Google Scholar]
- 8.Beckmann, C., J. D. Waggoner, T. O. Harris, G. S. Tamura, and C. E. Rubens. 2002. Identification of novel adhesins from group B streptococci by use of phage display reveals that C5a peptidase mediates fibronectin binding. Infect. Immun. 70:2869-2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bohnsack, J. F., S. Takahashi, S. Detrick, L. R. Pelinka, L. Hammitt, A. A. Aly, A. A. Whiting, and E. E. Adderson. 2001. Phylogenetic classification of serotype III group B streptococci on the basis of hylB gene analysis and DNA sequences specific to restriction digest pattern type III-3. J. Infect. Dis. 183:1694-1697. [DOI] [PubMed] [Google Scholar]
- 10.Bohnsack, J. F., A. A. Whiting, R. D. Bradford, B. K. Van Frank, S. Takahasi, and E. E. Adderson. 2002. Long-range mapping of the Streptococcus agalactiae phylogenetic lineage restriction digest pattern type III-3 reveals clustering of virulence genes. Infect. Immun. 70:134-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bryan, E. M., T. Bae, M. Kleerebezem, and G. M. Dunny. 2000. Improved vectors for nisin-controlled expression in gram-positive bacteria. Plasmid 44:183-190. [DOI] [PubMed] [Google Scholar]
- 12.Cheng, Q., D. Stafslien, S. S. Purushothaman, and P. P. Cleary. 2002. The group B streptococcal C5a peptidase is both a specific protease and an invasin. Infect. Immun. 70:2408-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cisar, J. O., E. L. Barsumian, R. P. Siraganian, W. B. Clark, M. K. Yeung, S. D. Hsu, S. H. Curl, A. E. Vatter, and A. L. Sandberg. 1991. Immunochemical and functional studies of Actinomyces viscosus T14V type 1 fimbriae with monoclonal and polyclonal antibodies directed against the fimbrial subunit. J. Gen. Microbiol. 137:1971-1979. [DOI] [PubMed] [Google Scholar]
- 14.Doran, K. S., J. C. Chang, V. M. Benoit, L. Eckmann, and V. Nizet. 2002. Group B streptococcal beta-hemolysin/cytolysin promotes invasion of human lung epithelial cells and the release of interleukin-8. J. Infect. Dis. 185:196-203. [DOI] [PubMed] [Google Scholar]
- 15.Framson, P. E., A. Nittayajarn, J. Merry, P. Youngman, and C. E. Rubens. 1997. New genetic techniques for group B streptococci: high-efficiency transformation, maintenance of temperature-sensitive pWV01 plasmids, and mutagenesis with Tn917. Appl. Environ. Microbiol. 63:3539-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glaser, P., C. Rusniok, C. Buchrieser, F. Chevalier, L. Frangeul, T. Msadek, M. Zouine, E. Couve, L. Lalioui, C. Poyart, P. Trieu-Cuot, and F. Kunst. 2002. Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol. Microbiol. 45:1499-1513. [DOI] [PubMed] [Google Scholar]
- 17.Harrison, L. H., J. A. Elliott, D. M. Dwyer, J. P. Libonati, P. Ferrieri, L. Billmann, and A. Schuchat. 1998. Serotype distribution of invasive group B streptococcal isolates in Maryland: implications for vaccine formulation. J. Infect. Dis. 177:998-1002. [DOI] [PubMed] [Google Scholar]
- 18.Li, T., I. Johansson, D. I. Hay, and N. Stromberg. 1999. Strains of Actinomyces naeslundii and Actinomyces viscosus exhibit structurally variant fimbrial subunit proteins and bind to different peptide motifs in salivary proteins. Infect. Immun. 67:2053-2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin, F. Y., J. D. Clemens, P. H. Azimi, J. A. Regan, L. E. Weisman, J. B. Philips III, G. G. Rhoads, R. A. Brenner, and P. Ferrieri. 1998. Capsular polysaccharide types of group B streptococcal isolates from neonates with early-onset systemic infection. J. Infect. Dis. 177:790-792. [DOI] [PubMed] [Google Scholar]
- 20.Lisitsyn, N., N. Lisitsyn, and M. Wigler. 1993. Cloning the differences between two complex genomes. Science 259:946-951. [DOI] [PubMed] [Google Scholar]
- 21.Musser, J. M., S. J. Mattingly, R. Quentin, A. Goudeau, and R. K. Selander. 1989. Identification of a high-virulence clone of type III Streptococcus agalactiae (group B Streptococcus) causing invasive neonatal disease. Proc. Natl. Acad. Sci. USA 86:4731-4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagano, Y., N. Nagano, S. Takahashi, K. Morono, K. Fujita, F. Taguchi, and Y. Okuwaki. 1991. Restriction endonuclease patterns of chromosomal DNA from group B β-hemolytic streptococci. J. Med. Microbiol. 35:297-303. [DOI] [PubMed] [Google Scholar]
- 23.Nyvad, B., and M. Kilian. 1987. Microbiology of the early colonization of human enamel and root surfaces in vivo. Scand. J. Dent. Res. 95:369-380. [DOI] [PubMed] [Google Scholar]
- 24.Peterson, M. D., and M. S. Mooseker. 1992. Characterization of the enterocyte-like brush border cytoskeleton of the C2BBe clones of the human intestinal cell line, Caco-2. J. Cell Sci. 102:581-600. [DOI] [PubMed] [Google Scholar]
- 25.Pritzlaff, C. A., J. C. Chang, S. P. Kuo, G. S. Tamura, C. E. Rubens, and V. Nizet. 2001. Genetic basis for the beta-haemolytic/cytolytic activity of group B Streptococcus. Mol. Microbiol. 39:236-247. [DOI] [PubMed] [Google Scholar]
- 26.Rubens, C. E., H. V. Raff, C. J. Jackson, E. Y. Chi, J. T. Bielitzki, and S. L. Hillier. 1991. Pathophysiology and histopathology of group B streptococcal sepsis in Macaca nemestrina primates induced after intraamniotic inoculation: evidence for bacterial cellular invasion. J. Infect. Dis. 164:320-330. [DOI] [PubMed] [Google Scholar]
- 27.Rubens, C. E., S. Smith, M. Hulse, E. Y. Chi, and G. van Belle. 1992. Respiratory epithelial cell invasion by group B streptococci. Infect. Immun. 60:5157-5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schneewind, O., K. F. Jones, and V. A. Fischetti. 1990. Sequence and structural characteristics of the trypsin-resistant T6 protein of group A streptococci. J. Bacteriol. 172:3310-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spellerberg, B., E. Rozdzinski, S. Martin, J. Weber-Heynemann, N. Schnitzler, R. Lutticken, and A. Podbielski. 1999. Lmb, a protein with similarities to the LraI adhesion family, mediates attachment of Streptococcus agalactiae to human laminin. Infect. Immun. 67:871-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.St. Geme, J. W., III. 2000. The pathogenesis of nontypeable Haemophilus influenzae otitis media. Vaccine 19:S41-S50. [DOI] [PubMed] [Google Scholar]
- 31.St. Geme, J. W., III, D. Cutter, and S. J. Barenkamp. 1996. Characterization of the genetic locus encoding Haemophilus influenzae type b surface fibrils. J. Bacteriol. 178:6281-6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahashi, S., E. E. Adderson, Y. Nagano, N. Nagano, M. R. Briesacher, and J. F. Bohnsack. 1998. Identification of a highly encapsulated, genetically related group of invasive type III group B streptococci. J. Infect. Dis. 177:1116-1119. [DOI] [PubMed] [Google Scholar]
- 33.Tamura, G. S., J. M. Kuypers, S. Smith, H. Raff, and C. E. Rubens. 1994. Adherence of group B streptococci to cultured epithelial cells: roles of environmental factors and bacterial surface components. Infect. Immun. 62:2450-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tamura, G. S., and A. Nittayajarn. 2000. Group B streptococci and other gram-positive cocci bind to cytokeratin 8. Infect. Immun. 68:2129-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamura, G. S., and C. E. Rubens. 1995. Group B streptococci adhere to a variant of fibronectin attached to a solid phase. Mol. Microbiol. 15:581-589. [DOI] [PubMed] [Google Scholar]
- 36.Tettelin, H., V. Masignani, M. J. Cieslewicz, J. A. Eisen, S. Peterson, M. R. Wessels, I. T. Paulsen, K. E. Nelson, I. Margarit, T. D. Read, L. C. Madoff, A. M. Wolf, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. T. DeBoy, A. S. Durkin, J. F. Kolonay, R. Madupu, M. R. Lewis, D. Radune, N. B. Fedorova, D. Scanlan, H. Khouri, S. Mulligan, H. A. Carty, R. T. Cline, S. E. Van Aken, J. Gill, M. Scarselli, M. Mora, E. T. Iacobini, C. Brettoni, G. Galli, M. Mariani, F. Vegni, D. Maione, D. Rinaudo, R. Rappuoli, J. L. Telford, D. L. Kasper, G. Grandi, and C. M. Fraser. 2002. Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc. Natl. Acad. Sci. USA 99:12391-12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wibawan, I. T., C. Lammler, and F. H. Pasaribu. 1992. Role of hydrophobic surface proteins in mediating adherence of group B streptococci to epithelial cells. J. Gen. Microbiol. 138:1237-1242. [DOI] [PubMed] [Google Scholar]
- 38.Yeung, M. K., and J. O. Cisar. 1990. Sequence homology between the subunits of two immunologically and functionally distinct types of fimbriae of Actinomyces spp. J. Bacteriol. 172:2462-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]