Abstract
Chromatin is disassembled and reassembled during DNA repair. To assay chromatin reassembly accompanying DNA double strand break repair, ChIP analysis can be used to monitor the presence of histone H3 near the lesion. The chromatin assembly factor Asf1p, as well as the acetylation of histone H3 lysine 56, have been shown to promote chromatin reassembly when DNA double strand break repair is complete. Using Gal-HO-mediated double strand break repair, we have tested each of the components of the nuclear Hat1p-containing type B histone acetyltransferase complex (NuB4) and have found that they can affect repair-linked chromatin reassembly but that their contributions are not equivalent. In particular, deletion of the catalytic subunit, Hat1p, caused a significant defect in chromatin reassembly. In addition, loss of the histone chaperone Hif1p, when combined with an allele of H3 that mutates lysines 14 and 23 to arginine, has a pronounced effect on chromatin reassembly that is similar to that observed in an asf1Δ. The role of Hat1p and Hif1p is at least partially redundant with the role of Asf1p. Consistent with a more prominent role for Hif1p in chromatin reassembly than either Hat1p or Hat2p, Hif1p exists in complex(es) independent of Hat1p and Hat2p and influences the activity of an H3-specific histone acetyltransferase activity. Our data directly demonstrate the role of the nuclear HAT1 complex (NuB4) components in DNA repair-linked chromatin reassembly.
Keywords: Chromatin, Chromatin Structure, DNA Repair, Histone Acetylase, Histone Modification, ASF1, HAT1, HIF1, Chromatin Assembly, Histone Chaperone
Introduction
The post-translational acetylation of the core histone NH2-terminal tails has been shown to be an important mechanism by which cells regulate the accessibility of chromatin (1). Histone acetylation plays an important role in the initial formation of chromatin structure, as well. A role for histone acetylation in chromatin assembly was first suggested by the observation that histones H3 and H4 are rapidly acetylated on their NH2-terminal tail domains in the cytoplasm following their synthesis (2, 3). Once incorporated into chromatin, this acetylation is removed during the process of chromatin maturation (4).
The acetylation of the NH2-terminal tails of newly synthesized histones H3 and H4 is an evolutionarily conserved phenomenon. The acetylation state of newly synthesized molecules of histone H4 has been analyzed in a diverse collection of eukaryotic organisms. In each case, new H4 is found to be diacetylated at lysine residues 5 and 12 (5–7). Whereas the presence of acetylation on the NH2-terminal tail of newly synthesized histone H3 has been conserved, distinct patterns of modification are found in different organisms (7, 8). In addition to the acetylation of the NH2-terminal tail domains of newly synthesized H3 and H4, it has recently been found that these molecules are also acetylated in their globular core domains. Histone H3 is acetylated on lysine 56 and histone H4 is acetylated on lysine 91 (9–12). These sites of acetylation have been observed in both yeast and mammalian cells and, hence, may also be evolutionarily conserved modifications (13–16).
The acetylation of newly synthesized histones is catalyzed by type B histone acetyltransferases. Type B histone acetyltransferases were originally defined as cytoplasmic enzymes that acetylate free, but not chromatin-associated, histones (17). The first type B histone acetyltransferase complex was isolated from Saccharomyces cerevisiae cytoplasmic extracts and consisted of two proteins, Hat1p and Hat2p (18, 19). Hat1p, the catalytic subunit, specifically acetylates lysine residues at positions 5 and 12 of free histone H4. Hat2p enhances the catalytic activity of Hat1p through facilitating the interaction of Hat1p with its histone substrate (18).
Although the acetylation of newly synthesized histones by type B histone acetyltransferases is presumed to play a role in replication-coupled histone deposition, HAT1 is a non-essential gene in yeast (18, 19). In vivo investigations into the function of Hat1p in S. cerevisiae demonstrated that loss of Hat1p resulted in defects in the telomeric silent chromatin structure and sensitivity to DNA damaging agents (20, 21). A role in DNA damage repair may be evolutionarily conserved as this phenotype is also seen in Schizosaccharomyces pombe and chicken DT40 cells that lack Hat1p (22, 23).
The involvement of Hat1p in DNA damage repair has been most extensively studied in S. cerevisiae. Cells lacking Hat1p are specifically sensitive to agents that generate DNA double strand breaks, such as high concentrations of MMS or induction of endonucleases (EcoRI or HO), due to a defect in recombinational repair. Interestingly, these phenotypes are only observed when hat1Δ is combined with mutations that alter specific sets of lysine residues in the histone H3 NH2-terminal tail (such as H3 K14R,K23R) suggesting that acetylation of the newly synthesized H3 and H4 may be functionally redundant (21).
Several observations suggest that Hat1p has a more extensive role in modulating chromatin structure than merely acetylating histone H4 in the cytoplasm. First, Hat1p is predominantly localized in the nucleus (24, 25). Second, when in the nucleus, Hat1p is found in a distinct complex (NuB4), in which Hat1p/Hat2p is joined by Hif1p (a H3/H4-specific histone chaperone) and histones H3 and H4 (24). Finally, both Hat1p and Hif1p have been shown to be directly recruited to chromatin within a relatively small domain surrounding a DNA double strand break (26).
The repair of damaged DNA occurs in a chromatin context and it has been shown that the removal of histones from DNA and their subsequent reassembly onto DNA accompanies DNA repair (27–29). The S. cerevisiae mating-type switching system has proven to be a valuable tool for the study of DNA double strand break repair (30). A GAL-inducible copy of the HO endonuclease is integrated into the genome. Switching of the mating type is initiated by the induction of the HO endonuclease, which then cuts at the MAT locus leaving a single double strand break. Sequences homologous to the HO cut site present at the silent mating loci, HML and HMR, are then used by the homologous recombination machinery to repair the break, resulting in a change of mating type (31).
The inducible HO system has been used to examine the reassembly of chromatin structure that occurs following repair of the double strand break (27, 29). By monitoring the presence of histone H3 by ChIP, it was demonstrated that histones are lost from near the site of an HO-induced double strand break as single strand DNA resection occurs. H3 levels then return following DNA repair. The involvement of chromatin assembly factors in this reassembly process was indicated by defects in restoration of histone H3 levels near the break site in the absence of the histone chaperone Asf1p or in the presence of mutations that prevent the acetylation of histone H3 lysine 56 (27).
To determine whether the nuclear Hat1p-containing type B histone acetyltransferase (NuB4) complex participates in the DNA double strand break repair-linked chromatin reassembly process, we employed the inducible GAL-HO endonuclease system and ChIP analysis to investigate the requirement of each of its components. We found that subunits of the NuB4 complex can affect repair-linked chromatin reassembly but that their contributions are not equivalent. Although loss of Hat1p causes a moderate defect in chromatin reassembly, deletion of the HIF1 gene, in combination with a mutation in the histone H3 tail, has a pronounced effect on chromatin reassembly that is similar to that seen in the absence of Asf1p. In addition, our results suggest that Hat1p complex components may function in a pathway that is distinct from Asf1p. Finally, biochemical evidence indeed indicated that Hif1p may exist in complexes distinct from the NuB4 complex and the presence of Hif1p is necessary for the activity of a histone H3-specific HAT.
EXPERIMENTAL PROCEDURES
Yeast Strains
Yeast culture and genetic manipulation were done by standard methods (32). Gene deletions were generated by PCR-mediated gene disruption with a nutritional marker. The genotypes of yeast strains used in this study are shown in Table 1.
TABLE 1.
Strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| SQY501 | MATα ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 ADE3::GAL10-HO HHF2-HHT2::LEU2 HHF1-HHT1::ADE3 (TRP1 CEN ARS)-HHF2-HHT2 | This study |
| ZGY101 | MATα ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ursa3Δ0 ADE3::GAL10-HO HHF2-HHT2::LEU2 HHF1-HHT1::ADE3 (TRP1 CEN ARS)-HHF2-HHT2 HAT1::URA3 | This study |
| ZGY102 | MATα ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 ADE3::GAL10-HO HHF2-HHT2::LEU2 HHF1-HHT1::ADE3 (TRP1 CEN ARS)-HHF2-HHT2 HAT2::URA3 | This study |
| ZGY103 | MATα ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 ADE3::GAL10-HO HHF2-HHT2::LEU2 HHF1-HHT1::ADE3 (TRP1 CEN ARS)-HHF2-HHT2 HIF1::URA3 | This study |
| ZGY104 | MATα ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 ADE3::GAL10-HO HHF2-HHT2::LEU2 HHF1-HHT1::ADE3 (TRP1 CEN ARS)-HHF2-HHT2 ASF1::URA3 | This study |
| ZGY105 | MATα ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 ADE3::GAL10-HO HHF2-HHT2::LEU2 HHF1-HHT1::ADE3 (TRP1 CEN ARS)-HHF2-HHT2 ASF1::URA3 HIF1::HIS3 | This study |
| ZGY106 | MATα ade2Δ::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 ADE3::GAL10-HO HHF2-HHT2::LEU2 HHF1-HHT1::ADE3 (TRP1 CEN ARS)-HHF2-HHT2 ASF1::URA3 HAT1::HIS3 | This study |
HO Endonuclease Sensitivity Assays
Yeast strains were grown overnight in rich medium with 2% raffinose. Cells were diluted and grown to an optical density at 600 nm (A600) of ∼0.8 and concentrated to an A600 of 1 to plate in 10-fold serial dilutions onto rich medium or medium with 2% galactose.
DNA Damage and Repair Analysis
Primers flanking the HO site in the MAT locus were used to determine the degree of cutting and repair of mating type by PCR amplification. Cells were grown overnight in rich medium containing 2% raffinose. Galactose and then glucose were added to 2% at the times indicated in the figure legends. The number of PCR cycles to produce amplification in the linear range was determined empirically. PCR products were resolved by agarose gel electrophoresis. Gels were stained with ethidium bromide and PCR products were quantitated with 1D image analysis software (Kodak).
Chromatin Immunoprecipitation Analyses
Cultures were grown overnight in rich medium containing 2% raffinose, diluted, and grown until the cells reached an A600 of ∼0.5. Galactose and then glucose were added to 2% at the times indicated in the figure legends. Samples were taken for chromatin immunoprecipitation (ChIP) analysis at the time points indicated in the figure legends and processed as described previously. Samples were analyzed using quantitative real time PCR in a multiplex reaction with primers and probes designed as described previously (27). All experiments were performed with two or three biological replicates.
Quantitative Real Time PCR Analysis
Real time PCR was used to quantitate fragments in the immunoprecipitated samples from the ChIP analyses, using the ABI 7300 sequence detector and TaqMan PCR Master Mix protocol. Each PCR was performed in sextuplet with cycling conditions as follows: 50 for 2 min, 95 for 10 min, and then 40 cycles, with 1 cycle consisting of 95 for 15 s and 60 for 1 min. The cycle threshold (CT) value was set so that the fluorescence signal was above the base line noise and as low as possible in the exponential amplification phase. The amount of change compared with the SMC2 control was calculated for each immunoprecipitation using the standard comparative CT method.
Whole Cell Extracts
Yeast whole cell extracts were prepared from overnight yeast culture in YPD as described previously. Briefly, cells were harvested at midlog phase and washed with cold H2O and extraction buffer (100 mm HEPES, pH 7.9, 245 mm KCl, 5 mm EGTA, 1 mm EDTA, 0.5 mm PMSF, and 0.3% β-mercaptoethanol). The cell pellets were passed through a 3-ml syringe into a 50-ml tube containing liquid N2. The frozen pellets were ground to a fine powder in the presence of liquid N2 and incubated with extraction buffer (150 μl of buffer/g of cells) on ice for 20 min, followed by centrifugation at 30,000 × g for 1 h at 4 °C. Supernatant was collected as whole cell extracts and dialyzed against dialysis buffer DN(50) (20 mm HEPES, pH 7.9, 50 mm NaCl, 5 mm MgCl2, 1 mm EDTA, 10% glycerol) before use.
Column Chromatography
Dialyzed yeast whole cell extracts were centrifuged at 10,000 × g for 10 min. The resulting clarified extracts were applied to a Mono Q column (GE Healthcare) equilibrated with DN(50). The column was washed with 10 column volumes, and proteins were eluted with a 20 column volume gradient from DN(50) to DN(1000) (25 mm Tris, pH 7.9, 1 m NaCl, 0.1 mm EDTA, and 10% glycerol). The elution profile of protein was determined by Western blot. The peak fractions containing Hif1p that were determined by Western blot were pooled and concentrated to a volume of 200 μl and then resolved by gel filtration chromatography (Superose 6 column, GE Healthcare) run with DN(300) buffer (25 mm Tris, pH 7.0, 0.1 mm EDTA, 10% glycerol, and 300 mm NaCl). The elution profiles of proteins were determined by Western blot.
Western Blotting
Western blots were performed and visualized using an ECL Plus chemiluminescent detection kit according to the manufacturer's instructions (Amersham Biosciences). The signal was detected by scanning on a Storm PhosphorImager.
Histone Acetyltransferase Assays
Liquid histone acetyltransferase assays were performed using free chicken histones as the substrate. Reactions were performed in a final volume of 50 μl in buffer DN(50) containing 0.1 μm [3H]acetyl-coenzyme A (6.1 Ci/mmol, ICN) and 1 mg/ml of chicken erythrocyte core histones. Chicken histones were purified as previously described. Reactions were incubated at 37 °C for 60 min. The assay mixture was resolved on an 18% SDS-PAGE and stained with Coomassie Blue. The gel was then incubated in fluorohance, dried, and exposed to x-ray film to identify the labeled histone.
HO Gene Expression Analysis
RT-PCR assays were performed as described previously (33). Briefly, total mRNA from 10 ml of yeast culture with an A600 of 0.8–1.0 was prepared using the purelink RNA minikit (Invitrogen). Reverse transcription is carried out using the high capacity cDNA reverse transcription kit (Applied Biosystems) following the manufacturer's protocol. PCR and real time PCR were then performed using synthesized first strand cDNA as template and the primer pairs targeted to the HO and ADH1 coding sequences (sequences available upon request).
RESULTS
Involvement of NuB4 Complex Components in DNA Repair-linked Chromatin Reassembly
The use of an inducible HO endonuclease has proven to be a valuable tool for the study of recombinational repair and the chromatin assembly and disassembly that must accompany it (27). To use this model system to look specifically at the role of the nuclear Hat1p (NuB4) complex in chromatin reassembly accompanying DNA double strand break repair, we integrated a copy of a galactose-inducible HO endonuclease gene into the genome that enables us to introduce a single double strand break at the MAT locus. We then individually deleted each of the NuB4 complex components to generate hat1Δ, hat2Δ, and hif1Δ strains. The strain background that was used for these studies has also been deleted for all of the endogenous genes encoding histones H3 and H4, which allows for the introduction of mutant alleles of H3 and H4 as the only copies of these histones (20). Previous studies have indicated that mutations in specific sets of lysine residues in the histone H3 NH2-terminal tail (such as H3 K14R,K23R) cause sensitivity to DNA damaging agents and that this sensitivity is increased by combining these mutations with mutations in the components of the NuB4 complex (21, 24).
To determine whether the NuB4 complex and histone H3 mutants were sensitive to a single double strand break at the MAT locus, we tested their ability to grow on medium containing galactose, which induced HO expression. As seen in Fig. 1, none of the single mutants (H3 K14R,K23R, hat1Δ, hat2Δ, or hif1Δ) showed a growth defect on galactose. Surprisingly, when the H3 K14R,K23R allele was combined with each of the NuB4 complex deletions, only the H3 K14R,K23R hif1Δ combination showed a significant synthetic sensitivity to HO induction suggesting that Hif1p may play a more prominent role in DNA repair or chromatin reassembly than Hat1p and Hat2p.
FIGURE 1.
Sensitivity of NuB4 complex and histone H3 mutants to an HO-induced DNA double strand break. 10-Fold serial dilutions of strains with the indicated genotypes were spotted on plates containing minimal media with either glucose or galactose as carbon source. Plates were incubated at 30 °C for 3 days and then photographed.
To assess the impact of these mutations on the reassembly of chromatin structure, we used ChIP analysis to monitor the presence of histone H3 near the HO cleavage site as the repair process proceeded (diagramed in Fig. 2A). As a positive control for these experiments, we generated an asf1Δ in this strain background as this histone chaperone has been shown to be necessary for chromatin reassembly following the repair of an HO-induced double strand break (27). Results from the asf1Δ strain are included in each of the figures for comparison.
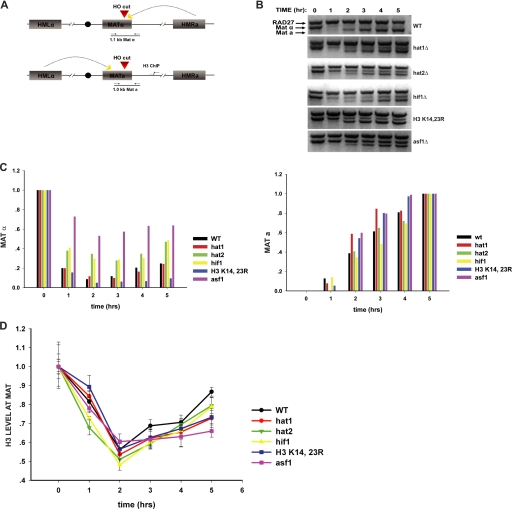
FIGURE 2.
Impact of NuB4 complex and histone H3 mutations on DNA repair-linked chromatin reassembly. A, schematic diagram of the inducible HO-mediated mating type switch system used to model recombinational DNA repair. Locations of primers used for PCR analysis of the double strand break at the MAT locus and for ChIP analysis of histone H3 disassembly and reassembly are indicated. B, introduction and repair of the double strand break at the MAT locus was monitored by PCR assay in the indicated strains. Galactose was added at the 0-h time point and glucose was added at the 2-h time point. Reaction products were resolved on a 1.5% agarose gel and visualized by ethidium bromide staining. The migration of the MATα- and MATa-specific bands is indicated. Amplification of a region of the RAD27 locus was used as a control. C, quantitation of double strand break formation and repair. Stained agarose gels were photographed and the MATα, MATa, and RAD27 bands were quantitated using one-dimensional Image Analysis software (Kodak). MATα and MATa fragments were normalized to the RAD27 fragment. The MATα and MATa fragments were plotted relative to the 0- and 5-h time points, respectively. D, ChIP analysis of the abundance of histone H3 at a site 600 bp from the double strand break at the MAT locus. The graph shows a comparison of the indicated NuB4 complex or histone H3 mutant to a wild type strain and an asf1Δ strain. Histone H3 levels were normalized to H3 levels at the SMC2 locus. Subsequent time points are normalized to the 0-h time point.
Cultures were grown to mid-log phase in raffinose, and galactose was added to induce expression of HO. After 2 h, glucose was added to repress HO and allow for repair of the HO-induced double strand break to proceed. The introduction of the double strand break at the MAT locus and its subsequent repair were monitored by a PCR reaction that spanned the HO cut site and that generates distinct fragments from MATa and MATα cells (see Fig. 2A). In a wild type strain (which starts as MATα), the HO site at MAT is efficiently cut in the presence of HO and then repaired following repression of HO (Fig. 2B). Similar kinetics of digestion and repair were also seen in hat1Δ, hat2Δ, hif1Δ, and H3 K14R,K23R strains (Fig. 2, B and C). Interestingly, we reproducibly observed inefficient generation of the HO-induced double strand break in asf1Δ cells (Figs. 2, B and C, and 4, B and C). One explanation is that Asf1p might be necessary for the proper transcriptional induction of the GAL-regulated HO gene (34, 35). However, we detected only minor differences in the kinetics and level of HO induction in asf1Δ cells (supplemental Fig. S1). Whether Asf1p plays a direct role in facilitating the HO-mediated cleavage of chromatin remains a possibility.
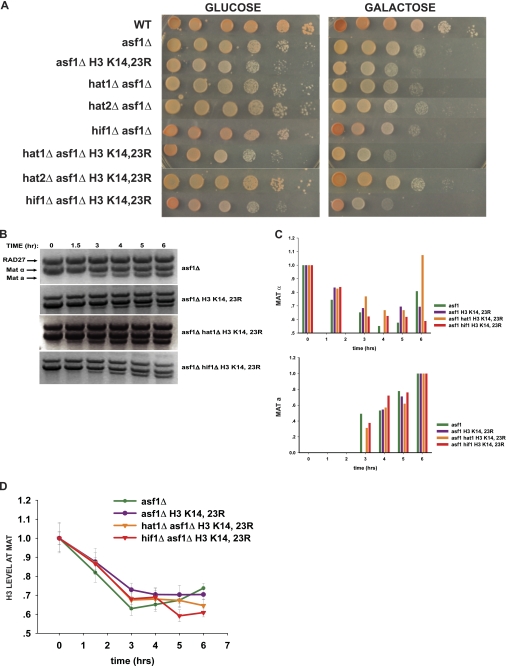
FIGURE 4.
Functional redundancy between NuB4 complex components, histone H3, and Asf1p in DNA repair-linked chromatin reassembly. A, 10-fold serial dilutions of the indicated strains were spotted on plates containing either glucose or galactose as carbon source. Plates were incubated at 30 °C for 3 days and then photographed. B, cutting and repair of the MAT locus in the indicated strains was monitored as described in the legend to Fig. 2. B, the levels of MATα and MATa fragments were quantitated as described in the legend to Fig. 2 except that the MATa levels are plotted relative to the 6-h time point. D, ChIP analysis of H3 levels near the MAT locus at the indicated times. Galactose was added to cultures at time 0. Glucose was added after 3 h. ChIP experiments were performed and analyzed as described in the legend to Fig. 2.
We then used ChIP to monitor the presence of histone H3 at a position 600 bp from the HO cut site. As reported previously, there is a loss of histone H3 as the MAT locus is cut and single strand resection occurs at the break site (27). The levels of histone H3 then returned as the recombinational repair proceeded and chromatin structure was reassembled (Fig. 2D). In the absence of Asf1p, there was also loss of H3 near the HO cut site. However, also as previously reported, there was a dramatic decrease in chromatin reassembly as indicated by the observation that there was only a slight increase in the levels of histone H3 near the break site following its repair (27). The hat1Δ and H3 K14R,K23R mutations also caused a significant defect in repair-linked chromatin reassembly that was reproducibly observed in multiple trials (Fig. 2D). The rate of reassembly in these mutants was intermediate between that in the wild type and asf1Δ cells. Loss of Hat2p and Hif1p resulted in a minor defect on chromatin reassembly (Fig. 2D). The fact that both the hat1Δ and H3 K14R,K23R mutations cause a clear defect in chromatin reassembly without an effect on cell viability suggests that the cells can tolerate a suboptimal level of chromatin reassembly during the DNA repair process.
We then examined chromatin reassembly when the H3 K14R,K23R allele was combined with the mutations in NuB4 complex components. Again, similar kinetics of repair were observed in each of the strains (Fig. 3, A and B). When the hat1Δ and H3 K14R,K23R mutations were combined, the level of chromatin reassembly was similar to that observed in the single mutants (Fig. 3C). The lack of an additive effect on chromatin reassembly suggests that the loss of Hat1p and the H3 K14R,K23R mutation may impact the same aspect of chromatin reassembly and is consistent with the observation that this mutant combination also does not cause a synthetic sensitivity to HO expression (see Fig. 1). Combining hat2Δ with the H3 K14R,K23R allele did not increase this reassembly defect. However, when hif1Δ was combined with the H3 K14R,K23R allele, there was a dramatic loss of chromatin reassembly (Fig. 3C). In fact, the hif1Δ/H3 K14R,K23R mutant had a defect in chromatin reassembly similar to that of the asf1Δ. The synthetic defect in chromatin reassembly with this combination of mutations mirrors the synthetic growth defect observed when these cells are grown on galactose (Fig. 1).
FIGURE 3.
Effect of combining the H3 K14R,K23R allele with mutations of the NuB4 complex components on DNA repair-linked chromatin reassembly. A, cutting and repair of the MAT locus in the indicated strains was monitored as described in the legend to Fig. 2. B, the levels of MATα and MATa fragments were quantitated as described in the legend to Fig. 2. C, ChIP analyses of H3 levels near the MAT locus in the indicated strains were performed and analyzed as described in the legend to Fig. 2.
Non-overlapping Action of Asf1p and NuB4 Complex Components in Chromatin Reassembly
Asf1p is an important histone chaperone that physically interacts with other histone chaperones that are involved in distinct pathways of chromatin assembly. For example, Asf1p interacts with both the CAF-1 complex and the Hir-Hpc complex that, respectively, are core components of the replication-coupled and replication-independent chromatin assembly pathways (36, 37). These interactions are thought to allow Asf1p to shuttle H3/H4 complexes into each of these assembly pathways (38). Recent evidence suggests that Asf1p also physically interacts with the NuB4 complex and that the NuB4 complex acts upstream of Asf1p in the process of chromatin assembly (38–40). Therefore, given that mutations in NuB4 complex components (and histone H3) cause defects in DNA repair-linked chromatin reassembly that are similar to those observed in an asf1Δ, we sought to determine whether the NuB4 complex and Asf1p act in concert or whether they function in distinct pathways to promote chromatin reassembly following DNA repair.
As expected from its importance in repair-linked chromatin reassembly, asf1Δ leads to a significant decrease in viability under HO-inducing conditions (galactose, Fig. 4A). When pairwise combinations of the hat1Δ, hat2Δ, hif1Δ, and H3 K14R,K23R alleles were constructed with an asf1Δ, only the H3 K14R,K23R allele showed a synthetic phenotype. The H3 K14R,K23R allele accentuated the slow growth phenotype of the asf1Δ strain on glucose and increased the sensitivity of asf1Δ to HO induction. When triple mutant combinations were examined, both the hat1Δ and hif1Δ increased the severity of the H3 K14R,K23R asf1Δ phenotypes with loss of Hif1p having a somewhat greater effect. Loss of Hat2p had no effect on any of the mutants (Fig. 4A).
The genetic interactions observed between ASF1 mutants and the HAT1, HIF1, and H3 mutants suggested that they are functioning in at least partially distinct pathways in the context of DNA repair-linked chromatin reassembly. To determine whether the synthetic sensitivity of these mutants to a double strand break resulted from a further decrease in chromatin reassembly activity, ChIP analysis was used to monitor chromatin structure at the MAT locus during repair. For these experiments the time course of HO induction was extended to 3 h to allow for more double strand break formation in the asf1Δ backgrounds (Fig. 4, B and C). As seen in Fig. 4D, following repression of HO synthesis, H3 levels gradually return during the course of repair in the asf1Δ mutant. However, in asf1Δ H3 K14R,K23R, asf1Δ H3 K14R,K23R hat1Δ, and asf1Δ H3 K14R,K23R hif1Δ mutants, there was no apparent restoration of histone H3 following repression of HO and, in fact, the levels of H3 continued to decrease. Intriguingly, the magnitude of the effect on chromatin reassembly correlated with the level of HO sensitivity of these mutants suggesting that the in vivo phenotype was a result of the chromatin reassembly defect.
Hif1p Is Present in High Molecular Weight Complexes Independent of Hat1p and Hat2p and Influences a Histone H3-specific HAT
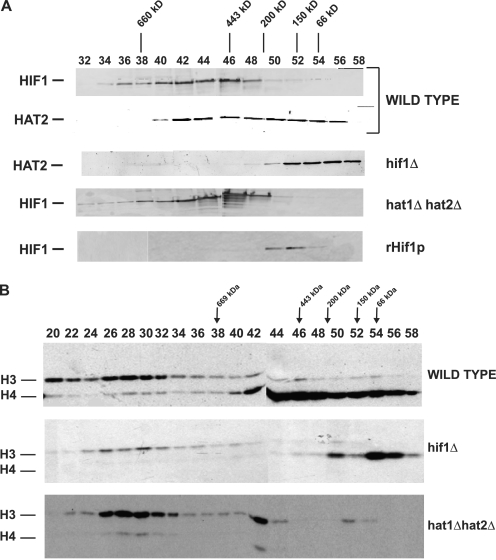
Hif1p was originally isolated and identified as a protein that interacts with the Hat1p/Hat2p complex in the nucleus (24, 25). The analysis of the role of these factors in DNA repair-mediated chromatin reassembly indicated that Hif1p plays a more significant role in this process than Hat1p and Hat2p suggesting that Hif1p may act independently of these factors. This led to the prediction that a portion of the native Hif1p in cells should exist in a form that is not physically associated with Hat1p/Hat2p. Therefore, to determine whether Hif1p was a component of complexes distinct from NuB4, we performed a biochemical characterization of the native protein isolated from yeast. Yeast whole cell extracts (made from a strain in which the endogenous Hif1p was fused to a Myc epitope tag) were fractionated over a Mono Q column and Hif1p containing fractions were concentrated and further fractionated by a gel filtration column (Superose 6) to resolve proteins and complexes by size. Hif1p eluted in a broad peak centered at ∼500 kDa (Fig. 5A). Hat2p eluted in two distinct peaks. The larger peak overlapped with the Hif1p peak and the molecular weight was consistent with that of the NuB4 complex (Hat1p/Hat2p/Hif1p) previously identified. The apparent molecular mass of the smaller peak, 150 to 200 kDa, closely matched the cytoplasmic Hat1p/Hat2p complex. In addition, these two peaks co-eluted with a strong histone H4-specific HAT activity (Fig. 5B).
FIGURE 5.
Hif1p is present in a high molecular weight complex independent of HAT1 and HAT2 and influences an H3-specific HAT activity. A, whole cell extracts were resolved by Superose 6 chromatography and the elution of Hif1p and Hat2p was determined by Western blot analysis as indicated. The elution position of size standards is indicated above the fraction numbers. The whole cell extracts were derived from strains with the genotypes indicated on the right. The bottom panel shows the elution pattern of rHif1p isolated from Escherichia coli. B, Superose 6 column fractions from the indicated strains were assayed for histone acetyltransferase activity using free histones [3H]acetyl-CoA as substrates. Reaction products were resolved by 18% SDS-PAGE and processed for fluorography. Position of radiolabeled histones was visualized by exposure of x-ray film. Migration of histones H3 and H4, as determined by Coomassie Blue staining, is indicated.
We then determined how the elution profile of Hat2p depended on the presence of Hif1p. As expected, deletion of the HIF1 gene caused a significant alteration in the Hat2p elution profile; with the loss of the higher molecular weight Hat2p peak and the corresponding H4-specific HAT activity (Fig. 5, A and B). This result strongly suggests that a significant fraction of Hat1p/Hat2p complex is associated with Hif1p.
Surprisingly, the converse was not true. Deletion of both HAT1 and HAT2 did not significantly affect the elution profile of Hif1p as the broad high molecular weight peak remained. This elution profile is not an intrinsic property of the protein as recombinant Hif1p eluted in a single narrow peak with an apparent molecular mass of ∼150 kDa. In addition, we observed a significant decrease in the level of an H3-specific HAT activity that elutes at >1 MDa in the hif1Δ cells that is not affected by the loss of Hat1p and Hat2p. Taken together, these observations suggest that native Hif1p is present in one, or more, high molecular weight complexes that are independent of Hat1p and Hat2p. In addition, whereas Hif1p does not appear to make a stable interaction with an H3-specific HAT complex, it is necessary for its full activity. These results confirm that Hif1p participates in Hat1p/Hat2p independent interactions as predicted by its more significant role in DNA repair-linked chromatin reassembly.
DISCUSSION
Although long hypothesized to be involved in chromatin assembly, directly linking Hat1p to this process has been problematic. One of the difficulties in studying the role of histone acetylation in chromatin assembly is that unlike transcription, which occurs at known places and times, chromatin assembly is a more fluid and transient phenomenon. An important advance in this area was reported in a recent study that exploited the HO-induced double strand break repair model system to monitor the disassembly and reassembly of the chromatin structure that occurs during the recombinational repair process (27, 30). This study demonstrated that Asf1p and the acetylation of histone H3 lysine 56 are required for the efficient reassembly of chromatin following repair of an HO-mediated double strand break at the MAT locus (27).
Several lines of evidence suggested that Hat1p might play a role in the reassembly of chromatin structure during the recombinational repair of a DNA double strand break. First, DNA damage sensitivity accompanies the loss of Hat1 activity in several eukaryotes (21–23). Second, the DNA damage sensitivity of S. cerevisiae hat1Δ cells is a result of defects in recombinational repair (21). Finally, Hat1p is recruited to chromatin at the MAT locus following an HO-induced double strand break (26). Therefore, we have used a similar strategy to determine whether Hat1p, as well as the other components of the NuB4 complex (Hat2p and Hif1p), also play a role in this repair-linked chromatin reassembly. We have demonstrated that there is a significant decrease in the rate at which chromatin is reformed near the site of a double strand break in hat1Δ cells providing direct evidence that Hat1p functions in a chromatin assembly process.
To date, yeast Hat1p has been found to be a component of two distinct complexes. In the cytoplasm, Hat1p is associated with Hat2p to form the HAT-B complex that is thought to acetylate newly synthesized histone H4 (18). In the nucleus, Hat1p is a subunit of a larger complex (termed the NuB4 complex) that contains Hat2p and the H3/H4-specific histone chaperone Hif1p (24, 25, 41). Both complexes have been found to be stably associated with histones H3 and H4 (24, 42). Surprisingly, the components of these complexes have diverse effects on the repair-linked chromatin reassembly process.
Based on both genetic and biochemical evidence, Hat2p has little influence on chromatin reassembly. Previous results have shown that the catalytic activity of Hat1p isolated from cells lacking Hat2p is decreased 10-fold (18). This suggests a number of possibilities. First, the role of Hat1p in the context of repair-linked chromatin reassembly may not be strongly dependent on its catalytic activity. Alternatively, Hat1p may have substrates other than histone H4 involved in chromatin reassembly and the ability of Hat1p to modify these proteins may not require Hat2p. Finally, in the context of the cell, Hat2p may not be as important for the catalytic activity of Hat1p as when the enzyme is assayed in vitro.
Conversely, the genetic and biochemical data indicate that Hif1p has functions in repair-linked chromatin reassembly that are independent of Hat1p. Hif1p may participate in multiple pathways of chromatin assembly of which only a subset involve Hat1p (or Hat2p). This is consistent with the more pronounced effect of hif1Δ on repair-linked chromatin reassembly. Alternatively, Hif1p may have both direct and indirect roles in chromatin reassembly. For example, in addition to acting directly in reassembly as a histone chaperone, Hif1p may indirectly affect chromatin reassembly through its influence on histone H3 acetylation. In the absence of Hif1p, there was a significant decrease in the activity of a histone H3-specific HAT despite the fact that Hif1p did not appear to form a stable complex with this enzyme activity (based on the lack of overlap in their elution profiles). The effect of Hif1p on this HAT activity is reminiscent of that between Asf1p and Rtt109p where Asf1p is required for Rtt109p activity in the absence of a stable association between them (40, 43–47). Alternatively, Hif1p may indirectly influence histone H3 acetylation, perhaps through transcriptional regulation of an H3-specific HAT. Although the H3-specific HAT activity that is affected by Hif1p has not been identified, if this activity targets residues other than H3 lysines 14 and 23, the synthetic interactions observed between the hif1Δ and the H3 K14R,K23R allele could be explained by a cumulative effect on H3 acetylation.
The possibility that Hif1p functions in pathways independent of Hat1p and Hat2p is consistent with the biochemical data presented here that indicated that Hif1p may be a component of a high molecular weight complex (or complexes) that does not contain Hat1p and Hat2p. This possibility is also consistent with results from Poveda and colleagues (25) where precipitation of epitope-tagged Hat1p co-precipitated only a small fraction of the Hif1p present in the extract. It will be interesting to determine whether the high molecular weight Hif1p-containing complexes are related to the multichaperone containing complexes from mammalian cells of which NASP, the human homolog of Hif1p, is a component (48–51).
Current models of chromatin assembly generally suggest that Asf1p plays a central role (38, 52). This histone chaperone is thought to participate in multiple pathways of chromatin assembly (as well as disassembly) by functioning to shuttle H3/H4 complexes into these pathways. This model is supported by the observation that Asf1p physically interacts with other histone chaperones that are specific for distinct chromatin assembly pathways, such as CAF-1 and the Hir-Hpc complex (36, 37). Hat1p and its associated factors are thought to act upstream of Asf1p as Asf1p has been found to be associated with newly synthesized histones that carry the acetylation pattern characteristic of Hat1p action (38, 53–55). In addition, a direct physical association between Asf1 and the NuB4 complex was recently reported suggesting the possibility that the NuB4 complex may directly transfer newly synthesized histones to Asf1p (40). Despite this biochemical data, the genetic results presented here indicated that mutations in HIF1, HAT1, and histone H3 show genetic interactions with mutations in ASF1 where combinations of these mutations generate increasing sensitivities to the HO-induced DNA double strand breaks and generate greater defects in chromatin reassembly. There are a number of interpretations for these results. First, Asf1p and NuB4 complex components may function in a common pathway but may be partially redundant for the optimal functioning of this pathway. For example, in the absence of Asf1p, the NuB4 complex may be capable of providing H3/H4 complexes to other chromatin assembly factors. Alternatively, the NuB4 complex may act in a chromatin assembly pathway that is distinct from Asf1p, perhaps exploiting the chromatin assembly activity of Hif1p.
The results presented here demonstrate that Hat1p and Hif1p function in the reassembly of chromatin that accompanies the recombinational repair of a DNA double strand break. It is not clear how this type of chromatin assembly relates to the replication-coupled and replication-independent pathways of chromatin assembly. It will be of interest to determine whether the NuB4 complex components influence these pathways of chromatin assembly, as well.
Supplementary Material
Footnotes
This work was supported, in whole or in part, by National Institutes of Health Grant GM62970 (to M. R. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
REFERENCES
- 1. Kouzarides T. (2007) Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 2. Ruiz-Carrillo A., Wangh L. J., Allfrey V. G. (1975) Science 190, 117–128 [DOI] [PubMed] [Google Scholar]
- 3. Jackson V., Shires A., Tanphaichitr N., Chalkley R. (1976) J. Mol. Biol. 104, 471–483 [DOI] [PubMed] [Google Scholar]
- 4. Annunziato A. T., Seale R. L. (1983) J. Biol. Chem. 258, 12675–12684 [PubMed] [Google Scholar]
- 5. Annunziato A. T., Hansen J. C. (2000) Gene Expr. 9, 37–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chicoine L. G., Schulman I. G., Richman R., Cook R. G., Allis C. D. (1986) J. Biol. Chem. 261, 1071–1076 [PubMed] [Google Scholar]
- 7. Sobel R. E., Cook R. G., Perry C. A., Annunziato A. T., Allis C. D. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 1237–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kuo M. H., Brownell J. E., Sobel R. E., Ranalli T. A., Cook R. G., Edmondson D. G., Roth S. Y., Allis C. D. (1996) Nature 383, 269–272 [DOI] [PubMed] [Google Scholar]
- 9. Masumoto H., Hawke D., Kobayashi R., Verreault A. (2005) Nature 436, 294–298 [DOI] [PubMed] [Google Scholar]
- 10. Ozdemir A., Spicuglia S., Lasonder E., Vermeulen M., Campsteijn C., Stunnenberg H. G., Logie C. (2005) J. Biol. Chem. 280, 25949–25952 [DOI] [PubMed] [Google Scholar]
- 11. Xu F., Zhang K., Grunstein M. (2005) Cell 121, 375–385 [DOI] [PubMed] [Google Scholar]
- 12. Ye J., Ai X., Eugeni E. E., Zhang L., Carpenter L. R., Jelinek M. A., Freitas M. A., Parthun M. R. (2005) Mol. Cell 18, 123–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang L., Eugeni E. E., Parthun M. R., Freitas M. A. (2003) Chromosoma 112, 77–86 [DOI] [PubMed] [Google Scholar]
- 14. Basu A., Rose K. L., Zhang J., Beavis R. C., Ueberheide B., Garcia B. A., Chait B., Zhao Y., Hunt D. F., Segal E., Allis C. D., Hake S. B. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 13785–13790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Das C., Lucia M. S., Hansen K. C., Tyler J. K. (2009) Nature 459, 113–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xie W., Song C., Young N. L., Sperling A. S., Xu F., Sridharan R., Conway A. E., Garcia B. A., Plath K., Clark A. T., Grunstein M. (2009) Mol. Cell 33, 417–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brownell J. E., Allis C. D. (1996) Curr. Opin. Genet. Dev. 6, 176–184 [DOI] [PubMed] [Google Scholar]
- 18. Parthun M. R., Widom J., Gottschling D. E. (1996) Cell 87, 85–94 [DOI] [PubMed] [Google Scholar]
- 19. Kleff S., Andrulis E. D., Anderson C. W., Sternglanz R. (1995) J. Biol. Chem. 270, 24674–24677 [DOI] [PubMed] [Google Scholar]
- 20. Kelly T. J., Qin S., Gottschling D. E., Parthun M. R. (2000) Mol. Cell. Biol. 20, 7051–7058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Qin S., Parthun M. R. (2002) Mol. Cell. Biol. 22, 8353–8365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Benson L. J., Phillips J. A., Gu Y., Parthun M. R., Hoffman C. S., Annunziato A. T. (2007) J. Biol. Chem. 282, 836–842 [DOI] [PubMed] [Google Scholar]
- 23. Barman H. K., Takami Y., Nishijima H., Shibahara K., Sanematsu F., Nakayama T. (2008) Biochem. Biophys. Res. Commun. 373, 624–630 [DOI] [PubMed] [Google Scholar]
- 24. Ai X., Parthun M. R. (2004) Mol. Cell 14, 195–205 [DOI] [PubMed] [Google Scholar]
- 25. Poveda A., Pamblanco M., Tafrov S., Tordera V., Sternglanz R., Sendra R. (2004) J. Biol. Chem. 279, 16033–16043 [DOI] [PubMed] [Google Scholar]
- 26. Qin S., Parthun M. R. (2006) Mol. Cell. Biol. 26, 3649–3658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen C. C., Carson J. J., Feser J., Tamburini B., Zabaronick S., Linger J., Tyler J. K. (2008) Cell 134, 231–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Osley M. A., Shen X. (2006) Trends Genet. 22, 671–677 [DOI] [PubMed] [Google Scholar]
- 29. Tsukuda T., Fleming A. B., Nickoloff J. A., Osley M. A. (2005) Nature 438, 379–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tsukuda T., Trujillo K. M., Martini E., Osley M. A. (2009) Methods 48, 40–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Haber J. E. (1995) Bioessays 17, 609–620 [DOI] [PubMed] [Google Scholar]
- 32. Adams A., Gottschling D. E., Kaiser C. A., Stearns T. (1997) Methods in Yeast Genetics, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 33. Durairaj G., Chaurasia P., Lahudkar S., Malik S., Shukla A., Bhaumik S. R. (2010) J. Biol. Chem. 285, 30472–30479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Adkins M. W., Howar S. R., Tyler J. K. (2004) Mol. Cell 14, 657–666 [DOI] [PubMed] [Google Scholar]
- 35. Korber P., Barbaric S., Luckenbach T., Schmid A., Schermer U. J., Blaschke D., Hörz W. (2006) J. Biol. Chem. 281, 5539–5545 [DOI] [PubMed] [Google Scholar]
- 36. Tyler J. K., Collins K. A., Prasad-Sinha J., Amiott E., Bulger M., Harte P. J., Kobayashi R., Kadonaga J. T. (2001) Mol. Cell. Biol. 21, 6574–6584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Green E. M., Antczak A. J., Bailey A. O., Franco A. A., Wu K. J., Yates J. R., 3rd, Kaufman P. D. (2005) Curr. Biol. 15, 2044–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Das C., Tyler J. K., Churchill M. E. (2010) Trends Biochem. Sci. 35, 476–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Campos E. I., Fillingham J., Li G., Zheng H., Voigt P., Kuo W. H., Seepany H., Gao Z., Day L. A., Greenblatt J. F., Reinberg D. (2010) Nat. Struct. Mol. Biol. 17, 1343–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fillingham J., Recht J., Silva A. C., Suter B., Emili A., Stagljar I., Krogan N. J., Allis C. D., Keogh M. C., Greenblatt J. F. (2008) Mol. Cell. Biol. 28, 4342–4353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ruiz-García A. B., Sendra R., Galiana M., Pamblanco M., Pérez-Ortín J. E., Tordera V. (1998) J. Biol. Chem. 273, 12599–12605 [DOI] [PubMed] [Google Scholar]
- 42. Mosammaparast N., Guo Y., Shabanowitz J., Hunt D. F., Pemberton L. F. (2002) J. Biol. Chem. 277, 862–868 [DOI] [PubMed] [Google Scholar]
- 43. Adkins M. W., Carson J. J., English C. M., Ramey C. J., Tyler J. K. (2007) J. Biol. Chem. 282, 1334–1340 [DOI] [PubMed] [Google Scholar]
- 44. Tsubota T., Berndsen C. E., Erkmann J. A., Smith C. L., Yang L., Freitas M. A., Denu J. M., Kaufman P. D. (2007) Mol. Cell 25, 703–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Han J., Zhou H., Li Z., Xu R. M., Zhang Z. (2007) J. Biol. Chem. 282, 28587–28596 [DOI] [PubMed] [Google Scholar]
- 46. Han J., Zhou H., Li Z., Xu R. M., Zhang Z. (2007) J. Biol. Chem. 282, 14158–14164 [DOI] [PubMed] [Google Scholar]
- 47. Selth L., Svejstrup J. Q. (2007) J. Biol. Chem. 282, 12358–12362 [DOI] [PubMed] [Google Scholar]
- 48. Tagami H., Ray-Gallet D., Almouzni G., Nakatani Y. (2004) Cell 116, 51–61 [DOI] [PubMed] [Google Scholar]
- 49. Lewis P. W., Elsaesser S. J., Noh K. M., Stadler S. C., Allis C. D. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 14075–14080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Campos E. I., Reinberg D. (2010) Genes Dev. 24, 1334–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Drané P., Ouararhni K., Depaux A., Shuaib M., Hamiche A. (2010) Genes Dev. 24, 1253–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. De Koning L., Corpet A., Haber J. E., Almouzni G. (2007) Nat. Struct. Mol. Biol. 14, 997–1007 [DOI] [PubMed] [Google Scholar]
- 53. Groth A., Corpet A., Cook A. J., Roche D., Bartek J., Lukas J., Almouzni G. (2007) Science 318, 1928–1931 [DOI] [PubMed] [Google Scholar]
- 54. Groth A., Ray-Gallet D., Quivy J. P., Lukas J., Bartek J., Almouzni G. (2005) Mol. Cell 17, 301–311 [DOI] [PubMed] [Google Scholar]
- 55. Jasencakova Z., Scharf A. N., Ask K., Corpet A., Imhof A., Almouzni G., Groth A. (2010) Mol. Cell 37, 736–743 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.