Abstract
Heparan sulfates (HS) are highly modified sugar polymers in multicellular organisms that function in cell adhesion and cellular responses to protein signaling. Functionally distinct, cell type-dependent HS modification patterns arise as the result of a conserved network of enzymes that catalyze deacetylations, sulfations, and epimerizations in specific positions of the sugar residues. To understand the genetic interactions of the enzymes during the HS modification process, we have measured the composition of HS purified from mutant strains of Caenorhabditis elegans. From these measurements we have developed a genetic network model of HS modification. We find the interactions to be highly recursive positive feed-forward and negative feedback loops. Our genetic analyses show that the HS C-5 epimerase hse-5, the HS 2-O-sulfotransferase hst-2, or the HS 6-O-sulfotransferase hst-6 inhibit N-sulfation. In contrast, hse-5 stimulates both 2-O- and 6-O-sulfation and, hst-2 and hst-6 inhibit 6-O- and 2-O-sulfation, respectively. The effects of hst-2 and hst-6 on N-sulfation, 6-O-sulfation, and 2-O-sulfation appear largely dependent on hse-5 function. This core of regulatory interactions is further modulated by 6-O-endosulfatase activity (sul-1). 47% of all 6-O-sulfates get removed from HS and this editing process is dependent on hst-2, thereby providing additional negative feedback between 2-O- and 6-O-sulfation. These findings suggest that the modification patterns are highly sensitive to the relative composition of the HS modification enzymes. Our comprehensive genetic analysis forms the basis of understanding the HS modification network in metazoans.
Keywords: Extracellular Matrix, Genetics, Glycosaminoglycan, Heparan Sulfate, Sulfotransferase
Introduction
Heparan sulfate proteoglycans are multifunctional components of animal cell membranes and extracellular matrices. They play important roles in the development and physiology of multicellular organisms (1, 2). Heparan sulfate (HS)2 glycans bind a diverse array of proteins involved in cellular adhesion, communication, motility, and organelle dynamics (3). These diverse binding capabilities are determined by distinct patterns of saccharide modifications. Cell type-specific modification patterns arise as the result of the action of an evolutionarily conserved network of enzymes that catalyze specific epimerization and sulfations of the sugar chain. How these enzymes interact during the HS modification process is the subject of this report.
HS is a modification product of a polymer of alternating glucuronic acid (GlcA) and N-acetylglucosamine (GlcNAc) (3). Polymer modification is not uniform nor does it go to completion, therefore the resulting glycans are chemically heterogeneous. Modification begins with the removal of acetyl groups from GlcNAc by the dual specificity enzyme N-deacetylase/sulfotransferase followed by N-sulfation of glucosamine by N-deacetylase/sulfotransferase (3). N-Sulfation occurs as a prerequisite for subsequent glucuronyl C-5 epimerization and O-linked sulfations (3). C-5 epimerization is the process of converting GlcA to isomeric iduronic acid (IdoA). The resultant pattern of modifications consists of continuous stretches of N-sulfated and O-sulfated domains (NS domains) interspersed between continuous stretches of unmodified domains (NA domains) with short transition zones of alternating modified and unmodified saccharides (NS/NA domains) between them (3). The processes regulating extent and distribution of these domains of modifications is not well understood.
HS modification patterns are dependent on cell type, developmental stage, and possibly the type of core protein (4–8). This suggests that modification patterns are regulated in a cell type-specific manner. The nature of this regulation is an important unanswered question regarding HS synthesis (3). Several HS biosynthetic and modification enzymes have been shown to physically interact or to exhibit co-dependent subcellular localization. For example, the heparan polymerizing enzymes EXT1 and EXT2 interact with each other and EXT2 interacts with N-deacetylase/sulfotransferase (9). The subcellular distribution of C-5 epimerase and Hs2st are mutually dependent suggesting assembly into a complex (10). These and other observations led to the hypothesis that heparan synthesis and modification occurs in the context of an organized quaternary complex named the “GAGosome” (9, 11). One prediction of the GAGosome concept is that the concentration and spatial organization of the enzymes within the Golgi should strongly influence HS modification patterns.
Genetic analyses of individual enzymes have been reported in a variety of experimental contexts. One general concept that has emerged from these studies is that loss of an individual enzyme by mutation affects modifications not catalyzed by that enzyme, for example, loss of the HS 2-O-sulfotransferase in mice or flies leads to an increase in 6-O-sulfation (12–14). These effects are not limited to modifications believed to occur after the step catalyzed by the mutant gene. For example, both C-5 epimerase and HS 2-O-sulfotransferase mutants display an increase in N-sulfation (14–16). Therefore, there exists a set of interactions between the modification genes that remains unexplained.
The control of heparan modifications undoubtedly results from a hierarchy of regulatory interactions, likely including a combination of substrate specificity and availability, physical interaction between the enzymes, trafficking of enzymes and substrates through the secretion pathway, transcription control of enzyme mRNA, translational control of protein production, and finally post-translational modification of the modification enzymes. To gain insight into the complex process of heparan modifications in vivo we have systematically perturbed HS modifications in Caenorhabditis elegans, using deletion and missense mutations in genes coding for proteins involved in HS modification. Using complete enzymatic depolymerization of the HS chains and separation of the resulting disaccharide mixture by reverse phase-ion pair liquid chromatography, we measured the HS disaccharide composition enabling us to determine the individual disaccharide fractions and the degree of sulfation for each mutant and mutant combination. From these measurements we have developed a comprehensive model of HS modification gene (referred throughout as HSMGs) interactions that forms the basis for understanding HS biosynthesis in metazoans.
EXPERIMENTAL PROCEDURES
Growth Conditions
All strains used for biochemical analysis were maintained at 20 °C on NGM plates seeded with streptomycin-resistant Escherichia coli OP50. For glycosaminoglycan production, worms were grown for 4 days in liquid culture.
Preparation of Glycosaminoglycans
One or two, 250-ml cultures of worms were lyophilized, then resuspended in 10 ml of acetone and disrupted with a Fisher brand Polytron for 2 min at half-power. The homogenate was de-lipidated in acetone, clarified by centrifugation, and lyophilized. 150–600 mg of acetone-dried worm powder was solubilized, and crude GAGs were purified as described by Toyoda et al. (17). The crude GAGs were frozen and stored at −80 °C.
Purification of Glycosaminoglycans
Frozen samples were thawed and incubated with 800 μl of DEAE for 1 h at 4 °C. The resin was collected by centrifugation and washed three times with 5 ml of 150 mm NaCl, 50 mm Tris-Cl, pH 8.0. Glycosaminoglycans were eluted by incubating in 800 μl of 1 m NaCl, 50 mm Tris-Cl, pH 8.0, for 10 min. The elution was repeated three times and the resulting fractions were pooled and desalted on a PD-10 column (GE Healthcare). Desalted glycosaminoglycans were lyophilized and resuspended in 50 μl of distilled water.
Enzymatic Depolymerization
The samples were digested with a mixture containing 1 mIU each of heparinase I, heparinase II, and heparinase III (Ibex, Montreal, Canada) in 20 mm Tris acetate, pH 7.0, 2 mm calcium acetate for 5 h at 37 °C. After digestion, the HS disaccharides were isolated by filtration using an YM-10 microcon filtration device (Fisher).
RP-IPA Chromatography
The protocol for determining the unsaturated disaccharide composition of heparan sulfate was essentially as described (17) with variations on the columns used. Briefly, the chromatographic equipment included a U3000 HPLC from Dionex, two single piston pumps from Eldex Laboratories, a RF2000 fluorescence detector from Dionex with a 12-μl flow cell volume, a dry reaction bath (FH-40) and a thermocontroller (TC-55) from Brinkman Instruments. The samples were analyzed using a Dionex C18 3-μm Acclaim 120 column (2.1 × 150 mm) or a Senshu Pak C22 3-μm Docosil column (2 × 100 mm) with comparable results. The flow rates were 0.300 ml/min for the U3000 gradient pump and 0.175 ml/min for the reaction pumps. The buffers were: Buffer A, water; Buffer B, 200 mm NaCl; Buffer C, 10 mm tetrabutylammonium hydrogen sulfate; and Buffer D, 50% acetonitrile, with an injection volume = 20 μl. Initial conditions were % B = 1.0, % C = 12.0, and % D = 17.0. The gradient was as follows: 1–55% B over 9.0 min, 55–70% B over 1.5 min, 70% B for 4 min, 1% B for 10 min. C and D were constant at 12 and 17%, respectively. To the effluent from the HPLC, 1.0% NaOH and 0.5% 2-cyanoacetamide, respectively, were added as reactants by the Eldex pumps at 0.175 ml/min each. The mixture was passed through a reaction coil (0.25 mm inner diameter × 15 m long) set in a dry reaction temperature controlled chamber set at 125 °C and monitored fluorometrically (excitation wavelength, 346 nm; emission wavelength, 410 nm). Peaks were identified based on the retention times of known unsaturated chondroitin and HS disaccharide standards (V-labs, Covington, LA). The relative amount of each disaccharide is expressed as a percent of the total disaccharides determined.
RESULTS
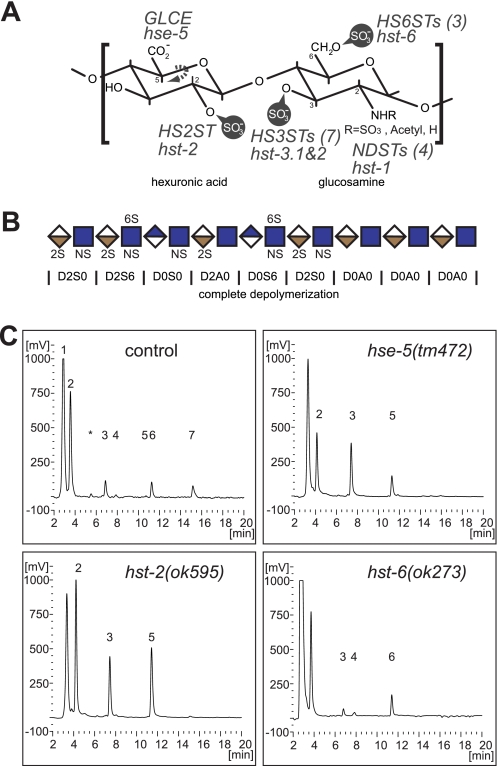
We have applied the method of Toyoda et al. (17) to analyze the heparinase-derived disaccharide composition of HS purified from C. elegans. The results of our analyses are presented in Table 1 (see supplemental Dataset S1 for all raw data). As previously reported (17–19), D0S0, D2S0, and D2S6 are the major sulfated HS disaccharides and D2A0 and D0S6 are minor (<1%) disaccharides found in C. elegans (Fig. 1, Table 1). The composition of disaccharides measured from control strains was 60% D0A0, 12% D0S0, 17% D2S0, and 11% D2S6 (Table 1 and supplemental Fig. S1). The total yield of HS/mg of dried tissue is 5.5 ng/mg and did not vary significantly between control and mutant strains (supplemental Table S1 and Dataset S1). Our findings in regard to HS composition and yield are in close agreement with the values reported by Toyoda et al. (17) (52% D0A0, 18% D0S0, 18% D2S0, 11% D2S6, 12 ng/mg yield).
TABLE 1.
Quantification of HS composition
| Genotype | n | D0A0a | D0S0 | D0S6 | D2S0 | D2S6 | N-Sulfation | 2-O-Sulfation | 6-O-Sulfation | Sulfates/100 disaccharides |
|---|---|---|---|---|---|---|---|---|---|---|
| % | ||||||||||
| Controlb | 16 | 60.0 ± 1.3 | 12.1 ± 0.5 | 0.2 ± 0.1 | 17.1 ± 1.3 | 10.7 ± 0.5 | 40.0 ± 1.3 | 27.5 ± 1.0 | 10.9 ± 0.6 | 78.5 ± 2.2 |
| Control | 5 | 62.5 ± 2.5 | 10.3 ± 0.8 | 0.3 ± 0.2 | 15.2 ± 1.0 | 11.7 ± 0.8 | 37.5 ± 2.5 | 26.9 ± 1.6 | 12.0 ± 1.0 | 76.4 ± 5.0 |
| hse-5(tm472) | 4 | 35.8 ± 2.1 | 44.8 ± 1.8 | 16.5 ± 0.4 | 2.6 ± 1.3 | 0.4 ± 0.4 | 64.2 ± 2.1 | 3.1 ± 1.7 | 17.9 ± 0.7 | 85.2 ± 2.6 |
| Control | 9 | 57.7 ± 1.6 | 13.2 ± 0.6 | 0.4 ± 0.1 | 18.2 ± 1.9 | 10.5 ± 0.8 | 42.3 ± 1.6 | 28.7 ± 1.5 | 10.9 ± 1.0 | 81.9 ± 2.9 |
| hst-2(ok595) | 6 | 41.7 ± 1.9 | 31.2 ± 2.1 | 27.1 ± 1.4 | 0.0 | 0.0 | 58.3 ± 1.9 | 0.0 | 27.1 ± 1.4 | 85.4 ± 3.3 |
| Control | 4 | 58.7 ± 1.0 | 12.0 ± 0.1 | 0.3 ± 0.2 | 16.7 ± 2.6 | 12.3 ± 1.9 | 41.3 ± 0.8 | 29.0 ± 0.7 | 12.6 ± 1.8 | 83.0 ± 3.2 |
| hst-6(ok273) | 5 | 54.3 ± 2.7 | 9.4 ± 0.6 | 0.0 ± 0.0 | 36.6 ± 2.2 | 0.0 ± 0.0 | 45.7 ± 2.7 | 36.5 ± 2.2 | 0.0 | 81.9 ± 4.9 |
| Control | 9 | 57.7 ± 1.6 | 13.2 ± 0.6 | 0.4 ± 0.1 | 18.2± | 10.5 ± 0.8 | 42.3 ± 1.6 | 28.7 ± 1.5 | 10.9 ± 1.0 | 81.9 ± 2.9 |
| sul-1(gk151) | 8 | 58.5 ± 1.0 | 15.1± | 0.6 ± 0.1 | 7.4 ± 1.0 | 18.4 ± 0.8 | 41.5 ± 1.0 | 25.9 ± 0.8 | 19.0 ± 0.9 | 86.5 ± 2.8 |
| Control | 5 | 62.5 ± 3.1 | 10.3 ± 1.0 | 0.3 ± 0.2 | 15.2 ± 1.2 | 11.7 ± 0.9 | 37.5 ± 3.1 | 26.9 ± 1.9 | 12.0 ± 1.1 | 76.4 ± 6.1 |
| hse-5(ot16) | 3 | 46.8 ± 4.2 | 31.8 ± 3.0 | 9.6 ± 0.5 | 4.2 ± 0.9 | 7.6 ± 0.6 | 53.2 ± 4.2 | 11.8 ± 1.5 | 17.2 ± 0.7 | 82.3 ± 6.0 |
| Control | 7 | 60.9 ± 2.7 | 11.5 ± 0.7 | 0.0 | 18.4 ± 2.6 | 9.2 ± 0.6 | 39.1 ± 2.7 | 27.6 ± 5.5 | 9.2 ± 0.6 | 75.9 ± 4.2 |
| hst-6(ot19)c | 4 | 56.1 ± 4.7 | 8.5 ± 1.1 | 0.0 | 35.6 ± 5.1 | 0.0 | 44.1 ± 4.9 | 35.6 ± 5.1 | 0.0 | 79.7 ± 10.1 |
| Control | 7 | 63.2 ± 1.3 | 10.9 ± 0.7 | 0.1 ± 0.1 | 14.3 ± 0.7 | 11.5 ± 0.6 | 36.8 ± 1.3 | 25.8 ± 1.2 | 11.5 ± 0.6 | 74.1 ± 2.8 |
| hse-5(tm472); hst-2(ok595) | 3 | 32.4 ± 4.7 | 48.5 ± 5.4 | 18.7 ± 0.9 | 0.0 | 0.0 | 67.2 ± 4.5 | 0.0 | 18.6 ± 0.9 | 85.8 ± 5.4 |
| hst-6(ok273) hst-2(ok595) | 2 | 32.4 ± 5.1 | 67.6 ± 5.1 | 0.0 | 0.0 | 0.0 | 67.6 ± 5.1 | 0.0 | 0.0 | 67.6 ± 5.1 |
| hse-5(tm472); hst-6(ok273) | 2 | 32.8 ± 4.9 | 59.3 ± 4.6 | 0.0 | 7.9 ± 0.3 | 0.0 | 67.2 ± 4.9 | 7.9 ± 0.3 | 0.0 | 75.1 ± 5.2 |
| hse-5(tm472);hst-6(ok273) hst-2(ok595) | 3 | 28.1 ± 5.3 | 71.9 ± 5.3 | 0.0 | 0.0 | 0.0 | 71.9 ± 5.3 | 0.0 | 0.0 | 71.9 ± 5.3 |
| Control | 9 | 57.7 ± 1.6 | 13.2 ± 0.6 | 0.4 ± 0.1 | 18.2± | 10.5 ± 0.8 | 42.3 ± 1.6 | 28.7 ± 1.5 | 10.9 ± 1.0 | 81.9 ± 2.9 |
| sul-1(gk151); hst-2(ok595) | 3 | 38.8 ± 0.7 | 32.5 ± 1.1 | 28.7 ± 0.5 | 0.0 | 0.0 | 61.2 ± 0.7 | 0.0 | 28.7 ± 0.5 | 89.9 ± 1.1 |
a D0A0 refers to a systematic nomenclature to describe disaccharides in glycosaminoglycans (44). D signifies the Δ4,5-unsaturated uronic acid with the following number denoting the uronic acid O-sulfation. The second letter represents the hexosamine descriptor with A indicating N-acetylated and S indicating N-sulfated glucosamine. The last number describes the hexosamine O-sulfation.
b Except where indicated all strains contain the evIs82b transgene (Is[unc-129::GFP]). The HS composition of evIs82b is comparable to HS from wild type N2 (supplemental Dataset S1).
c This strain harbors the otIs35; mgIs18 transgenes (Is[ttx-3::kal-1] and Is[ttx-3::GFP]). The HS composition of otIs35; mgIs18 is comparable to both evIs82b and wild type N2 (supplemental Dataset S1). Values ± the standard error of the mean are given. Each mutant was assayed on at least two different days and the average value is compared to the value of the control strain on the same days.
FIGURE 1.
Disaccharide analyses of HS from C. elegans single deletion mutants. A, schematic of the HS disaccharide. The positions in the hexuronic and glucosamine residues that can be modified are indicated together with the gene names that encode the respective enzymatic activities (italic uppercase, vertebrate gene names with the number of vertebrate genes indicated in parentheses; italic lowercase, C. elegans gene names). GLCE, HS C-5 glucuronic acid epimerase; HS6STs, HS 6-O-sulfotransferases; HS2ST, HS 2-O-sulfotransferase; HS3STs, HS 3-O-sulfotransferases; NDSTs, N-deacetylase-N-sulfotransferases. B, schematic of the HS polymer. Blue diamonds indicate glucuronic acid, tan, iduronic acid, and blue squares, N-acetylglucosamine. Relevant modifications are indicated. D0A0 refers to a systematic nomenclature to describe disaccharides in glycosaminoglycans (44). The first letter D signifies the Δ4,5-unsaturated uronic acid with the following number denoting the uronic acid O-sulfation. The second letter represents the hexosamine descriptor with A indicating N-acetylated, and S indicating N-sulfated glucosamine. The last number describes the hexosamine O-sulfation. C, chromatograms of disaccharides from control, hse-5(tm472), hst-2(ok595), or hst-6(ok273) mutant animals. Chromatogram peaks are: 1) unbound peak; 2) D0A0; 3) D0S0; 4) D2A0; 5) D0S6; 6) D2S0; 7) D2S6.
To systematically interrogate and quantify the contribution of individual HSMGs to the genetic control of HS modifications, we removed individual HSMGs and analyzed the resulting HS composition. We first analyzed HS from animals with the hse-5(tm472) deletion allele, predicted to result in complete loss of function of the sole HS C-5 glucuronic acid epimerase hse-5 in C. elegans (20). The HS disaccharide composition of the hse-5(tm472) allele differs from control HS in three principal ways. First, the amount of N-sulfated disaccharides increases from 38% in the control to 64% in the hse-5 mutant (Fig. 1C, Table 1, and supplemental Fig. S1A). Second, the amount of 2-O-sulfated HS disaccharides decreases from 27% in control animals to 3% in the mutant. Finally, the amount of 6-O-sulfated disaccharides increases from 12% in the control to 18% in the mutant. These results indicate that hse-5 exerts a stimulatory effect on 2-O-sulfation and inhibits 6-O-sulfation.
We next analyzed HS from animals carrying the deletion alleles hst-2(ok595) (20, 21) or hst-6(ok273) (20), both predicted to represent complete loss of function alleles of the HS 2-O-sulfotransferase hst-2 or the sole C. elegans HS 6-O-sulfotransferase hst-6, respectively. HS from hst-2(ok595) mutant animals contains no detectable 2-O-sulfated HS disaccharides and the amount of N-sulfated disaccharides and 6-O-sulfated disaccharides increase from 42% in the control to 58% in the mutant and, 11% in the control to 27% in the mutant, respectively (Fig. 1C, Table 1, and supplemental Fig. S1B). Furthermore, we find that HS from the hst-6(ok273) mutant animals completely lack 6-O-sulfation and exhibit an increase in N-sulfation from 41% in the control strain to 46% in the mutant as well as an increase in 2-O-sulfation from 29% in the control strain to 37% in the mutant (Fig. 1C, Table 1, and supplemental Fig. S1C). Thus, hst-2 and hst-6 each limit N-sulfation as well as 6-O-sulfation and 2-O-sulfation, respectively.
A characteristic feature of the 6-O-sulfate group in HS is that it can be enzymatically removed by the action of a 6-O-endosulfatase (22–24). Whereas vertebrate genomes encode two 6-O-endosulfatase genes (Sulf1 and Sulf2) (22), only a single ortholog (sul-1) encoding a 6-O-endosulfatase can be identified in C. elegans (supplemental Fig. S2A). HS from animals harboring a large deletion in the sul-1 locus, predicted to result in complete loss of function (supplemental Fig. S2, A and B) display no changes in N-sulfation (42 versus 42%) or 2-O-sulfation (29 versus 26%) compared with HS from control animals (Table 1 and supplemental Fig. S1D). However, we find an increase in 6-O-sulfation in the sul-1 mutant (19 versus 11% in the control). Therefore the inhibitory input of sul-1 to the steady state 6-O-sulfate phenotype is 8 sulfates/100 disaccharides. Furthermore, the increase in 6-O-sulfation occurs only on the tri-sulfated disaccharide D2S6 and not on the D0S6 disaccharide, suggesting that 6-O-sulfation in C. elegans occurs exclusively in NS domains. In summary, removing any of the individual HSMGs, hse-5, hst-2, or hst-6, in C. elegans results in HS with increased N-sulfation, and 6-O-sulfation (if hse-5 or hst-2 are removed) or 2-O-sulfation (if hst-6 is removed). Furthermore, genetic removal of the only HS 6-O-endosulfatase sul-1 results in increased 6-O-sulfation to a similar extent (47%) as observed in mice (25). We conclude that the basic regulatory principles of HS biosynthesis are conserved from worms to vertebrates.
To understand the genetic network governing HS modification in vivo, we conducted double and triple mutant analyses. We first determined, whether individual HSMGs can act independently in controlling N-sulfation. Because the hse-5(tm472);hst-6(ok273)hst-2(ok595) triple mutant is viable (26) in C. elegans, the individual contributions of each HSMG can be quantified directly by comparing the N-sulfation phenotype of the triple mutant to the three possible double mutants involving these genes. We find that HS from the hse-5(tm472);hst-6(ok273)hst-2(ok595) triple mutant is more extensively N-sulfated than control HS (72 versus 37% N-sulfation in the control (Table 1)). Interestingly, HS from each of the double mutants analyzed, i.e. hst-2(ok595)hst-6(ok273), hse-5(tm472);hst-6(ok273), or hse-5(tm472);hst-2(ok595), had marginally reduced N-sulfation compared with the triple mutant (67, 68, or 67 versus 72% in the triple (Table 1)) suggesting that hse-5, hst-2, or hst-6 all have a minor role in restricting N-sulfation that is independent of the other HSMG genes. This interpretation is further corroborated by the following observations. First, both hse-5(tm472);hst-2(ok595) and hse-5(tm472);hst-6(ok273) have a slightly higher N-sulfation (68 and 67%, Table 1) than the hse-5(tm472) single mutant (64%, Table 1). Thus the minor independent effects of hst-2 or hst-6 on N-sulfation can be detected by both reintroducing the activity of either gene into the triple mutant and removal of either gene from the hse-5(tm472) single mutant. Second, the amount of N-sulfation in the hst-2(ok595)hst-6(ok273) double mutant (67%) is greater than either mutant alone (58% in hst-2(ok595) and 46% in hst-6(ok273), Table 1) demonstrating that both hst-2 and hst-6 can act independently to restrict the extent of N-sulfation.
In a complementary manner, we assessed and quantified the ability of the HSMGs to interact with each other to limit the extent of N-sulfation. The hse-5(tm472);hst-2(ok595) double deletion mutant has an N-sulfation content of 67% compared with 64 or 58% in the hse-5(tm472) or hst-2(ok595) mutants, respectively (Table 1 and supplemental Fig. S3, A-C), demonstrating that hst-2 has a larger inhibitory effect (18%) in the presence of hse-5 than in the absence (3%). A qualitatively similar relationship between hst-6 and hse-5 can be observed from comparing the hse-5(tm472);hst-6(ok273) double mutant (N-sulfation is 67%) to the hse-5(tm472) and hst-6(ok273) single mutants (64 and 46%, respectively) (Table 1 and supplemental Fig. S3, A–C). Thus hst-6 has a greater influence on N-sulfation in the presence of hse-5 than in the absence. In summary, our findings indicate that hse-5 interact with hst-2 and hst-6 to limit the extent of N-sulfation (Fig. 2A).
FIGURE 2.
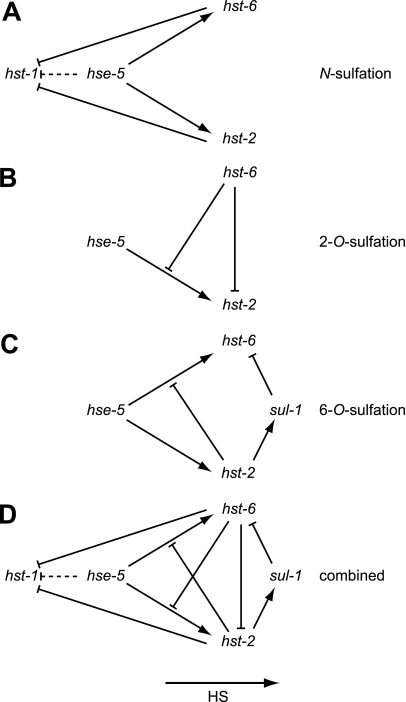
Models of the heparan modification network. Shown are the most parsimonious genetic models governing N-sulfation (A), 2-O-sulfation (B), and 6-O-sulfation (C), with D depicting the combined negative and positive genetic interactions. For simplicity, some known interactions likely also present in C. elegans have been omitted such as positive interactions between hst-1 and hse-5, hst-2, or hst-6 (3). The dashed line indicates a minor function of hse-5 in the absence of both hst-2 and hst-6. The interactions as depicted represent the qualitative relationships in the steady state of C. elegans.
We next asked how HSMGs interact to control 2-O-sulfation. Individual contributions of hse-5 and hst-6 to the 2-O-sulfation phenotype can be determined and quantified by comparing the 2-O-sulfation content of the hse-5(tm472);hst-6(ok273) double mutant to the respective single mutants. Hse-5 has a strong stimulatory effect on 2-O-sulfation (8% in the hse-5(tm472);hst-6(ok273) double mutant versus 37% in hst-6(ok273) single mutant). The effect of hse-5 on 2-O-sulfation is greater in the absence of hst-6 than in its presence indicating an inhibitory effect of hst-6 on 2-O-sulfation. Interestingly, the fraction of 2-O-sulfated disaccharides in the hse-5(tm472);hst-6(ok273) double mutant (8% 2-O-S) is greater than that of the hse-5 single mutant (3% 2-O-S), indicating that the inhibitory effects of hst-6 on 2-O-sulfation are the sum of hse-5 dependent and independent effects (Fig. 2B).
To determine how HSMGs interact to control 6-O-sulfation, we first focused on sul-1 encoding the only 6-O-endosulfatase. Biochemical and cell culture experiments suggest a role for 2-O-sulfation in the function of sulf (27, 28). To address this question in vivo, we constructed a sul-1(gk151)hst-2(ok595) double null mutant and analyzed the HS composition. We find that HS from animals lacking both hst-2 and sul-1 display a disaccharide composition nearly identical to the hst-2 single mutant (N-sulfation 58% in hst-2(ok595) versus 61% in the double mutant and 6-O-sulfation 27% in hst-2(ok595) versus 29% in the double mutant (Table 1 and supplemental Fig. S3D)). These results show that, in vivo all sul-1 activity is dependent on hst-2. Intriguingly, the extent of 6-O-sulfation is 9 sulfates/100 disaccharides more in the hst-2 mutant than in the sul-1 mutant demonstrating that hst-2 inhibits 6-O-sulfation by both sul-1-dependent and -independent mechanisms. To further determine whether the inhibitory effects of hst-2 on 6-O-sulfation are dependent or independent on hse-5, we compared HS from the hse-5(tm472);hst-2(ok595) double with the hse-5(tm472) and hst-2(ok595) single mutants. HS from the hse-5(tm472);hst-2(ok595) double deletion mutant has a 6-O-sulfation content that is nearly identical to that of the hse-5(tm472) single mutant (19 versus 18%), but different from the 6-O-sulfation content of the hst-2(ok595) deletion mutant (27%). Thus hse-5 is epistatic to hst-2 indicating that all inhibitory effects of hst-2 on 6-O-sulfation are dependent on hse-5. The observations that the hse-5 6-O-sulfation phenotype is epistatic to hst-2 and, that hse-5 and sul-1 single mutants share the same 6-O-sulfation phenotype (18 versus 19%), support the conclusion that hse-5, hst-2, and sul-1 act in a linear pathway to limit 6-O-sulfation and that the net effect of this regulatory pathway is to reduce the steady-state level of 6-O-sulfation by 9 sulfates/100 disaccharides. Additionally, our results indicate that hse-5 stimulates 6-O-sulfation by a mechanism that can be inhibited by hst-2 (Fig. 2C).
We next analyzed the HS composition of two strains carrying missense mutations in the hse-5 epimerase and the hst-6 6-O-sulfotransferase (20). We first analyzed HS from the hse-5(ot16) allele that encodes a G610E missense mutation (supplemental Fig. S4). Based on the penetrance of neuroanatomical defects in the hse-5(tm472) deletion and hse-5(ot16) mutant, the ot16 missense allele is inferred to be a strong, if not a complete loss of function allele (20). We find that HS from the hse-5(ot16) allele displays a smaller increase in the amount of N-sulfated HS disaccharides from 37% in the control to 53%, compared with 64% in HS of the hse-5(tm472) deletion allele (Table 1 and supplemental Fig. S3E). The amount of 2-O-sulfated HS disaccharides from the hse-5(ot16) missense allele was 12% compared with 3% in the hse-5(tm472) deletion allele and 26% in HS from the control strain (Table 1 and supplemental Fig. S3E). Last, 6-O-sulfation increased from 12% in the control strain to 17% in hse-5(ot16) compared with 18% in HS from the hse-5(tm472) deletion allele. Thus, the 6-O-sulfation phenotype of the hse-5(ot16) allele is indistinguishable from the hse-5(tm472) null allele. In contrast, both the N-sulfation and 2-O-sulfation phenotypes are less severe in the hse-5(ot16) allele compared with the hse-5(tm472) null allele. One possible explanation is that hse-5(ot16) results in a partial loss of catalytic activity and that 6-O-sulfation is more affected than 2-O- or N-sulfation. Alternatively, the hse-5(ot16) allele may reveal functions that are independent of catalytic activity. Consistent with such a scenario, recent in vitro studies suggest protein-dependent effects of the C-5 epimerase on HS2st substrate specificity (29). Further experiments are required to resolve this question in vivo.
Last, we analyzed HS from the hst-6(ot19) missense allele. The hst-6(ot19) allele encodes a H85Y mutation and is predicted to affect PAPS binding. Genetically, hst-6(ot19) is indistinguishable from the hst-6(ok273) deletion allele suggesting that it represents a strong, if not complete loss of function allele (30). In accord with this conclusion, we detect no 6-O-sulfated HS disaccharides in HS from hst-6(ot19) suggesting that the encoded enzyme is catalytically dead (Table 1 and supplemental Fig. S3F). The effects of the hst-6(ot19) allele on HS composition are not significantly different from the hst-6(ok273) deletion allele, consistent with the interpretation that the effects of hst-6 on hst-2 may require catalytic activity.
DISCUSSION
Modification patterns encode the physical and chemical information that control the diverse functions of HS. The modification process is the result of a network of conserved enzymes that catalyze specific sulfations and epimerization of the saccharide subunits. To gain insight into the events that control the modification process in vivo, we have systematically perturbed the network of modification genes with defined deletion and missense mutations and measured the HS composition phenotype in C. elegans. From these measurements we have developed a genetic model of HS modification in C. elegans (Fig. 2D). We find the interaction network to be highly recursive, that is, each member of the network has multiple inputs from and outputs to other members of the network. The logic is characterized by positive feed-forward and negative feedback loops. This small network of enzymes is remarkably complex in that each chemical product can encode significance individually and in combination with virtually every other. Specifically, hst-1 acts first and stimulates the activity of subsequent modification steps controlled by hse-5, hst-2, and hst-6 (reviewed in Ref. 3). hse-5 acts next and stimulates the steps controlled by hst-2 and hst-6. Hse-5, hst-2, and hst-6 inhibit the extent of the reactions catalyzed by hst-1. hst-2 and hst-6 have a mutually inhibitory interaction. The modification pattern that arises from the core regulatory interactions of hst-1, hse-5, hst-2, and hst-6 is modulated by an inhibitory input involving hst-2 and the 6-O-endosulfatase sul-1.
Genetic analysis of the heparan modification process has been limited to the removal of a single enzyme (13–15, 31, 32). Here we report the effects of double and triple mutant analyses for most of the enzymatic steps of the HS modification pathway as well as of missense mutations in hse-5 and hst-6. At least three important implications arise from these experiments. First, hst-2 and hst-6 act in parallel (the hst-2 hst-6 double mutant N-sulfation phenotype is greater than either of the single mutant phenotypes) to limit the extent of N-sulfation and, both of these inhibitory pathways are strongly dependent on hse-5. Additionally, hse-5 has a slight effect on N-sulfation independent of hst-2 and hst-6 although most of the inhibition requires the O-sulfotransferases. These interpretations result from the facts that (i) the hse-5 phenotype is epistatic to the hst-2 and hst-6 N-sulfation phenotypes, (ii) all three double mutants involving hse-5, hst-2, and hst-6 have the same phenotype with regard to N-sulfation, and (iii) the triple mutant phenotype is slightly more severe than each of the double mutant phenotypes. The extent of N-sulfation is an important focus of regulation because of its role in establishing the overall organization and the extent of heparan sulfation. Inhibition of N-sulfation by epimerase and O-sulfotransferases seems to be a universal attribute of HS modifying networks, as this effect has been seen in mouse and fly mutants (12, 14, 15). Second, hse-5 activity stimulates 6-O-sulfation. This interpretation follows from the fact that the degree of 6-O-sulfation is less in the hse-5;hst-2 double mutant than in the hst-2 mutant. Finally, the 6-O-sulfation phenotype of hst-2 is epistatic to that of sul-1. Therefore, the hst-2 function is required for 6-O-sulfate removal. It should be noted that a limitation of our analysis is that all of the measurements and thus the interactions that they reveal are from whole worms and represent the net effects at steady state. The network in individual cells may differ quantitatively, and possibly qualitatively.
One important mechanism that governs the interaction between heparan modifying enzymes is the substrate requirements of the individual reactions. For example, C-5 epimerization requires the flanking GlcN on the non-reducing end of GlcA to be N-sulfated, whereas there is no preference for N-sulfation of the GlcN linked at the reducing end of GlcA (33). C-5 epimerization is blocked by 2-O-sulfation or 6-O-sulfation on an adjacent GlcN (33). Additionally, Hs2st has a strong preference for iduronic acid substrates and is blocked by an adjacent 6-O-sulfate (34, 35). Therefore, 6-O-sulfation acts to block epimerization and 2-O-sulfation at the level of substrate requirement. Two lines of evidence support the hypothesis that the inhibition of 2-O-sulfation by hst-6 reported here could be at least in part the result of substrate specificity. First, the slight but reproducible decrease in the D0S0 disaccharide observed in hst-6(ok273) mutants and, second the results with the hst-6(ot19) missense mutation implying a requirement for catalytic activity (Table 1). One interesting question raised by this study regards the substrate requirements of the 6-O-sulfotransferase. Whereas we observe HST-6 clearly exhibiting a strong preference for an adjacent 2-O-sulfate in vivo (as there is almost no D0S6 detected from HS purified from worms), it is completely capable of sulfating GlcNS in the hst-2(ok595) null mutant. Human HS6st-2 and -3 were shown by Jemth et al. (36) to have a strong preference for 2-O-sulfated substrates in vitro. One possibility is that the 6-O-sulfotransferase reaction may be coupled to the 2-O-sulfotransferase reaction, for example, due to physical interactions between the enzymes. Alternatively, but not mutually exclusive, the 2-O-sulfated substrate may be kinetically favored over the un-sulfated substrate.
The effect of hst-2 on 6-O-sulfation has several interesting mechanistic implications. Our double mutant analysis indicates that the inhibitory effect of hst-2 on 6-O-sulfation results from at least two mechanisms, one that is sul-1 dependent and one that is sul-1 independent. This interpretation is based on the observation that the degree of 6-O-sulfation in the hst-2 sul-1 double null mutant is greater than in the sul-1 mutant alone. Also, there is more 6-O-sulfate in the hst-2(ok595) null mutant than in wild type animals or in hse-5(tm472) null animals. Importantly, this increase in 6-O-sulfation is lost in the hse-5(tm472);hst-2(ok595) double deletion mutant. In other words, hst-6 activity is up-regulated by the loss of hst-2 activity and this up-regulation is hse-5 dependent. Because the stereochemistry of the hexuronic acid is lost during lyase digestion, our analytical method does not directly measure HSE-5 catalytic activity. Thus, mechanistically, the dependence on hse-5 could be due to catalytic activity, i.e. a preference of the sulfotransferases for IdoA versus GlcA. Alternatively, this effect could be independent of IdoA as epimerase activity is decreased by 50% in HS2st mutant CHO cells (10). One possible mechanism for this effect could be that the HST-6 protein physically interacts with the HSE-5 protein. This interaction could have any number of functional consequences for each protein. For example, HST-6 could be stabilized in the hst-2(ok595) deletion mutant by binding to the HSE-5 protein. It has been observed that HS2st always co-localized with C-5 epimerase, whereas the converse was not true (10). Thus, C-5 epimerase can traffic to subcellular compartments independently of HS2st perhaps in connection with HST-6. These interpretations are consistent with our genetic data showing that almost all hst-2 functions are dependent on hse-5, whereas not all hse-5 functions are dependent on hst-2. It should be pointed out that to date no evidence has been found that HS2ST and any of the HS6STs physically interact or show mutually dependent subcellular localization (37).
We show here that in C. elegans, all 6-O-sulfation occurs in NS domains and that 6-O-endosulfatase activity requires hst-2. This contrasts with the situation in mammals. For example, mice encode multiple isoforms of Hs6st (Hs6st1–3 in mammals). The substrate specificities of the three 6-O-sulfotransferases have been analyzed in vitro and have been shown to exhibit subtle differences (29, 38, 39). The Hs6st1 isoform had a strong preference for iduronic acid containing substrates that lacked 2-O-sulfate (29), whereas HS6st2 has a preference for IdoA(2S)-GlcNS (38). These in vitro findings have been extended in vivo by measuring the HS composition from Hs6st1 null mice (31). In these mice there was no significant decrease in the tri-sulfated disaccharide, IdoA(2S)-GlcNS(6S), however, there was a marked decrease in GlcNAc(6S) and HexAGlcNS(6S). These results suggest that Hs6st2 preferentially catalyzes the synthesis of 6-O-sulfate within NS domains, whereas Hs6st1 and possibly Hs6st3 catalyze NS/NA transition zone 6-O-sulfation. Similarly, vertebrate genomes encode two 6-O-endosulfatases (Sulf1, Sulf2). HS composition and domain organization were analyzed from mouse embryonic fibroblast mutants for Sulf1, Sulf2, or both (25). The activities of the two genes exhibit a measurable degree of preference for substrate structure. Sulf1 activity exhibited a slight preference for 6-O-sulfates within NS domains, and Sulf2 exhibited a slight preference for 6-O-sulfates within NS/NA transition zones (25). Thus, comparison of the 6-O-sulfation pathway of C. elegans to mammals suggests that there are at least two sets of paired activities for the addition and removal of 6-O-sulfates, one for NS domains and one for NS/NA domains.
The results reported here show that the HS modification network is highly interconnected causing the output to be highly sensitive to the amount of individual HS modifying enzymes (Fig. 2D). Based on the highly recursive nature of interactions between different HS modification enzymes, we suggest that the HS composition and by inference HS modification patterns are dependent on the relative composition of HS modification enzymes in a given cell. That is, changes in the relative expression of a single gene will affect the activity of essentially all other genes involved in creating HS modification patterns. We have previously reported that ectopic overexpression of hst-6 in the worm hypodermis leads to defects in motor neuron axon projections (26). These anatomical defects are accompanied by decreases in the degree of N-sulfation and 2-O-sulfation (supplemental Dataset S1) consistent with the models presented here (Fig. 2).
Finally, an additional layer of regulation may be provided by the availability of the sulfate donor PAPS. Our laboratory and others have shown that the activity of the selective PAPS transporter PAPST1 controls both the overall degree of sulfation and the composition of HS in vivo (40–42). These results were anticipated based on studies perturbing PAPS synthesis with PAPS synthase inhibitors (43). Thus, it may be possible that cell-specific HS modification patterns are “self-assembling” and require no additional regulatory factors. Regardless of the ultimate validity of this hypothesis, the model presented here forms the basis of understanding the HS modification process in vivo.
Supplementary Material
Acknowledgments
We thank members of the Bülow laboratory for their criticisms and suggestions during the course of this study. We are indebted to S. Almo for allowing use of his equipment. Additional thanks to T. Gesteira Ferreira and U. Lindahl for comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants 5R01HD055380 and RC1GM090825 (to H. E. B.) and 5T32NS07098 (to R. A. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4, Table S1, and Data S1.
- HS
- heparan sulfate
- D0A0
- 2-acetamido-2-deoxy-4-O-(4-deoxy-α-l-threo-hex-enepyranosyluronic acid)-d-glucose (ΔUA-GlcNAc)
- D0S0
- 2-deoxy-2-sulfamido-4-O-(4-deoxy-α-l-threo-hex-enepyranosyluronic acid)-d-glucose (ΔUA-GlcNS)
- D0S6
- 2-deoxy-2-sulfamido-4-O-(4-deoxy-2-O-sulfo-α-l-threo-hex-enepyranosyluronic acid)-6-O-sulfo-d-glucose (ΔUA-GlcNS6S)
- D2S0
- 2-deoxy-2-sulfamido-4-O-(4-deoxy-2-O-sulfo-α-l-threo-hex-enepyranosyluronic acid)-d-glucose (ΔUA2S-GlcNS)
- D2S6
- 2-deoxy-2-sulfamido-4-O-(4-deoxy-2-O-sulfo-α-l-threo-hex-enepyranosyluronicacid)-6-O-sulfo-d-glucose (ΔUA2S-GlcNS6S)
- PAPS
- 3′-phosphoadenosine-5′-phosphosulfate
- HSMG
- HS modification gene
- NS domain
- N-sulfated domain.
REFERENCES
- 1. Bishop J. R., Schuksz M., Esko J. D. (2007) Nature 446, 1030–1037 [DOI] [PubMed] [Google Scholar]
- 2. Bülow H. E., Hobert O. (2006) Annu. Rev. Cell Dev. Biol. 22, 375–407 [DOI] [PubMed] [Google Scholar]
- 3. Lindahl U., Li J. P. (2009) Int. Rev. Cell Mol. Biol. 276, 105–159 [DOI] [PubMed] [Google Scholar]
- 4. Li F., Shi W., Capurro M., Filmus J. (2011) J. Cell Biol. 192, 691–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shworak N. W., Kojima T., Rosenberg R. D. (1993) Haemostasis 23, Suppl. 1, 161–176 [DOI] [PubMed] [Google Scholar]
- 6. Tveit H., Dick G., Skibeli V., Prydz K. (2005) J. Biol. Chem. 280, 29596–29603 [DOI] [PubMed] [Google Scholar]
- 7. Zako M., Dong J., Goldberger O., Bernfield M., Gallagher J. T., Deakin J. A. (2003) J. Biol. Chem. 278, 13561–13569 [DOI] [PubMed] [Google Scholar]
- 8. Kato M., Wang H., Bernfield M., Gallagher J. T., Turnbull J. E. (1994) J. Biol. Chem. 269, 18881–18890 [PubMed] [Google Scholar]
- 9. Presto J., Thuveson M., Carlsson P., Busse M., Wilén M., Eriksson I., Kusche-Gullberg M., Kjellén L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 4751–4756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pinhal M. A., Smith B., Olson S., Aikawa J., Kimata K., Esko J. D. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 12984–12989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Esko J. D., Selleck S. B. (2002) Annu. Rev. Biochem. 71, 435–471 [DOI] [PubMed] [Google Scholar]
- 12. Kamimura K., Koyama T., Habuchi H., Ueda R., Masu M., Kimata K., Nakato H. (2006) J. Cell Biol. 174, 773–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stanford K. I., Wang L., Castagnola J., Song D., Bishop J. R., Brown J. R., Lawrence R., Bai X., Habuchi H., Tanaka M., Cardoso W. V., Kimata K., Esko J. D. (2010) J. Biol. Chem. 285, 286–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Merry C. L., Bullock S. L., Swan D. C., Backen A. C., Lyon M., Beddington R. S., Wilson V. A., Gallagher J. T. (2001) J. Biol. Chem. 276, 35429–35434 [DOI] [PubMed] [Google Scholar]
- 15. Li J. P., Gong F., Hagner-McWhirter A., Forsberg E., Abrink M., Kisilevsky R., Zhang X., Lindahl U. (2003) J. Biol. Chem. 278, 28363–28366 [DOI] [PubMed] [Google Scholar]
- 16. Ledin J., Staatz W., Li J. P., Götte M., Selleck S., Kjellén L., Spillmann D. (2004) J. Biol. Chem. 279, 42732–42741 [DOI] [PubMed] [Google Scholar]
- 17. Toyoda H., Kinoshita-Toyoda A., Selleck S. B. (2000) J. Biol. Chem. 275, 2269–2275 [DOI] [PubMed] [Google Scholar]
- 18. Lawrence R., Olson S. K., Steele R. E., Wang L., Warrior R., Cummings R. D., Esko J. D. (2008) J. Biol. Chem. 283, 33674–33684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yamada S., Van Die I., Van den Eijnden D. H., Yokota A., Kitagawa H., Sugahara K. (1999) FEBS Lett. 459, 327–331 [DOI] [PubMed] [Google Scholar]
- 20. Bülow H. E., Hobert O. (2004) Neuron 41, 723–736 [DOI] [PubMed] [Google Scholar]
- 21. Kinnunen T., Huang Z., Townsend J., Gatdula M. M., Brown J. R., Esko J. D., Turnbull J. E. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 1507–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Morimoto-Tomita M., Uchimura K., Werb Z., Hemmerich S., Rosen S. D. (2002) J. Biol. Chem. 277, 49175–49185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dhoot G. K., Gustafsson M. K., Ai X., Sun W., Standiford D. M., Emerson C. P., Jr. (2001) Science 293, 1663–1666 [DOI] [PubMed] [Google Scholar]
- 24. Ai X., Do A. T., Lozynska O., Kusche-Gullberg M., Lindahl U., Emerson C. P., Jr. (2003) J. Cell Biol. 162, 341–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lamanna W. C., Baldwin R. J., Padva M., Kalus I., Ten Dam G., van Kuppevelt T. H., Gallagher J. T., von Figura K., Dierks T., Merry C. L. (2006) Biochem. J. 400, 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bülow H. E., Tjoe N., Townley R. A., Didiano D., van Kuppevelt T. H., Hobert O. (2008) Curr. Biol. 18, 1978–1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guimond S. E., Puvirajesinghe T. M., Skidmore M. A., Kalus I., Dierks T., Yates E. A., Turnbull J. E. (2009) J. Biol. Chem. 284, 25714–25722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Frese M. A., Milz F., Dick M., Lamanna W. C., Dierks T. (2009) J. Biol. Chem. 284, 28033–28044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Smeds E., Feta A., Kusche-Gullberg M. (2010) Glycobiology 20, 1274–1282 [DOI] [PubMed] [Google Scholar]
- 30. Bülow H. E., Berry K. L., Topper L. H., Peles E., Hobert O. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 6346–6351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Habuchi H., Nagai N., Sugaya N., Atsumi F., Stevens R. L., Kimata K. (2007) J. Biol. Chem. 282, 15578–15588 [DOI] [PubMed] [Google Scholar]
- 32. Bai X., Esko J. D. (1996) J. Biol. Chem. 271, 17711–17717 [DOI] [PubMed] [Google Scholar]
- 33. Jacobsson I., Lindahl U., Jensen J. W., Rodén L., Prihar H., Feingold D. S. (1984) J. Biol. Chem. 259, 1056–1063 [PubMed] [Google Scholar]
- 34. Kobayashi M., Habuchi H., Habuchi O., Saito M., Kimata K. (1996) J. Biol. Chem. 271, 7645–7653 [DOI] [PubMed] [Google Scholar]
- 35. Jacobsson I., Lindahl U. (1980) J. Biol. Chem. 255, 5094–5100 [PubMed] [Google Scholar]
- 36. Jemth P., Smeds E., Do A. T., Habuchi H., Kimata K., Lindahl U., Kusche-Gullberg M. (2003) J. Biol. Chem. 278, 24371–24376 [DOI] [PubMed] [Google Scholar]
- 37. Nagai N., Habuchi H., Esko J. D., Kimata K. (2004) J. Cell Sci. 117, 3331–3341 [DOI] [PubMed] [Google Scholar]
- 38. Habuchi H., Tanaka M., Habuchi O., Yoshida K., Suzuki H., Ban K., Kimata K. (2000) J. Biol. Chem. 275, 2859–2868 [DOI] [PubMed] [Google Scholar]
- 39. Habuchi H., Miyake G., Nogami K., Kuroiwa A., Matsuda Y., Kusche-Gullberg M., Habuchi O., Tanaka M., Kimata K. (2003) Biochem. J. 371, 131–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bhattacharya R., Townley R. A., Berry K. L., Bülow H. E. (2009) J. Cell Sci. 122, 4492–4504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dejima K., Murata D., Mizuguchi S., Nomura K. H., Izumikawa T., Kitagawa H., Gengyo-Ando K., Yoshina S., Ichimiya T., Nishihara S., Mitani S., Nomura K. (2010) J. Biol. Chem. 285, 24717–24728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Clément A., Wiweger M., von der Hardt S., Rusch M. A., Selleck S. B., Chien C. B., Roehl H. H. (2008) PLoS Genet. 4, e1000136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Safaiyan F., Kolset S. O., Prydz K., Gottfridsson E., Lindahl U., Salmivirta M. (1999) J. Biol. Chem. 274, 36267–36273 [DOI] [PubMed] [Google Scholar]
- 44. Lawrence R., Lu H., Rosenberg R. D., Esko J. D., Zhang L. (2008) Nat. Methods 5, 291–292 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.