Abstract
We made use of EXLX1, an expansin from Bacillus subtilis, to investigate protein features essential for its plant cell wall binding and wall loosening activities. We found that the two expansin domains, D1 and D2, need to be linked for wall extension activity and that D2 mediates EXLX1 binding to whole cell walls and to cellulose via distinct residues on the D2 surface. Binding to cellulose is mediated by three aromatic residues arranged linearly on the putative binding surface that spans D1 and D2. Mutation of these three residues to alanine eliminated cellulose binding and concomitantly eliminated wall loosening activity measured either by cell wall extension or by weakening of filter paper but hardly affected binding to whole cell walls, which is mediated by basic residues located on other D2 surfaces. Mutation of these basic residues to glutamine reduced cell wall binding but not wall loosening activities. We propose domain D2 as the founding member of a new carbohydrate binding module family, CBM63, but its function in expansin activity apparently goes beyond simply anchoring D1 to the wall. Several polar residues on the putative binding surface of domain D1 are also important for activity, most notably Asp82, whose mutation to alanine or asparagine completely eliminated wall loosening activity. The functional insights based on this bacterial expansin may be extrapolated to the interactions of plant expansins with cell walls.
Keywords: Carbohydrate-binding Protein, Cell Wall, Plant, Protein Domains, Protein Motifs, Protein Structure, Cellulose, Expansin, Site-directed Mutagenesis, Structure-Function
Introduction
Expansins were discovered in the 1990s as proteins that induce extension and stress relaxation of plant cell walls at low pH (1). Their biological functions have meanwhile been extended by numerous molecular, genomic, and genetic studies implicating them in diverse physiological and developmental processes, including cell enlargement, fruit softening, pollination, leaf abscission, and leaf primordium emergence (2–15). Two expansin families, named α-expansin and β-expansin, are found in all groups of land plants (16, 17) and evidently have different targets in the plant cell wall (1, 18).
The action of expansin on the cell wall is not yet understood in molecular detail. The leading hypothesis is that it loosens plant cell walls by disrupting the noncovalent binding of matrix polysaccharides to cellulose (19–21), resulting in physical effects, such as polymer creep and stress relaxation of extended (stretched) cell walls (20, 22). The crystal structures of two β-expansins have been solved (Protein Data Bank codes 2HCZ and 1N10), showing them to consist of two compact domains, D1 and D2, tightly packed onto each other with an open, highly conserved surface spanning the two domains. This putative polysaccharide binding surface (PPBS)3 is lined with aromatic and polar residues suitable for polysaccharide binding (19). The functions of the two domains and of specific PPBS residues have not yet been tested experimentally because heterologous expression of plant expansins has proved difficult, thus limiting detailed structure-function analysis.
Opening a potential route around this impasse, a Bacillus subtilis protein, named YOAJ or EXLX1, was found to possess structural features and wall extension activities characteristic of plant expansins (21). Related proteins are found in phylogenetically diverse bacteria that infect plants and cause vascular wilt disease. Gene knock-out studies indicate that these “bacterial expansins” promote plant infection or root surface colonization by bacteria, possibly by modifying plant cell walls (21, 23).
Similar to plant expansins, EXLX1 has a mass of 23 kDa and consists of two compact domains, D1 and D2, connected by a 4-amino acid linker and packed closely against each other. Domain D1 forms a six-stranded double-ψ β-barrel with high structural similarity to D1 of plant expansins, much lower similarity to GH-45 glycoside hydrolases, and even more distant similarity to GH-102 peptidoglycan lytic transglycosylases (21). Aspartic acid residue Asp-82 in EXLX1 corresponds to an aspartic acid residue in the catalytic sites of GH-45 and GH-102 enzymes, but other residues essential for these catalytic activities are missing in EXLX1 as well as in plant expansins. Consistent with this structure, no bona fide lytic activity has been detected in EXLX1 or other expansins (19–21, 24). Domain D2 in EXLX1 is structurally related to domain D2 of plant expansins, forming an Ig-like β-sandwich. An open, nearly planar PPBS, ∼50 Å long, spans the two EXLX1 domains, formed by Asp-82 and other polar residues in domain D1 and by three aromatic residues (Trp-125, Trp-126, and Tyr-157) in D2 (21).
In this study, we exploited the ease of EXLX1 expression in Escherichia coli to create protein variants to assess the roles of the two domains for plant cell wall loosening and binding activities. Additionally, we modified conserved residues on the PPBS to assess their importance for wall loosening and binding activities with results that may be extrapolated to plant expansin function.
EXPERIMENTAL PROCEDURES
Polysaccharides
Avicel (FMC BioPolymer, PH-101), fibrous cotton fibers (Sigma, C-6288) and filter paper (Whatman No. 3 and VWR 413) were used as cellulose substrates. Phosphoric acid-swollen cotton fibers and phosphoric acid-swollen Avicel were prepared as described (25). Insoluble arabinoxylan from wheat flour was purchased from Megazyme (lot number 20301).
Plant Materials
Wheat coleoptiles (Triticum aestivum L. cv. Pennmore) were prepared as described (26). Fresh celery (Apium graveolens) was purchased from a local market. Parenchyma strips (10 × 1.5 × 0.5 mm) for wall extension assays were prepared from the apical 5-cm region of the inner five to six petioles.
Wheat coleoptile cell walls were prepared by an adapted protocol (27, 28). Five grams of wheat coleoptiles (1–1.5 cm long, abraded) were ground to fine powder in liquid N2 and washed five times with 50 ml of 1 m NaCl at 4 °C for 2 h by inversion. The residues (mostly cell walls) were vacuum-dried for 2 h and stored at room temperature. For binding studies, cell walls were resuspended in 25 mm HEPES, pH 7.5. For sequential extractions, cell walls were first resuspended by inversion in 50 ml of 50 mm CDTA in 20 mm potassium phosphate, pH 7 overnight at room temperature and washed five times with deionized H2O for 1 h. CDTA was used to solubilize pectins. The wall residues were then resuspended in 50 ml of 0.1 m KOH + 0.3% NaBH4, inverted overnight at room temperature to solubilize loosely bound hemicelluloses, and then washed five times with deionized H2O for 1 h. Finally, to solubilize tightly bound hemicelluloses, residues were resuspended in 50 ml of 4 m KOH + 4% H3BO3 + 0.5% NaBH4, inverted overnight at room temperature, and washed five times with deionized H2O for 1 h. NaN3 (3 mm) was included in steps longer than 2 h to prevent microbial growth.
Bacterial Materials
Gluconacetobacter xylinus was grown as described (29). In brief, G. xylinus cultures were grown in Hestrin-Schramm medium (30) with 2% (w/v) glucose at 30 °C for 72 h under static conditions. Cellulose pellicles were extensively washed with deionized H2O and stored in 3 mm NaN3 at 4 °C. Pellicle strips (10 × 0.5 × 0.5 mm) were prepared for extension assays.
Cloning and Expression of Wild Type EXLX1 and EXLX1 Variants
EXLX1 was amplified from B. subtilis genomic DNA by PCR using 5′-GGTTCCATGGCATATGACGACCTGCATGAAGG-3′ and 5′-CAGCTCGAGTTATTCAGGAAACTGAAC-3′ as primers. Subsequently, EXLX1 was cloned between NcoI and XhoI sites of pET22b (Novagen). The original signal peptide of EXLX1 was substituted with pelB, and a methionine was added at the N terminus of the mature EXLX1. D1 and D2 domains were amplified by PCR using primers 5′-GCAGCATATGGACGACCTGCATG-3′ and 5′-CAGCTCGAGTTAGACAACACGCCATTTAAT-3′ and primers 5′-CAGCATATGAATTTCACGTACCGGATC-3′ and 5′-CAGCTCGAGTTATTCAGGAAACTGAAC-3′, respectively. D1 and D2 were cloned into pET22b between the NdeI and XhoI sites. EXLX1 variants were generated by site-directed mutagenesis (Stratagene QuikChange kit). The primers used to generate EXLX1 variants for structure-function analysis are listed in supplemental Table S1. All modifications were confirmed by sequencing.
EXLX1 and variants were expressed in E. coli strain BL21 (DE3-pLys). Cultures were grown to A600 = 0.6 at 37 °C and then induced with 0.1 mm isopropyl 1-thio-β-d-galactopyranoside for 4 h at room temperature.
Protein Purification
Wild type EXLX1 and all EXLX1 variants (except D1) were purified as described (21) except that 50 mm HEPES, pH 7.5 was used instead of 50 mm Tris·HCl, pH 8.0. The purity of 10-μg samples was visualized by 10% SDS-PAGE (15% for domains D1 and D2) and staining with Coomassie Brilliant Blue R-250.
Domain D1 Purification
The cell pellet from a 1-liter culture was resuspended in 40 ml of buffer containing 50 mm HEPES, pH 7.5, 5 mm DTT, 1% CHAPS, 1 mm EDTA, and ½ pellet of protease inhibitors (SIGMAFAST, Sigma-Aldrich). Proteins were precipitated by addition of 24 ml of 3.5 m ammonium sulfate, mixed at 4 °C for 15 min, and then centrifuged at 18,000 × g for 15 min. The pellet was resuspended in 25 mm HEPES, pH 7.5 to a final volume of 2.5 ml and desalted on a PD-10 desalting column (GE Healthcare). The sample was filtered through a 0.2-μm Whatman GD/X polyether sulfone (PES) filter and loaded onto a HiPrep Sephacryl S-100 column (GE Healthcare) using 25 mm HEPES, pH 7.5 + 0.15 m NaCl as the mobile phase at a flow rate of 0.5 ml/min. The purified protein was desalted and concentrated with an Amicon 10-kDa filter (Millipore). In contrast to EXLX1 and D2, D1 precipitated when stored at 4 °C for more than 3 days. Therefore, D1 was stored at −80 °C until use.
Binding Assays
Binding of EXLX1 and variants to cellulose, insoluble arabinoxylans, and wheat coleoptile cell walls, including sequentially extracted cell walls, was analyzed by depletion isotherms. In brief, variable amounts of EXLX1 were added to buffer containing a fixed amount of binding substrate. The mixture was shaken on a Thermomixer R (Eppendorf) set at 1100 rpm and 25 °C until equilibrium was reached (1 h). The samples were centrifuged at 14,000 × g for 10 min to pellet the binding substrate. Protein in the supernatant was quantified by the Bradford assay (Pierce) using BSA for calibration. Soluble protein was subtracted from the protein initially added to obtain the protein bound to the insoluble polysaccharides. Dissociation constants (Kd) and binding capacities were calculated by fitting the data to a single site Langmuir isotherm with Origin 7 (OriginLab).
Wall Extension Assays with Various Substrates
Wheat coleoptiles, celery, and G. xylinus strips were clamped in a constant force extensometer at 25-, 12.5-, and 20-g force, respectively, in 25 mm HEPES, pH 7.5. Specimen length was recorded at 30-s intervals before and after addition of wild type EXLX1 or EXLX1 variants as described (1, 18). To weaken wheat coleoptiles, the specimen was incubated in 70 mm NaOH for 20 min, washed extensively with deionized H2O, and finally incubated in 25 mm HEPES, pH 7.5 or other buffer as indicated in the text.
Circular Dichroism (CD) Spectra
Proteins were dialyzed extensively against 5 mm sodium phosphate, pH 7.0. CD spectra of EXLX1 and variants were collected on a Jasco 810 spectropolarimeter from 190 to 240 nm with a step of 1.0 nm and a 2-s averaging time. Four replicates per EXLX1 variant were averaged. After subtraction of buffer spectra, CD spectra were smoothed by the means-movement method and analyzed using the DichroWeb on-line server (31, 32). The CONTINLL program was used to deconvolute CD spectra using SP175 as a reference data set (33–35). The secondary structure of EXLX1 and variants was compared with the actual secondary structure of EXLX1 as determined from Protein Data Bank code 3D30 structure (21).
Paper Strength Determination
Whatman filter paper No. 3 was cut into 10 × 1.5-mm strips. Ten strips were incubated with 120 μg/ml EXLX1 and variants in 1 ml of 25 mm HEPES, pH 7.5. The samples were gently inverted for 4 h at 25 °C. Strips were clamped onto an extensometer and extended at 1.5 mm/min until breakage. The force at the point of breakage was recorded.
Labeling EXLX1 with Alexa Fluor and Staining of Cellulose and Cell Wall
Amino acid residue Asp-96 in EXLX1 was changed to a cysteine to enable the protein to be labeled with Alexa Fluor 488 C5-maleimide (Invitrogen). The change was introduced by site-directed mutagenesis (Stratagene QuikChange kit) with 5′-CGGCAATATGAAATGCGGAAAAATCAATATTAAATG-3′ and 5′-CATTTAATATTGATTTTTCCGCATTTCATATTGCCG-3′ as primers. Asp-96 is located in domain D1, but it is not part of the PPBS. D96C (92 μm) was incubated in 1.2 ml of 50 mm HEPES, pH 7.5, 1 mm tris(2-carboxyethyl)phosphine, and 1 mm Alexa Fluor 488 for 2 h at room temperature in the dark. Alexa Fluor 488-labeled D96C was separated on a PD-10 desalting column (GE Healthcare) three times to remove all traces of free Alexa Fluor 488. Labeled protein was concentrated in 25 mm HEPES, pH 7.5 on an Amicon 10-kDa filter. Labeled protein (5 μg/ml) was incubated with 10 mg/ml Avicel or 0.5 mg/ml fresh wheat coleoptile cross-sections in 0.3 ml of 25 mm HEPES, pH 7.5 for 1 h on a Thermomixer R (Eppendorf) set at 1100 rpm and 25 °C. Labeled samples were washed five times with 1 ml of 25 mm HEPES, pH 7.5 for 1 h with shaking, mounted onto glass slides, and viewed on a Zeiss Axioplan fluorescence microscope with a GFP filter set.
Protein Alignment
Protein alignment was done by using the MUSCLE server (36). Sequence logos were generated by WebLogo server (37).
Protein Models
Protein structures were visualized with University of California San Francisco Chimera software (38).
RESULTS
Optimizing Wall Loosening Assays for EXLX1 Structure-Function Analysis
Wall loosening by expansins may be measured by clamping cell wall specimens in a constant force extensometer and measuring the rate of wall extension before and after addition of expansin (1, 18). Because EXLX1 exhibits relatively low specific activity in such “creep assays” compared with plant expansins (21), we first sought to increase its activity for our structure-function analysis. To that end, we tested walls of differing composition, including cell walls from wheat coleoptiles and celery parenchyma as well as cellulose pellicles from G. xylinus. EXLX1 induced wall creep with all these materials (supplemental Fig. S1, A–C), but activity was weak, variable, or both. The best results were obtained with wheat coleoptile walls pretreated briefly with 70 mm NaOH, which increased EXLX1 response ∼5-fold (supplemental Fig. S1, A and D). Additionally, this pretreatment reduced sample-to-sample variability. The sensitivity and reproducibility gained by this pretreatment made structure-function analysis of EXLX1 feasible.
The rate of wall extension depended on EXLX1 concentration, increasing steeply in the 0–25 μg/ml range and approaching saturation at concentrations >50 μg/ml (Fig. 1A). At >200 μg/ml, the wall samples tended to break after a short time, making accurate estimation of the maximum rate of wall extension difficult. For our analysis (below), EXLX1 variants were routinely tested for activity at 30 μg/ml. Higher protein amounts were used for EXLX1 variants with little or no activity.
FIGURE 1.
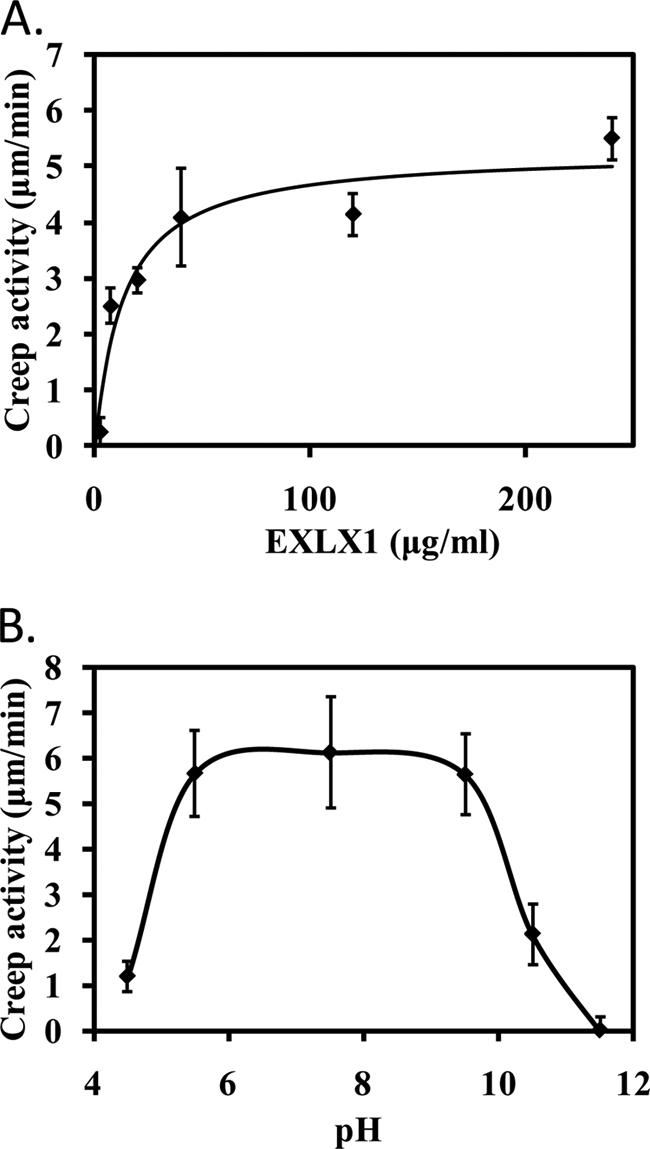
Characterization of EXLX1 wall extension activity (creep) with alkali-pretreated wheat coleoptiles. A, concentration dependence. B, pH dependence using 150 μg/ml EXLX1. The buffers used to stabilize the pH were 25 mm NaOAc, pH 4.5; 25 mm NaOAc, pH 5.5; 25 mm HEPES, pH 7.5; 25 mm Tris, pH 9.5; 25 mm borate, pH 10.5; 25 mm borate, pH 11.5; 25 mm sodium phosphate buffer, pH 10.5; and 25 mm sodium phosphate buffer, pH 11.5. Error bars indicate S.E. (4 ≤ n ≤ 20).
EXLX1 induced cell wall creep at pH values from 5.5 to 9.5 (Fig. 1B); at pH = 4.5, the activity was very low, whereas at lower pH values, EXLX1 precipitated. Activity was reduced at pH 10.5 and was not detected at pH 11.5. Two different buffers were tested at pH 10.5 and 11.5 with similar results, reducing the possibility of buffer-specific inhibition. Based on these results, the activity of EXLX1 variants was routinely assayed in 25 mm HEPES, pH 7.5.
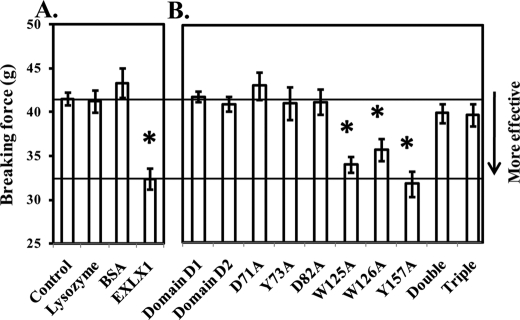
In addition to these wall creep assays, we tested the ability of EXLX1 to weaken Whatman filter paper (a network of pure cellulose) by extending uniform strips under increasing force until they broke (39). Preincubation of strips with EXLX1 reduced the breakage force by 20% compared with preincubation with buffer (Fig. 2A). Although this effect was reproducible, it was not sensitive enough to be used alone for structure-function analysis of EXLX1. Instead, we used it as a supplemental assay to test EXLX1 variants with little or no cell wall extension activity.
FIGURE 2.
Filter paper weakening activity of EXLX1 and selected EXLX1 variants. Activities of wild type EXLX1 (A) and selected EXLX1 variants (B) were tested at a concentration of 120 μg/ml. Equimolar amounts of lysozyme and BSA were used as negative controls. HEPES (25 mm), pH 7.5 was used as buffer control. Error bars indicate S.E. (8 ≤ n ≤ 10). Double and Triple variants correspond to W125A/W126A and W125A/W126A/Y157A, respectively. * denotes p ≤ 0.05 (two-tailed t test).
Production and Purification of EXLX1 Variants
We designed protein variants consisting of single domains or full-length proteins with specific PPBS residues modified by site-directed mutagenesis. EXLX1 variants were expressed in E. coli, purified by cation exchange chromatography, and tested for wall loosening activities or for wall binding. Protein purity was evaluated by SDS-PAGE (supplemental Fig. S2). The secondary structure of EXLX1 variants, including single domain variants, was assessed by circular dichroism spectra to ensure that the proteins were not denatured or seriously misfolded (supplemental Table S2).
Roles of Domains D1 and D2
To test the hypothesis that wall loosening activity is mediated solely by one of the domains, each EXLX1 domain was expressed as described above and tested for wall creep activity. No activity was detected for either domain at concentrations as high as 100 μg/ml (Fig. 3). On a molar basis, this concentration is ∼26-fold higher than an EXLX1 concentration of 7.5 μg/ml, which induced a substantial creep response (Fig. 1A). Likewise, a mixture of D1 and D2 together did not result in activity (Fig. 3). These results indicate that D1 and D2 by themselves have negligible wall extension activity and need to be connected for effective wall extension activity.
FIGURE 3.
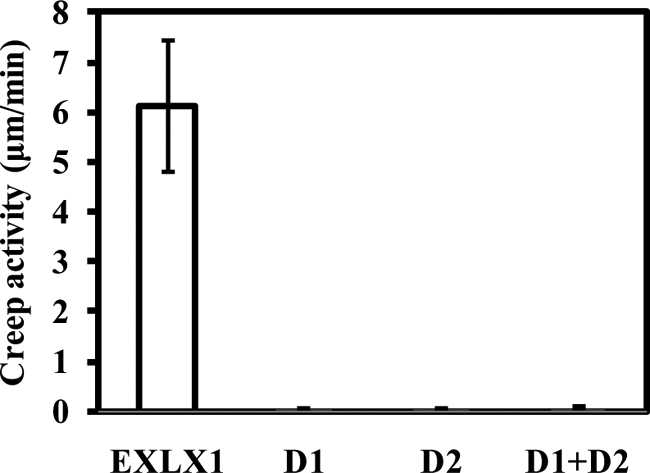
Creep activity of domains D1 and D2 and full-length EXLX1. Domain D1 (100 μg/ml), domain D2 (100 μg/ml), a mixture of D1 and D2 (100 μg/ml each), and full-length EXLX1 (200 μg/ml) were tested for creep activity with alkali-pretreated wheat coleoptiles. Error bars indicate S.E. (4 ≤ n ≤ 8).
D1 Residues Needed for Wall Extension Activity
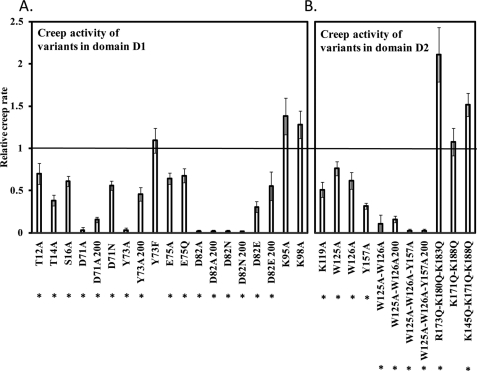
Thr-12 and Asp-82 in domain D1 (shown in Fig. 4) are absolutely conserved among expansins as well as in some distantly related enzymes (19, 21). Despite its strict conservation, Thr-12 was not essential for activity because the T12A variant exhibited ∼70% wall extension activity compared with wild type EXLX1 (Fig. 5A). Therefore, the role of Thr-12 may be subtle or not related to wall loosening. In contrast, neither D82A nor D82N showed detectable creep activity even at 200 μg/ml (Fig. 5A), whereas the D82E variant had 30% activity compared with wild type EXLX1 (Fig. 5A). These results suggest that the carboxyl group of Asp-82 is crucial for wall extension activity.
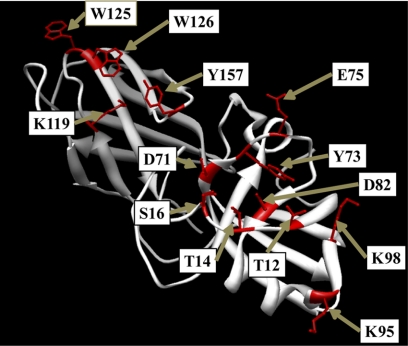
FIGURE 4.
Location of key EXLX1 amino acid residues modified by site-directed mutagenesis.
FIGURE 5.
Creep activity of EXLX1 variants. A, creep activity of amino acid variants in domain D1 relative to wild type EXLX1. B, creep activity of amino acid variants in domain D2 relative to wild type EXLX1. Alkali-pretreated wheat coleoptiles were used as substrate. When activity was tested at 200 instead of 30 μg/ml, the number 200 appears next to the name of the EXLX1 variant. Error bars indicate S.E. (4 ≤ n ≤ 24). * denotes p ≤ 0.05 (two-tailed t test).
We also mutated other residues near Asp-82 that are part of the PPBS on the D1 domain (Thr-14, Ser-16, Asp-71, Tyr-73, Glu-75, Lys-95, and Lys-98) (Fig. 5A). D71A showed greatly reduced activity, whereas D71N had moderate activity (∼50%) compared with wild type EXLX1 (Fig. 5A). Thus, the carboxyl group at Asp-71 is important but not essential for wall creep activity. Y73A activity was severely reduced, whereas a more conservative substitution, Y73F, gave good activity (Fig. 5A), suggesting that the aromatic ring of Tyr-73, but not its hydroxyl group, is important for activity. The roles of Thr-14 and Ser-16 were studied by altering them to alanine. Thr-14 is close to Asp-82 (3.6 Å) and could form H-bonds with Asp-82 or with a polysaccharide ligand. Indeed, T14A showed an ∼60% reduction in activity compared with wild type (Fig. 5A). S16A was only moderately reduced in activity (39%; Fig. 5A). Likewise, E75A and E75Q activities were reduced by 30–35% (Fig. 5A), indicating that Glu-75 plays a moderate role in wall creep activity. Finally, alanine substitutions of Lys-95 and Lys-98 (Fig. 4) did not affect activity. Collectively, these results show that many residues on the conserved D1 surface contribute to activity with mutation of Asp-82 resulting in complete loss of activity, whereas mutations of other residues resulted in reduced activity.
D2 Residues Needed for Wall Extension Activity
Because Lys-119 along with the aromatic amino acids Trp-125, Trp-126, and Tyr-157 are part of the PPBS spanning D1 and D2, their importance for wall creep activity was tested by mutagenesis. K119A, W125A, W126A, and Y157A variants showed reduced activity by ∼50, ∼25, ∼35, and ∼65%, respectively, compared with wild type EXLX1 (Fig. 5B). The double change in W125A/W126A almost entirely eliminated activity at 30 μg/ml and led to ∼80% less activity at 200 μg/ml compared with wild type (Fig. 5B). The triple variant W125A/W126A/Y157A had no activity at 200 μg/ml (Fig. 5B). The results indicate that these residues on the conserved D2 surface are important for EXLX1 creep activity.
Activity of EXLX1 Variants on Pure Cellulose
Domain D1 or D2 alone did not weaken paper (Fig. 2B) in agreement with the corresponding cell wall creep results. Likewise, the inactive variants D71A, D82A, and Y73A did not weaken filter paper (Fig. 2B). In contrast, the activities of the variants W125A, W126A, and Y157A were indistinguishable from wild type EXLX1 (Fig. 2B). One possibility is that these aromatic residues are not important for activity against cellulose. Another possibility is that the paper strength assay is not sensitive enough to distinguish wild type EXLX1 from these moderately inferior variants. The second possibility is more likely because W125A/W126A and W125A/W126A/Y157A variants were much less competent than wild type EXLX1 in weakening filter paper (Fig. 2B).
These results indicate that the PPBS, defined by Asp-71, Tyr-73, and Asp-82 in domain D1 and by Trp-125, Trp-126, and Tyr-157 in domain D2, is crucial for activity with pure cellulose. Additionally, D1 and D2 need to be connected for weakening paper. Thus, there is a good correspondence in the results of the wall creep assay and paper strength assay, implicating cellulose as a key target in both assays.
Protein Features Important for Wall Binding
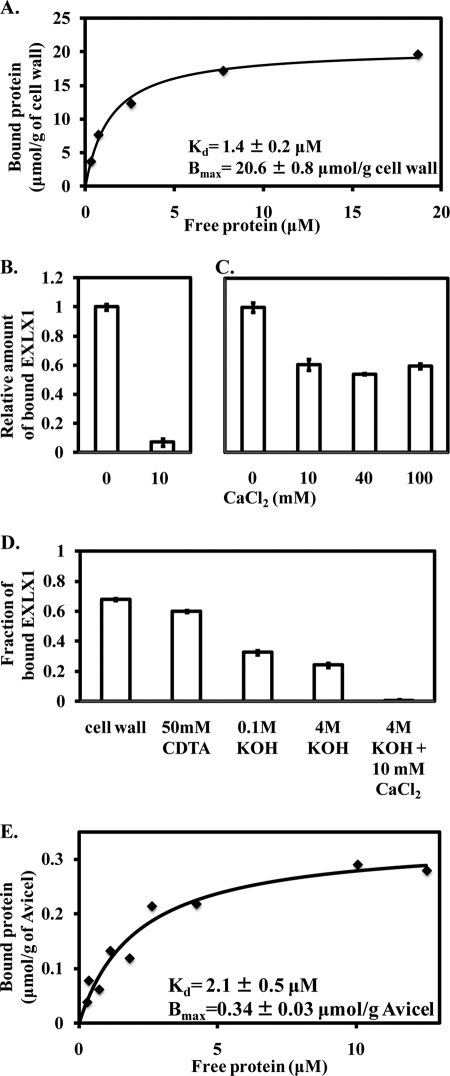
To identify the potential targets of EXLX1, the binding of EXLX1 variants to plant cell walls and to selective wall components was studied and compared with wall creep activity. We first analyzed the binding of wild type EXLX1 to cell walls. Binding was consistent with a single binding site in a Langmuir isotherm model with an affinity for EXLX1 of Kd = 1.4 μm and a binding capacity of Bmax = 20.6 μmol/g of cell wall (Table 1 and Fig. 6A). Binding was also visualized with EXLX1 fluorescently labeled with Alexa Fluor 488 (Fig. 7, A and B). Binding to wheat coleoptile cross-sections was seen in all cell walls but most abundantly at cell corners flanking the intercellular spaces, a pattern similar to homogalacturonan labeling (40), suggesting binding of EXLX1 to pectin. Because EXLX1 is a basic protein (pI = 9.3), electrostatic binding to pectin may be expected. Supporting this idea, inclusion of 10 mm CaCl2 in the binding assay eliminated >90% of the binding to whole cell walls (Fig. 6B). Pretreatment of cell walls with 70 mm NaOH, used in the creep assay, did not affect EXLX1 binding (supplemental Fig. S3A).
TABLE 1.
Characterization of binding of EXLX1 variants to Avicel, wheat coleoptile cell walls, and insoluble arabinoxylan
HEPES (25 mm), pH 7.5 was used as a buffer. Values shown are mean ± S.E. (5 ≤ n ≤ 10).
| Substrate | Bmax | Kd |
|---|---|---|
| μmol/g substrate | μm | |
| Avicel | ||
| Wild type | 0.34 ± 0.03 | 2.12 ± 0.51 |
| D1 | a | a |
| D2 | 0.32 ± 0.02 | 2.23 ± 0.42 |
| K119A | 0.26 ± 0.02 | 3.53 ± 1.02 |
| W125A | 0.14 ± 0.03 | 5.72 ± 1.21 |
| W126A | 0.15 ± 0.03 | 4.62 ± 1.58 |
| Y157A | 0.25 ± 0.03 | 5.61 ± 1.53 |
| W125/W126A | 0.13 ± 0.03 | 10.54 ± 2.61 |
| W125/W126A/Y157A | 0.10 ± 0.03 | 9.95 ± 2.73 |
| K171Q/K188Q | 0.29 ± 0.02 | 1.99 ± 0.28 |
| R173Q/K180Q/K183Q | 0.29 ± 0.02 | 3.48 ± 0.59 |
| Wheat coleoptile cell walls | ||
| Wild type | 20.6 ± 0.8 | 1.41 ± 0.22 |
| D1 | a | a |
| D2 | 30.1 ± 1.8 | 1.79 ± 0.18 |
| K119A | 13.5 ± 1 | 4.43 ± 0.92 |
| W125/W126A/Y157A | 19.1 ± 1.6 | 3.19 ± 0.77 |
| K171Q | 13.3 ± 3.2 | 4.91 ± 2.25 |
| K188Q | 16.7 ± 1.5 | 6.47 ± 1.24 |
| K171Q/K188Q | a | a |
| R173Q/K180Q/K183Q | a | a |
| K145Q/K171Q/K188Q | a | a |
| Insoluble arabinoxylan | ||
| Wild type | 3.3 ± 0.4 | 4.72 ± 1.61 |
| R173Q/K180Q/K183Q | a | a |
| K145Q/K171Q/K188Q | a | a |
a Too low to determine accurately.
FIGURE 6.
Binding of EXLX1 to wheat coleoptile cell walls and cellulose. A, binding isotherm of EXLX1 to cell walls. HEPES (25 mm), pH 7.5 was used as buffer. The effect of CaCl2 on EXLX1 binding to wheat coleoptile cell walls (B) and Avicel (C) is shown. In both cases, 12 μg/ml EXLX1 was incubated in 0.3 ml of 25 mm HEPES, pH 7.5 containing 0.5 mg/ml cell walls or 10 mg/ml Avicel. Error bars indicate S.E. (3 ≤ n ≤ 6). D, binding of EXLX1 to cell wall residues after sequential extraction of matrix polysaccharides. E, binding isotherm of EXLX1 to Avicel. HEPES (25 mm), pH 7.5 was used as buffer. Error bars indicate S.E. (4 ≤ n ≤ 10).
FIGURE 7.
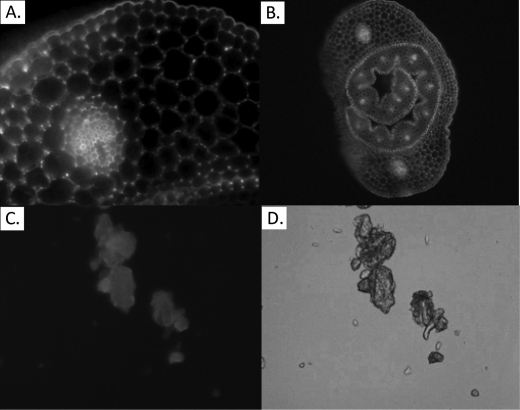
Visualization of Alexa Fluor 488-labeled EXLX1 binding to wheat coleoptile cross-sections (A and B) and Avicel (C) by fluorescence microscopy is shown. The wheat coleoptile cross-section in B includes the young leaf in addition to the surrounding coleoptile. D, visualization of Avicel by light microscopy.
To identify the major binding target of EXLX1, we measured its binding to walls that were sequentially extracted to remove pectins and hemicelluloses. The results indicate that EXLX1 binds predominantly to pectins and hemicelluloses (Fig. 6D). As a further test, we measured EXLX1 binding to insoluble arabinoxylan from wheat flour, which is similar to glucuronoarabinoxylan, the dominant hemicellulose in grass coleoptiles (28). EXLX1 bound with an affinity of Kd = 4.7 μm and a binding capacity of Bmax = 3.3 μmol/g of insoluble arabinoxylan (Table 1). This binding is also mediated by polar or electrostatic forces because inclusion of 10 mm CaCl2 during the binding assay eliminated binding (supplemental Fig. S3B). Collectively, our results indicate that EXLX1 binds primarily to the matrix polysaccharides of wheat coleoptile cell walls through electrostatic or polar forces.
EXLX1 Binding to Cellulose
Because EXLX1 induced creep of G. xylinus pellicle strips and weakened filter paper (both consisting of pure cellulose networks), EXLX1 binding to cellulose was studied. EXLX1 bound to several forms of cellulose, including cotton fibers, phosphoric acid-swollen fibrous cotton fibers, Avicel, phosphoric acid-swollen Avicel, and filter paper (supplemental Fig. S3C). EXLX1 binding to Avicel was consistent with a single binding site having an affinity of Kd = 2.1 μm for EXLX1 and a binding capacity of Bmax = 0.34 μmol/g of cellulose (Fig. 6E and Table 1). Binding was qualitatively confirmed by Alexa Fluor 488-labeled EXLX1, which bound to Avicel in a diffuse pattern (Fig. 7, C and D). In contrast to the largely electrostatic binding of EXLX1 to pectins and hemicelluloses, much of EXLX1 binding to Avicel is through hydrophobic forces as evidenced by the relatively modest reduction of binding to Avicel by CaCl2 addition (Fig. 6C).
It is notable that the binding capacity of Avicel for EXLX1 (0.34 μmol/g) is 10- and 60-fold lower than the binding capacities of wheat arabinoxylan (3.3 μmol/g) and wheat coleoptile cell walls (20.6 μmol/g), respectively. Thus, EXLX1 binding to the cellulosic portion of wheat coleoptile cell walls might be difficult to detect because total binding is dominated by EXLX1 binding to matrix polysaccharides. EXLX1 binding to cell walls pre-extracted with 4 m KOH is largely through electrostatic or polar forces as evidenced by reduced binding when CaCl2 was included in the binding assay (Fig. 6D) in contrast with the largely hydrophobic binding of EXLX1 to cellulose (Fig. 6C). Thus, EXLX1 binding to the extracted wheat coleoptile cell wall is most likely due to binding to residual matrix polysaccharides that were not extracted rather than EXLX1 binding to the cellulosic portion of the cell wall.
Binding of EXLX1 Domains and Variants to Cell Walls
Domain D1 did not bind to wheat coleoptile cell walls or to cellulose (Table 1), whereas domain D2 bound to both materials in a manner similar to that of full-length EXLX1 (Table 1). These results indicate that binding of EXLX1 to cellulose and to whole cell walls is mediated almost entirely by D2.
To identify the specific D2 residues participating in binding, EXLX1 variants in the D2 domain were studied. Binding to cellulose was reduced by half in W125A and W126A with even greater effects found in the double and triple aromatic mutants, whereas it was only mildly reduced in Y157A and K119A (Table 1). These results indicate that EXLX1 binding to cellulose is mediated primarily by the aromatic residues Trp-125 and Trp-126 and secondarily by Lys-119 and Tyr-157. Figs. 5B and 2B show that Lys-119, Trp-125, Trp-126, and Tyr-157 are likewise important for creep activity with wheat coleoptiles and for filter paper weakening. We therefore conclude that binding of EXLX1 to cellulose is closely linked to its cell wall loosening activities.
In analyzing D2 binding to whole cell walls, we found quite different results. Cell wall binding in the triple variant W125A/W126A/Y157A was not substantially affected, whereas binding was mildly reduced in K119A (Table 1). This result shows that EXLX1 binding to whole cell walls is mediated by D2 residues other than the aromatic residues important for cellulose binding (Trp-125, Trp-126, and Tyr-157).
It is possible that binding to whole cell walls, which are negatively charged due to acidic wall polysaccharides, is mediated by positively charged residues in D2, which has a pI of 9.66. To test this possibility, five EXLX1 variants were generated, K171Q, K188Q, K171Q/K188Q, K145Q/K171Q/K188Q, and R173Q/K180Q/K183Q, in which 1, 1, 2, 3, and 3 positively charged residues, respectively, were replaced by glutamine (see distribution in supplemental Fig. S4). The variants exhibited reduced binding to cell walls in proportion to the number of positive charges that were replaced with K145Q/K171Q/K188Q and R173Q/K180Q/K183Q showing essentially no binding (Table 1). Likewise, K145Q/K171Q/K188Q and R173Q/K180Q/K183Q showed virtually no binding to insoluble arabinoxylan. Thus, EXLX1 binding to whole cell walls is dominated by electrostatic and polar interaction of the basic D2 domain with matrix polysaccharides.
The wall extension activities of K145Q/K171Q/K188Q and R173Q/K180Q/K183Q were also tested. Both variants were more active than wild type EXLX1 (Fig. 5B). This result indicates that EXLX1 binding to pectins and hemicelluloses is not needed for wall creep activity and indeed may compete with the weaker binding, most likely to cellulose, that leads to cell wall creep.
DISCUSSION
The functional roles of the two domains of expansin and its conserved amino acid residues have been a matter of speculation but until now have not been tested experimentally. By comparing the binding and wall loosening activities of recombinant variants of the bacterial expansin EXLX1, we assessed the functional significance of specific protein features. One clear conclusion is that domain D2 is the principle determinant of binding to cell walls with different residues involved in binding to cellulose and binding to matrix polysaccharides. Although EXLX1 binding to the matrix is quantitatively much greater than its binding to cellulose, binding to the matrix could be strongly reduced in EXLX1 variants without diminishing its wall loosening activity. In contrast, reduction of cellulose binding greatly reduced wall loosening activity, whether measured as cell wall creep or weakening of filter paper, but hardly affected binding to the whole cell wall. Furthermore, we found that the two protein domains had to be connected for wall loosening activity and that many of the conserved residues on the PPBS contribute to wall loosening activity, most notably Asp-82, whose carboxyl group was essential for wall loosening activity.
Roles of Domain D2
The essential role of D2 for wall loosening may be partly explained by the fact that D2 almost exclusively mediates EXLX1 binding to cellulose and to matrix polysaccharides. Its binding to cellulose is mediated primarily through the conserved aromatic residues Trp-125, Trp-126, and Tyr-157, whereas its binding to matrix polysaccharides is mediated through nonconserved basic residues that are not part of the PPBS. These binding properties, as well as EXLX1 loosening of filter paper and G. xylinus pellicles, point to bare cellulose as a key target of EXLX1 action in its wall loosening activity. The nonproductive binding to acidic residues may be useful in limiting diffusion of the protein in the wall or concentrating it to specific wall regions. Similar 2-fold binding properties are likely to be found in other expansins with a basic pI, such as the β-expansins in maize pollen (26), and potentially in other cell wall-modifying proteins.
Domain D2 as Carbohydrate Binding Module (CBM)
CBMs are non-catalytic modules that bind to carbohydrates (41). Most often, CBMs are linked to catalytic modules, such as glycoside hydrolases and pectin lyases, where they potentiate activity by targeting and proximity effects (41–43). The idea that expansins might contain a CBM first emerged from the observation that conserved aromatic resides near the C terminus of α-expansin sequences were spaced like those found in some CBMs (44). Molecular modeling of β-expansin proteins, prior to publication of the first crystal-based structures, led to the hypothesis that the C terminus of β-expansins might resemble family-3 CBMs, possibly with two cellulose binding surfaces (45, 46). However, comparison of the crystal-based structures of expansin domain D2 with CBM3 structures does not support this idea; although both structures are β-sandwiches, they have different β-strand topologies and lack significant sequence similarity. Furthermore, these ideas were based on speculative interpretations of structure rather than experimental assessments of binding activities. Thus, our results are important in substantiating the idea that domain D2 indeed serves a binding function and in identifying the roles of specific residues in cell wall binding.
EXLX1 domain D2 is a β-sandwich with an Ig-like fold, which is a feature of many CBM families. The affinity of EXLX1 and D2 with Avicel (Kd = 2.1 μm) is similar to that of other CBMs (0.13–6 μm) (47–54). We found that D2 mediates binding of EXLX1 to cellulose mainly through Trp-125 and Trp-126 and secondarily through Tyr-157 and Lys-119. The side chains of Trp-125, Trp-126, and Tyr-157 are arranged in a line with a spacing consistent with binding the n, n + 2, and n + 4 pyranose rings of a glucan chain. This arrangement is generally similar to the aromatic surface in several families of CBMs (families 1, 5, 10, and 29) (41, 55).
Careful inspection of the EXLX1 structure (Protein Data Bank code 3D30) reveals that the side chains of Trp-125, Trp-126, and Tyr-157 are not quite planar but are twisted clockwise, suggesting that D2 binds to a single glucan chain rather than a flat, highly crystalline cellulose surface. The low binding capacity of Avicel for EXLX1 and D2 (∼0.3 μmol/g of Avicel) compared with the majority of CBMs (range of 0.2–10 μmol/g of Avicel) (47–54) suggests that most cellulose surfaces are not structured appropriately for EXLX1 binding. The exact structure of the EXLX1 cellulosic target merits further investigation.
The Carbohydrate-Active Enzymes (CAZy) database currently classifies CBMs into 62 families based on structural and sequence relatedness. In surveying available CBM structures, we found that D2 shows general structural similarities to CBM families 9, 31, 34, 39, and 41, but its sequence similarity with proteins in these families is not statistically significant. Also, there is no reason to believe that D2 is related to these other families in terms of evolutionary origin, binding specificity, or functional role. We therefore propose it as the founding member of a new CBM family, CBM63 (with agreement of Dr. Bernard Henrissat, founder of the CAZy database). CBM63 should include highly similar D2 domains from other putative expansins from bacteria and fungi (supplemental Fig. S5). Additionally, it should contain D2 domains of plant expansins in the α-expansin and β-expansin families (supplemental Fig. S5) (56). However, the binding specificities and roles of additional members of CBM63 need to be experimentally determined before a complete picture of the family is formed.
D2 is rather atypical among CBMs because it is not loosely linked to a catalytic domain by a flexible linker (41, 42), but it is tightly packed against D1. The crystal structures for three different expansins (Protein Data Bank codes 3D30, 1N10, and 2HCZ) all show the same spatial alignment and tight fit of the two expansin domains; thus, this close spatial configuration of the two domains may be important for activity. The slightly twisted aromatic surface of D2 extends the open, largely polar surface of D1, forming the conserved PPBS. We suggest that, in mediating cell wall loosening, D1 and D2 act cooperatively rather than independently; in other words, D2 function may extend beyond simple anchoring of the D1 domain to the wall. This idea is supported by the fact that domain D1 alone did not exhibit wall extension activity, whereas by comparison, most catalytic domains linked to CBMs retain >10% activity upon CBM truncation (57–65). Indirect support can also be seen in the fact that K119A and Y157A (closest to D1) affect creep activity more than W125A and W126A, whereas these latter residues are more significant for cellulose binding. Thus, D2 may contribute to the wall loosening action of EXLX1 not only by binding to a β1,4-d-glucan target but also by participating directly in reconfiguring polysaccharides at the cellulose-matrix interface, leading to cell wall creep.
Role of D1 Domain
We estimate that the PPBS of EXLX1, running 48 Å from Thr-12 in domain D1 to Trp-125 in domain D2, could accommodate an oligosaccharide backbone with nine sugar rings. In contrast to the D2 surface, which is dominated by aromatic residues that can interact with the hydrophobic center of sugar rings, the PPBS on D1 is dominated by polar residues. These could interact with a polysaccharide by H-bonds with hydroxyl groups on the periphery of sugar rings. In support of this idea, we found that several of the polar residues on the D1 PPBS, including Thr-12, Thr-14, Ser-16, and Glu-75, made partial contributions to EXLX1 wall loosening activity. Larger effects were found with Asp-71 and Tyr-73, whereas the carboxyl group of Asp-82 proved to be essential for wall loosening activity.
This last result is particularly striking because Asp-82 is highly conserved among expansins, and a corresponding aspartic acid residue is also highly conserved in GH-45 endoglucanases where it is thought to serve as the catalytic proton donor for the hydrolysis of β1,4-d-glucans (66). However, GH-45 catalysis by this mechanism requires a second aspartic acid residue acting as a catalytic base, and EXLX1 lacks such a residue. We considered the possibility that Glu-75 might function in this role, but, arguing against this, the E75A variant retained most of its wall loosening activity (Fig. 5A). In some cases, the catalytic base of inverting glycoside hydrolases has been elusive. It is possible that a remote amino acid may activate the nucleophilic water molecule via a string of solvent molecules in a Grotthus-type mechanism used by GH-6 and GH-124 glycoside hydrolases (67, 68). Regardless of the identity of the base, the carboxylic group of Asp-82 needs to be protonated to act as a catalytic acid. However, unlike plant expansins that have an acidic pH optimum (1, 26), EXLX1 activity at pH 9.5 was as high as that at pH 5.5 or 7.5, arguing against Asp-82 acting as a catalytic acid. It is theoretically possible that the environment around Asp-82 elevates the pKa of Asp-82 (expected to be ∼3.9). However, an increase of pKa by 5–6 units, particularly for a residue that is exposed to the surface, would be extraordinary. These results, combined with those of a previous study showing that EXLX1 did not hydrolyze various wheat coleoptile cell walls or cellulose (21), argue against hydrolytic activity in EXLX1.
Another structure with distant similarity to domain D1 of EXLX1 is a GH-102 peptidoglycan lytic transglycosylase from E. coli called EcMltA (21). EcMltA utilizes only one acidic amino acid residue to cleave β1,4-glycosidic bonds in bacterial cell wall peptidoglycan (70). Structural alignment of EXLX1 and EcMltA showed that Asp-82 corresponded to the sole catalytic residue of EcMltA (21). However, this mechanism requires the carboxylic group of Asp-82 to be protonated. Additionally, the presence of the N-acetyl group of the muramoyl residue in peptidoglycan helps to cleave the β1,4-glycosidic bond in EcMltA (69, 71, 72). Such a group does not exist in a pure glucan chain, such as cellulose. Finally, EcMltA requires an additional domain, not present in EXLX1, to stabilize the glycan chain and potentiate enzymatic action. Collectively, these results suggest that EXLX1 is unlikely to act enzymatically by a mechanism similar to that of glycoside hydrolases or lytic transglycosylases.
Model of Expansin Action
The prevailing model of plant expansin action proposes that expansins loosen plant cell walls by targeting cellulose-hemicellulose junctions that link cellulose microfibrils into a strong but resilient network (2). According to this idea, expansin binding to a polysaccharide junction weakens it sufficiently to enable chain movement (slippage) under the action of cell wall stress.
Our data show that residues Asp-71, Tyr-73, and Asp-82 on domain D1, along with the slightly twisted aromatic surface on domain D2, are crucial for creep activity by EXLX1. EXLX1 may induce wall creep when these residues bind a glucan that is part of the load-bearing network in the cell wall, distorting its shape and allowing physical slippage of the junction if the wall is in tension. Our results point to cellulose as a key target of the loosening action of EXLX1, implicating cellulose-cellulose junctions as possible load-bearing junctions within the plant cell wall.
As the structure of EXLX1 is very similar to that of plant expansins (21) and most of the important residues identified here correspond to similar residues in plant expansins, we anticipate that the same principles of wall binding and loosening that were discerned here from analysis of EXLX1 will also apply to plant expansins. Key differences may be in the specific targets of action, the pH dependence of activity, and the higher wall loosening activity by plant expansins.
Supplementary Material
Acknowledgments
We are grateful to Daniel Durachko and Edward Wagner for technical assistance, Phil Bevilacqua and Melissa Mullen (Department of Chemistry, Pennsylvania State University) for help with the spectropolarimeter measurements, and Bernard Henrissat (Laboratoire d'Architecture et de Fonction des Macromolécules Biologiques, UMR6098 CNRS, Universités Aix-Marseille) for constructive comments.
This work was supported by United States Department of Energy Grant DE-FG02-84ER13179 from the Office of Basic Energy Sciences.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5 and Tables S1 and S2.
- PPBS
- putative polysaccharide binding surface
- CBM
- carbohydrate binding module
- CDTA
- 1,2-cyclohexylenedinitrilotetraacetic acid.
REFERENCES
- 1. McQueen-Mason S., Durachko D. M., Cosgrove D. J. (1992) Plant Cell 4, 1425–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cosgrove D. J. (2005) Nat. Rev. Mol. Cell Biol. 6, 850–861 [DOI] [PubMed] [Google Scholar]
- 3. Pien S., Wyrzykowska J., McQueen-Mason S., Smart C., Fleming A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 11812–11817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cho H. T., Cosgrove D. J. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 9783–9788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Choi D., Lee Y., Cho H. T., Kende H. (2003) Plant Cell 15, 1386–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zenoni S., Reale L., Tornielli G. B., Lanfaloni L., Porceddu A., Ferrarini A., Moretti C., Zamboni A., Speghini A., Ferranti F., Pezzotti M. (2004) Plant Cell 16, 295–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cho H. T., Kende H. (1997) Plant Cell 9, 1661–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Reinhardt D., Wittwer F., Mandel T., Kuhlemeier C. (1998) Plant Cell 10, 1427–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brummell D. A., Harpster M. H., Dunsmuir P. (1999) Plant Mol. Biol. 39, 161–169 [DOI] [PubMed] [Google Scholar]
- 10. Vriezen W. H., De Graaf B., Mariani C., Voesenek L. A. (2000) Planta 210, 956–963 [DOI] [PubMed] [Google Scholar]
- 11. Lee Y., Kende H. (2001) Plant Physiol. 127, 645–654 [PMC free article] [PubMed] [Google Scholar]
- 12. Wu Y., Thorne E. T., Sharp R. E., Cosgrove D. J. (2001) Plant Physiol. 126, 1471–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cho H. T., Cosgrove D. J. (2002) Plant Cell 14, 3237–3253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Valdivia E. R., Sampedro J., Lamb J. C., Chopra S., Cosgrove D. J. (2007) Plant Physiol. 143, 1269–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Valdivia E. R., Stephenson A. G., Durachko D. M., Cosgrove D. (2009) Sex Plant Reprod. 22, 141–152 [DOI] [PubMed] [Google Scholar]
- 16. Sampedro J., Cosgrove D. J. (2005) Genome Biol. 6, 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li Y., Darley C. P., Ongaro V., Fleming A., Schipper O., Baldauf S. L., McQueen-Mason S. J. (2002) Plant Physiol. 128, 854–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cosgrove D. J., Bedinger P., Durachko D. M. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 6559–6564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yennawar N. H., Li L. C., Dudzinski D. M., Tabuchi A., Cosgrove D. J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 14664–14671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McQueen-Mason S. J., Cosgrove D. J. (1995) Plant Physiol. 107, 87–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kerff F., Amoroso A., Herman R., Sauvage E., Petrella S., Filée P., Charlier P., Joris B., Tabuchi A., Nikolaidis N., Cosgrove D. J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 16876–16881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cosgrove D. J. (2000) Nature 407, 321–326 [DOI] [PubMed] [Google Scholar]
- 23. Jahr H., Dreier J., Meletzus D., Bahro R., Eichenlaub R. (2000) Mol. Plant-Microbe Interact. 13, 703–714 [DOI] [PubMed] [Google Scholar]
- 24. McQueen-Mason S., Cosgrove D. J. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 6574–6578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hall M., Bansal P., Lee J. H., Realff M. J., Bommarius A. S. (2010) FEBS J. 277, 1571–1582 [DOI] [PubMed] [Google Scholar]
- 26. Li L. C., Bedinger P. A., Volk C., Jones A. D., Cosgrove D. J. (2003) Plant Physiol. 132, 2073–2085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Carpita N. C., Defernez M., Findlay K., Wells B., Shoue D. A., Catchpole G., Wilson R. H., McCann M. C. (2001) Plant Physiol. 127, 551–565 [PMC free article] [PubMed] [Google Scholar]
- 28. Carpita N. C. (1984) Plant Physiol. 76, 205–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Whitney S. E., Gidley M. J., McQueen-Mason S. J. (2000) Plant J. 22, 327–334 [DOI] [PubMed] [Google Scholar]
- 30. Hestrin S., Schramm M. (1954) Biochem. J. 58, 345–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Whitmore L., Wallace B. A. (2004) Nucleic Acids Res. 32, W668–W673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Whitmore L., Wallace B. A. (2008) Biopolymers 89, 392–400 [DOI] [PubMed] [Google Scholar]
- 33. Provencher S. W., Glöckner J. (1981) Biochemistry 20, 33–37 [DOI] [PubMed] [Google Scholar]
- 34. van Stokkum I. H., Spoelder H. J., Bloemendal M., van Grondelle R., Groen F. C. (1990) Anal. Biochem. 191, 110–118 [DOI] [PubMed] [Google Scholar]
- 35. Lees J. G., Miles A. J., Wien F., Wallace B. A. (2006) Bioinformatics 22, 1955–1962 [DOI] [PubMed] [Google Scholar]
- 36. Edgar R. C. (2004) Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. (2004) Genome Res. 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 39. Kim E. S., Lee H. J., Bang W. G., Choi I. G., Kim K. H. (2009) Biotechnol. Bioeng. 102, 1342–1353 [DOI] [PubMed] [Google Scholar]
- 40. Willats W. G., Orfila C., Limberg G., Buchholt H. C., van Alebeek G. J., Voragen A. G., Marcus S. E., Christensen T. M., Mikkelsen J. D., Murray B. S., Knox J. P. (2001) J. Biol. Chem. 276, 19404–19413 [DOI] [PubMed] [Google Scholar]
- 41. Boraston A. B., Bolam D. N., Gilbert H. J., Davies G. J. (2004) Biochem. J. 382, 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gilbert H. J. (2010) Plant Physiol. 153, 444–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hervé C., Rogowski A., Blake A. W., Marcus S. E., Gilbert H. J., Knox J. P. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 15293–15298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cosgrove D. J. (1996) BioEssays 18, 533–540 [DOI] [PubMed] [Google Scholar]
- 45. Shoseyov O., Shani Z., Levy I. (2006) Microbiol. Mol. Biol. Rev. 70, 283–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Barre A., Rougé P. (2002) Biochem. Biophys. Res. Commun. 296, 1346–1351 [DOI] [PubMed] [Google Scholar]
- 47. McLean B. W., Bray M. R., Boraston A. B., Gilkes N. R., Haynes C. A., Kilburn D. G. (2000) Protein Eng. 13, 801–809 [DOI] [PubMed] [Google Scholar]
- 48. Murashima K., Kosugi A., Doi R. H. (2005) J. Bacteriol. 187, 7146–7149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Goldstein M. A., Doi R. H. (1994) J. Bacteriol. 176, 7328–7334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Goldstein M. A., Takagi M., Hashida S., Shoseyov O., Doi R. H., Segel I. H. (1993) J. Bacteriol. 175, 5762–5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pagès S., Gal L., Bélaïch A., Gaudin C., Tardif C., Bélaïch J. P. (1997) J. Bacteriol. 179, 2810–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Linder M., Salovuori I., Ruohonen L., Teeri T. T. (1996) J. Biol. Chem. 271, 21268–21272 [DOI] [PubMed] [Google Scholar]
- 53. Henshaw J. L., Bolam D. N., Pires V. M., Czjzek M., Henrissat B., Ferreira L. M., Fontes C. M., Gilbert H. J. (2004) J. Biol. Chem. 279, 21552–21559 [DOI] [PubMed] [Google Scholar]
- 54. Boraston A. B., Chiu P., Warren R. A., Kilburn D. G. (2000) Biochemistry 39, 11129–11136 [DOI] [PubMed] [Google Scholar]
- 55. Charnock S. J., Bolam D. N., Nurizzo D., Szabó L., McKie V. A., Gilbert H. J., Davies G. J. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 14077–14082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kende H., Bradford K., Brummell D., Cho H. T., Cosgrove D., Fleming A., Gehring C., Lee Y., McQueen-Mason S., Rose J., Voesenek L. A. (2004) Plant Mol. Biol. 55, 311–314 [DOI] [PubMed] [Google Scholar]
- 57. Tomme P., Van Tilbeurgh H., Pettersson G., Van Damme J., Vandekerckhove J., Knowles J., Teeri T., Claeyssens M. (1988) Eur. J. Biochem. 170, 575–581 [DOI] [PubMed] [Google Scholar]
- 58. Bolam D. N., Ciruela A., McQueen-Mason S., Simpson P., Williamson M. P., Rixon J. E., Boraston A., Hazlewood G. P., Gilbert H. J. (1998) Biochem. J. 331, 775–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Boraston A. B., Kwan E., Chiu P., Warren R. A., Kilburn D. G. (2003) J. Biol. Chem. 278, 6120–6127 [DOI] [PubMed] [Google Scholar]
- 60. Charnock S. J., Bolam D. N., Turkenburg J. P., Gilbert H. J., Ferreira L. M., Davies G. J., Fontes C. M. (2000) Biochemistry 39, 5013–5021 [DOI] [PubMed] [Google Scholar]
- 61. Ali M. K., Hayashi H., Karita S., Goto M., Kimura T., Sakka K., Ohmiya K. (2001) Biosci. Biotechnol. Biochem. 65, 41–47 [DOI] [PubMed] [Google Scholar]
- 62. Zverlov V. V., Volkov I. Y., Velikodvorskaya G. A., Schwarz W. H. (2001) Microbiology 147, 621–629 [DOI] [PubMed] [Google Scholar]
- 63. Gilkes N. R., Warren R. A., Miller R. C., Jr., Kilburn D. G. (1988) J. Biol. Chem. 263, 10401–10407 [PubMed] [Google Scholar]
- 64. Maglione G., Matsushita O., Russell J. B., Wilson D. B. (1992) Appl. Environ. Microbiol. 58, 3593–3597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hall J., Black G. W., Ferreira L. M., Millward-Sadler S. J., Ali B. R., Hazlewood G. P., Gilbert H. J. (1995) Biochem. J. 309, 749–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Davies G. J., Tolley S. P., Henrissat B., Hjort C., Schülein M. (1995) Biochemistry 34, 16210–16220 [DOI] [PubMed] [Google Scholar]
- 67. Koivula A., Ruohonen L., Wohlfahrt G., Reinikainen T., Teeri T. T., Piens K., Claeyssens M., Weber M., Vasella A., Becker D., Sinnott M. L., Zou J. Y., Kleywegt G. J., Szardenings M., Ståhlberg J., Jones T. A. (2002) J. Am. Chem. Soc. 124, 10015–10024 [DOI] [PubMed] [Google Scholar]
- 68. Brás J. L., Cartmell A., Carvalho A. L., Verzé G., Bayer E. A., Vazana Y., Correia M. A., Prates J. A., Ratnaparkhe S., Boraston A. B., Romão M. J., Fontes C. M., Gilbert H. J. (2011) Proc. Natl. Acad. Sci. U.S.A. 108, 5237–5242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Scheurwater E., Reid C. W., Clarke A. J. (2008) Int. J. Biochem. Cell Biol. 40, 586–591 [DOI] [PubMed] [Google Scholar]
- 70. van Straaten K. E., Dijkstra B. W., Vollmer W., Thunnissen A. M. (2005) J. Mol. Biol. 352, 1068–1080 [DOI] [PubMed] [Google Scholar]
- 71. Reid C. W., Blackburn N. T., Legaree B. A., Auzanneau F. I., Clarke A. J. (2004) FEBS Lett. 574, 73–79 [DOI] [PubMed] [Google Scholar]
- 72. Reid C. W., Legaree B. A., Clarke A. J. (2007) FEBS Lett. 581, 4988–4992 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.