Abstract
Voltage-gated sodium channel (VGSC) activity has previously been reported in endothelial cells (ECs). However, the exact isoforms of VGSCs present, their mode(s) of action, and potential role(s) in angiogenesis have not been investigated. The main aims of this study were to determine the role of VGSC activity in angiogenic functions and to elucidate the potentially associated signaling mechanisms using human umbilical vein endothelial cells (HUVECs) as a model system. Real-time PCR showed that the primary functional VGSC α- and β-subunit isoforms in HUVECs were Nav1.5, Nav1.7, VGSCβ1, and VGSCβ3. Western blots verified that VGSCα proteins were expressed in HUVECs, and immunohistochemistry revealed VGSCα expression in mouse aortic ECs in vivo. Electrophysiological recordings showed that the channels were functional and suppressed by tetrodotoxin (TTX). VGSC activity modulated the following angiogenic properties of HUVECs: VEGF-induced proliferation or chemotaxis, tubular differentiation, and substrate adhesion. Interestingly, different aspects of angiogenesis were controlled by the different VGSC isoforms based on TTX sensitivity and effects of siRNA-mediated gene silencing. Additionally, we show for the first time that TTX-resistant (TTX-R) VGSCs (Nav1.5) potentiate VEGF-induced ERK1/2 activation through the PKCα-B-RAF signaling axis. We postulate that this potentiation occurs through modulation of VEGF-induced HUVEC depolarization and [Ca2+]i. We conclude that VGSCs regulate multiple angiogenic functions and VEGF signaling in HUVECs. Our results imply that targeting VGSC expression/activity could be a novel strategy for controlling angiogenesis.
Keywords: Calcium, Endothelium, ERK, Sodium Channels, Sodium Calcium Exchange, Angiogenesis, ERK1/2, VEGF
Introduction
Angiogenesis (the development of new blood vessels) is a process of fundamental biological importance, being essential for several normal functions, including embryonic development and tissue modeling and repair (1). Importantly, angiogenesis also manifests itself pathophysiologically, as in tumor progression and atherosclerosis (1, 2). Therapeutic angiogenesis may provide an alternative strategy for salvaging the ischemic myocardium (3), and several proangiogenic factors are in clinical trials with mixed results (4). Consequently, there is a need to further elucidate the complex molecular mechanisms controlling angiogenic activities of endothelial cells (ECs).6
The vascular endothelial growth factor (VEGF) is essential for EC functioning under normal and pathophysiological conditions (5). Binding of VEGF to its major receptor, VEGFR2 (Flk1/KDR), activates at least three parallel intracellular signaling cascades involving phospholipase Cγ1 (PLCγ1), Src kinase, and phosphoinositide 3-kinase (PI3K) (5–7). The final result is mobilization of downstream effectors, such as protein kinase C (PKC), endothelial nitric-oxide synthase, mitogen-activated protein kinases (MAPKs), and focal adhesion kinase (5). Additionally, VEGF-activated ECs exhibit a transient increase in intracellular Ca2+ concentration ([Ca2+]i) (8). Currently, more than 20 agents targeting VEGF or its receptors are in clinical trials for anti-cancer therapy (9), and VEGF is being evaluated for the treatment of myocardial ischemia (4).
Voltage-gated sodium channels (VGSCs) are plasma membrane proteins that allow influx of Na+ upon membrane depolarization. VGSCs comprise an α subunit (VGSCα), associated with one or more auxiliary β subunits (VGSCβs) (10). In mammals, nine VGSCα isoforms have been identified: Nav1.1–Nav1.9. Based on their sensitivity to the selective VGSC blocker tetrodotoxin (TTX), the isoforms are defined as TTX-resistant (TTX-R; Nav1.5, Nav1.8, and Nav1.9; IC50 in the μmol/liter range) and TTX-sensitive (TTX-S; Nav1.1–Nav1.4, Nav1.6, and Nav1.7; IC50 in the nmol/liter range). VGSC expression is not restricted to neurons and cardiac muscle cells; a number of “non-excitable” cells, including ECs, also express functional VGSCs (11–14).
We and others previously demonstrated up-regulation of functional VGSC expression in metastatic tumor cells, and VGSC activity was found to enhance cellular invasiveness in a variety of human cancers, including prostate (15, 16), breast (17), and colon (18). Based on the parallels between the processes involved in angiogenesis and tumor cell invasion (19) and the similarities between endothelial and neuronal guidance (20), we hypothesized that VGSC activity may be involved in the angiogenic properties of ECs. VGSC expression has been reported in human umbilical vein endothelial cells (HUVECs) (11, 14). However, the subtype(s) of channel present has not been determined, and the status of β-subunits, which can significantly influence VGSC function and act independently as cell adhesion molecules (21), is not known. Finally, although it has been speculated that ion channel activity may be involved in angiogenesis (22), the specific functional roles of the VGSCs expressed in ECs (and non-excitable cells generally) and the associated molecular signaling mechanism(s) are unclear.
Here, we show, for the first time, that functional VGSCs expressed in HUVECs exert significant control upon the cells' angiogenic activities and provide insights into underlying mechanisms. Molecular expression analyses and electrophysiology revealed consistently that the main functional VGSC isoforms in HUVECs are Nav1.5 (TTX-R) and Nav1.7 (TTX-S). VGSC activity increased VEGF-induced proliferation, chemotaxis, and tubular differentiation and decreased adhesion to substrate; TTX-R and TTX-S VGSC activities controlled different aspects of the angiogenic cascade. Furthermore, we report that TTX-R VGSC activity modulates the membrane potential of HUVECs in response to VEGF and subsequent Ca2+ signaling events, namely PKCα and ERK1/2 activation.
EXPERIMENTAL PROCEDURES
Materials
U73122, BAPTA-AM, EGTA-AM, GF109203X, l-NAME, PP2, PD98059, SKF96059, Nifedipine, and LY294002 were purchased from Calbiochem. DCB, VEGF-A, phorbol 12-myristate 13-acetate (PMA) and ionomycin were purchased from Sigma. TTX was purchased from Tocris. The suppliers of all other reagents are indicated throughout. HUVECs were from pooled donors (TCS Cellworks, Buckingham, UK) and were cultured according to the supplier's instructions.
Cell Culture and Protein Extraction
HUVECs were maintained in large vessel EC growth medium with the addition of large vessel endothelial growth supplements (TCS Cellworks) and grown in a humidified incubator at 37 °C and 5% CO2 in air. For the VEGF stimulation assays, cells were seeded in 35-mm dishes at a density of ∼105 cells/dish in complete HUVEC medium. The next day, the medium was aspirated, and the cells were washed twice with PBS and then serum-starved for 1 h in a serum-free physiological buffer (S/F), containing 144 mm NaCl, 5.4 mm KCl, 2.5 mm CaCl2, 1 mm MgCl2, 5.6 mm d-glucose, and 5 mm Tris-HCl, pH 7.4, at 37 °C. TTX was added 10 min before the beginning of the study. Cells were activated by the addition of 50 ng/ml VEGF-A (Sigma) and maintained for the indicated times at 37 °C, and the process was terminated by washing the cells once with ice-cold PBS, placing them on ice, and immediately applying 100 μl of cold lysis buffer containing 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1% (v/v) Triton X-100, 0.5 mm DTT, 1 mm PMSF, 1% (v/v) protease inhibitor mixture (Sigma), 1 mm NaF, 5 mm bpVphen (Calbiochem), 5 μm fenvalerate (Calbiochem), 1 mm Na3VO4, and 1% (v/v) phosphatase inhibitor mixtures I and II (Sigma). The cell homogenate was incubated for 10 min on ice and then centrifuged at 5,000 × g for 10 min at 4 °C. The supernatant was aliquoted and stored at −80 °C until further use. Protein concentrations were determined using the bicinchoninic acid assay (BCA; Sigma), with bovine serum albumin (BSA; Sigma) as the protein standard. The protein concentration of the HUVEC samples was calculated from the linear region of the standard curve, freshly prepared for each experiment. In the present study, HUVECs were used between passages 3 and 9.
Reverse Transcription-“TaqMan” Real-time PCR (RT-PCR)
Total RNA was extracted from HUVECs using Stratagene Miniprep kits in general (15) or TRIzolTM (Invitrogen) in siRNA experiments, according to the manufacturers' instructions. The quality of the RNA preparation was assessed by measuring the ratio of the absorbencies at 260 and 280 nm and by agarose electrophoresis. In all of the experiments, the RNAs used had absorbance ratios in the range 1.8–2.0. The purified RNA was stored at −20 °C until required. cDNA was synthesized from 1 μg of the purified RNA as described before (15). RT-PCR was performed on cDNAs using the DNA Engine Opticon system (MJ Research). PCRs were carried out in a 20-μl final volume. The reaction mixture contained 1 μl of cDNA, a 0.5 μm concentration of each specific primer, and 1× SYBR Green PCR mix (Qiagen). Amplification started with an initial denaturation step at 95 °C for 15 min, with subsequent three-step cycling of 95 °C for 30 s, 59 °C for 30 s, and 72 °C, followed by reading the plate. The program was repeated for 49 more cycles. Finally, a melting curve step was performed from 65 to 95 °C in order to verify product composition. Additionally, the PCR products were separated on a 1% agarose gel by electrophoresis in order to confirm their sizes. Each PCR was carried out in triplicate for the target gene and the normalizing gene. Additionally, blank reactions without added cDNA were also performed to control for any contamination or false amplification due to primer dimerization. The PCR primers used to determine the relative expression of all VGSCα and VGSCβ isoforms are summarized in supplemental Table 1. The efficiencies of the primers were tested as described (15) and found to be similar (not shown). In the case of the VGSCα isoforms, the primers were designed to amplify products spanning conserved introns between D1:S2 and D1:S5/S6, thus also controlling for any genomic DNA contamination (15). The relative gene expression data were analyzed by the 2−ΔΔCT method (23).
Western Blots
Cell lysates (10–15 μg of protein) were subjected to SDS-PAGE using the NuPAGE electrophoresis system (Invitrogen) under reducing conditions as described (24). Individual protein bands were electrotransferred to an activated PVDF membrane with 0.2-μm pores (Invitrogen) over 2 h at 30 V constant voltage at 4 °C in NuPAGE transfer buffer (Invitrogen). The membrane was then washed with TBST (50 mm Tris-HCl pH 8, 150 mm NaCl, 0.1% (v/v) Tween 20), blocked for 1 h with 5% nonfat dried milk (Marvel) in TBST, and then probed with the appropriate primary antibody in TBST containing 5% (w/v) nonfat dried milk and 0.5 mg/ml BSA at 4 °C overnight. The following day, the membrane was washed with TBST and probed with the appropriate secondary antibody for 1 h at room temperature. After a final wash step, the protein bands were visualized with an ECLplus chemiluminescence detection kit (Amersham Biosciences). The following antibodies were used: rabbit pan-VGSC from Upstate, rabbit anti-phospho-p44/p42 MAPK (Thr-202/Tyr-204) from Cell Signaling, rabbit p44/p42 (total ERK) from Cell Signaling, rabbit anti-phospho-PLCγ1 (Tyr-783) from Cell Signaling, mouse anti-PLCγ1 from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), mouse anti-NCX1 from Swant, rabbit anti-phospho-PKD/PKCμ (Ser-744/Ser-748) from Cell Signaling, rabbit anti-PKD/PKCμ from Cell Signaling, mouse anti-GAPDH from Abcam Laboratories, and peroxidase-conjugated secondary antibody (Cell Signaling).
Immunoprecipitation
Protein A-agarose beads (Roche Applied Science) were washed twice in ice-cold PBS and then incubated with 1 μg of anti-pan-VGSC or anti-NCX1 antibodies for 1 h at 4 °C. The beads were then washed twice in ice-cold PBS and added to the corresponding cell lysates. Beads with no conjugated antibody were used as a negative control. Following overnight incubation at 4 °C, the beads were washed twice with lysis buffer and once with PBS and solubilized in sample buffer at 95 °C for 5 min. Proteins were visualized as described above.
Electrophysiology and Pharmacology
Details of the patch pipettes, solutions and the whole-cell voltage clamp/current-recording protocols were as described previously (15, 17). In brief, patch pipettes (tip resistances, 5–10 megaohms) were filled with a solution designed to block the outward K+ currents as follows: 5 mm NaCl, 145 mm CsCl, 2 mm MgCl2, 1 mm CaCl2, 10 mm HEPES, and 11 mm EGTA, adjusted to pH 7.4 with 1 m CsOH. Cells were bathed in a physiological saline solution containing 144 mm NaCl, 5.4 mm KCl, 1 mm MgCl2, 2.5 mm CaCl2, 5 mm HEPES, and 5.6 mm glucose (pH 7.3). Whole-cell membrane currents were recorded from cells that appeared “isolated” in culture, using an Axopatch 200B amplifier (Axon Instruments). Analog signals were filtered at 10 kHz using a low pass Bessel filter, and series resistance errors were compensated by >80%. Electrophysiological signals were sampled at 50 kHz and digitized using a Digidata 1200 interface. Data acquisition and analysis of whole-cell currents were performed using pClamp software (Axon Instruments). Currents were generated by applying 5-mV steps, from −70 to +70 mV, from a holding potential of −100 mV, with an interpulse interval of 2 s. Experiments on HUVECs were performed on at least three separate dishes that had been cultured for 1–3 days. TTX was applied in bath solution in a range of concentrations up to 10 μm. Data were analyzed as means ± S.E. (n ≥ 4).
Transwell Migration Assay
The migration assay was done as described previously (25). Briefly, 1–1.5 × 106 HUVECs (∼70% confluent) in a T75 flask were washed with PBS and then incubated for 1 h in MCDB-131 medium containing 0.3% (v/v) FCS (Serotex) and 0.5 mg/ml hydrocortisone (Sigma) in a humidified incubator at 37 °C in 5% CO2 air. After 1 h, the cells were labeled with 1 μm CellTracker green 5-chloromethylfluorescein diacetate (Molecular Probes) and incubated for a further 1 h at 37 °C in 5% CO2 air. The cells were then washed with PBS, trypsinized, and placed into the upper chamber of a 3-μm pore FluoroblokTM insert (BD Biosciences) containing 350 μl of serum-free medium with or without TTX at different concentrations (up to 10 μm). Approximately 3 × 104 cells were plated onto each filter. After adding 800 μl of medium containing 50 ng/ml VEGF-A (Sigma), the assay plates were incubated at 37 °C in 5% CO2 for 16 h. Basal migration of HUVECs due to the presence of 0.3% (v/v) FCS in the migration medium was assessed by seeding 3 × 104 HUVECs in the upper chamber without adding the chemoattractant (VEGF-A) in the lower chamber. The numbers of migrating cells under these conditions were subtracted from the number of the migrating cells in the presence of VEGF-A to assess directed chemotaxis. Images of the migrated cells in the lower chamber were obtained with an inverted fluorescent microscope (Olympus LX70, Middlesex, UK) and a digital camera (CoolSNAP-Pro color, Media Cybernetics) under a ×10 magnification. Typically, images were acquired from five fields of view. The numbers of migrated cells were counted using Image Pro-Plus 5.0 software (Media Cybernetics). Data were expressed as percentage migration relative to untreated (control) cells.
Tubulogenesis Assay
The assay was carried out essentially as described previously (24, 25). Briefly, cells were plated at a density of 3 × 104 cells/well in 24-well plates precoated with 300 μl of Matrigel (BD Biosciences) in complete HUVEC medium, with or without TTX. Cells were incubated for periods of 4 h before images were obtained and quantified for tubule length using Image Pro-Plus 5.0 software.
Single-cell Adhesion Assay
A single-cell adhesion measurement apparatus was used (26). Suction was applied onto individual cells via a glass micropipette (tip sizes, 19–22 μm outer diameter). The detachment negative pressure was recorded on-line using a digital manometer and measured in kilopascals. Adhesion was measured 24 h after treating the cells with TTX (20 nm to 10 μm). For each concentration, at least 50 cells were measured from each of a minimum of five dishes, each assayed within 20 min.
Proliferation Assay
HUVECs were seeded into 96-well plates (Nunc) at a density of 3 × 103 cells/well in complete EC medium. After 24 h, the medium was aspirated, and 100 μl of MCDB-131 medium containing 0.3% FCS and 1 μg/ml hydrocortisone was added. Where appropriate, the medium contained 50 ng/ml VEGF and different concentrations of TTX. Each condition was tested in triplicate. After incubation for 72 h at 37 °C in a humidified incubator at 37 °C and 5% CO2 in air, the relative number of cells in each well was determined by the alkaline phosphatase assay. Briefly, the medium was removed, and 100 μl of 3 mg/ml 4-nitrophenyl phosphate (Sigma) in 0.1 m sodium acetate, 0.1% (v/v) Triton X-100 was added, and the cells were further incubated at 37 °C for 2 h. The reaction was stopped by adding 50 μl of 1 m NaOH, and absorbance at 405 nm was measured by using a SPECTRAmax 340PC plate reader (Molecular Devices).
Gene Silencing
siRNA was used to knock down the expression of the two predominant VGSCα isoforms detected in HUVECs, Nav1.5 and Nav1.7. The siRNA duplexes (siGENOME SMARTpools) used (Dharmacon) had the following accession numbers: NM_000335, NM_198056 (Nav1.5), and NM_0002977 (Nav1.7). Lamin A/C (accession number NM_005572) was used to determine transfection efficiency; a non-targeting siRNA pool, also from Dharmacon, was the control. Transfections were carried out according to the manufacturer's instructions, essentially as detailed previously (24) with minor modifications. Briefly, HUVECs were plated in T25 cell culture flasks and allowed to settle for 24 h in complete medium. The following day, subconfluent cultures (∼40–50%, 2 × 105 cells) were transfected at 37 °C with siRNA using Oligofectamine (Invitrogen) according to the manufacturer's instructions. The final concentration of siRNA was 100 nm in Opti-MEM (Invitrogen). The transfection was for 4 h at 37 °C and was terminated by the addition of 2 volumes of endothelial complete medium (TCS Cellworks). Transfected HUVECs were maintained for 72 h prior to downstream applications. Typically, one flask from each experimental condition was used for RNA extraction for RT-PCR, protein extraction for Western blots, and Transwell migration assays, as described above. Control experiments, in parallel, included the following: 1) transfection with non-targeting pooled siRNA; 2) transfection with Oligofectamine alone; 3) transfection with lamin A/C siRNA duplex, serving as a positive control of transfection efficiency; and 4) untreated cells.
Immunohistochemistry
To investigate VGSC expression in ECs in vivo (mouse aorta), we used a commercially available pan-VGSC (Upstate) antibody. Male NCr nude mice (25 g, n = 7) were humanely killed according to Home Office guidelines. Their aortas were excised, washed in PBS, and immediately frozen in liquid nitrogen. The aortas were sectioned at 4-μm thickness using a cryostat (Leica), and sections were collected onto polylysine-coated slides (VWR). The staining procedure was as described previously (27). Briefly, the sections were fixed using 4% paraformaldehyde in PBS (pH 7.4) for 1 h and washed three times for 10 min each in TBS (50 mm Tris-HCl, pH 8, 150 mm NaCl). The endogenous peroxidase activity was removed by using 2% (v/v) H2O2 (Sigma) in TBS for 30 min, followed by three 10-min washes in TBS and incubation in blocking solution (0.2% (w/v) BSA in TBS) for 1 h at room temperature. Following this, the sections were incubated in the pan-VGSC antibody (1:50) overnight at room temperature. Subsequently, the aortas were washed three times for 10 min each in TBS, followed by a 4-h incubation in biotinylated donkey anti-rabbit IgG 1:200 (Jackson Immunochemicals). After a further three 10-min washes in TBS and a 3-h incubation in ABC solution (Vector) followed by three 10-min TBS washes and then two 10-min washes in 50 mm Tris-HCl, pH 8, buffer (TB), the reaction was visualized using a DAB kit (Vector Immunochemicals). The reaction was stopped by washing in TB, and the sections were dehydrated using 10-min washes in 50, 70, 90, 95, and 100% (v/v) ethanol, washed 2 × 10 min in inhibisol (BDH), and mounted under a coverslip using DPX mountant (BDH)., The sections were visualized with a Zeiss Axioskop microscope.
Immunocytochemistry
For immunofluorescence studies, HUVECs were plated on glass coverslips and subsequently fixed with 4% paraformaldehyde in 0.1 m PBS for 10 min. Steps were as described previously (24, 27). The pan-VGSC antibody was visualized using Alexa Fluor® 488-conjugated goat anti-rabbit IgG (1:500; Molecular Probes). For dual labeling, following staining for VGSC protein (using the pan-VGSC antibody), some coverslips were incubated with mouse anti-NCX1 (1:50; Swant) for 12 h, followed by Alexa Fluor® 568-conjugated donkey anti-mouse preabsorbed in mouse (1:500; Molecular Probes) for 3 h. Finally, the sections were washed in PBS three times for 10 min each and mounted under coverslips using Vectamount for Fluorescence (Vector Laboratories) and observed using a scanning confocal microscope model SP1 (Leica), using the appropriate lasers and filter sets. In order to ensure that no cross-reactivity occurred, double or single labeling was undertaken with the omission of either one or both primary antibodies.
B-Raf Activity Assay
HUVECs in 10-cm dishes (∼1 × 106 cells) were serum-starved for 1 h in S/F medium and then stimulated with 50 ng/ml VEGF for 10 min in the presence of various concentrations of TTX as described above. The cells were lysed in 1 ml of ice-cold lysis buffer, incubated for 10 min on ice, centrifuged for 10 min at 5,000 × g, and then assayed for B-Raf activity using a B-Raf kinase cascade kit (Upstate) according to the manufacturer's instructions. Briefly, 5 μl of anti-B-Raf kinase cascade validated rabbit antibody (Upstate) was complexed with 20 μl of Protein A/G PLUS-agarose beads (Santa Cruz Biotechnology, Inc.) per sample for 1 h at 4 °C. The beads were then washed three times with ice-cold PBS, resuspended in 50 μl of lysis buffer, and applied to 0.95 ml of the assay sample, containing ∼1 mg of total protein. As an additional control, 20 μl of Protein A/G PLUS-agarose beads with no added antibody were applied to 0.95 ml of VEGF-stimulated HUVEC cell lysate and were used as the blank. The cell lysate was incubated overnight at 4 °C with the anti-B-Raf Ab-complexed agarose beads on a rotating device. The following day, the agarose beads were pelleted in a microcentrifuge and washed three times with ice-cold lysis buffer and once with assay buffer containing 20 mm MOPS, pH 7.2, 25 mm β-glycerol phosphate, 5 mm EGTA, 1 mm sodium orthovanadate, and 1 mm DTT (Upstate). The B-Raf immunoprecipitates were resuspended in 20 μl of assay buffer supplemented with 2 μg of inactive MEK-1 (Upstate) and 1 μg of inactive MAPK-2/Erk-2 (Upstate). The reaction was initiated by the addition of a 10-μl ATP mixture containing 75 mm MgCl2 and 500 μm ATP (Upstate) in assay buffer and proceeded for 30 min at 30 °C. A blank sample containing no beads and a positive control containing 0.1 μg of constitutively active B-Raf (Δ1–415, Upstate) were also assayed. The reaction was terminated by removing 6 μl of the supernatant and placing it on ice. 4 μl of the activated MAPK-2/Erk-2 was applied to 30 μl of assay buffer containing 50 μg of myelin basic protein (Upstate), 150 μm ATP, and 1 μCi of [γ-32P]ATP (specific activity 3000 Ci/mmol; PerkinElmer Life Sciences). The reaction proceeded for 15 min at 30 °C and was terminated by spotting 25 μl of the reaction mixture on a P81 phosphocellulose filter. The filters were washed three times for 10 min in 75 mm H3PO4, and the myelin basic protein-associated 32P radioactivity bound to the filters was quantified by scintillation counting, using Optima GoldTM (PerkinElmer Life Sciences) as the scintillant.
PKCα Translocation Assay
PKCα translocation to the plasma membrane was determined as described previously (28) with minor modifications. Briefly, HUVECs in 10-cm dishes (∼1 × 106 cells) were serum-starved for 1 h in S/F medium and then stimulated with 50 ng/ml VEGF in the presence of various concentrations of TTX for 10 min. The cells were suspended in 1 ml of ice-cold lysis buffer without Triton X-100, homogenized by passing 20 times through a 21-gauge needle, sonicated three times for 20 s with 1-min intervals on ice, and finally centrifuged for 10 min at 5,000 × g. The supernatant was ultracentrifuged for 1 h at 100,000 × gav in a Ti-32 rotor (Beckman). The supernatant was collected as the cytosolic fraction, and the membrane fraction was resuspended in sample buffer and heated for 10 min at 95 °C. PKCα translocation to the membrane fraction was then determined by Western blot.
[Ca2+]i Measurement
Measurements of [Ca2+]i were carried out essentially as described previously (29). Briefly, HUVECs were seeded at a density of 1 × 104 cells/well in a sterile flat clear-bottomed black-walled 96-well microtiter plate (Corning Glass). Cells were allowed to attach to the plate and recover for 24 h in a humidified incubator (5% CO2, in air, 37 °C). The following day, the medium was removed; cells were washed twice with PBS and loaded with the Ca2+ dye indicator Fluo-4NW (Invitrogen) for 45 min at 37 °C in the dark. The Fluo-4NW solution was freshly prepared prior to each experiment by adding 10 ml of Hanks' balanced salt solution (Invitrogen), supplemented with 20 mm HEPES, pH 7.4, and 100 μl of 250 mm probenicid stock solution (Component B, Molecular Devices), to one bottle of Component A (Fluo-4NW dye mix; Molecular Devices). The dye was dissolved by vigorous vortexing. The probenicid final concentration during dye loading was 2.5 mm. Following the incubation period, 50 μl of Hanks' balanced salt solution containing the appropriate concentrations of inhibitors or vehicle was added, and the cells were incubated for a further 15 min. The plate was then transferred in the assay chamber of a FLIPR plate reader (Molecular Devices), and HUVECs were challenged with 50 ng/ml VEGF in Hanks' balanced salt solution for 400 s at 37 °C. The [Ca2+]i was monitored immediately after the addition of stimulant, as a measure of changes in fluorescence intensity at 37 °C on a FLIPR plate reader (Molecular Devices) with excitation of 485 nm and emission wavelength of 525 nm (cut-off, 515 nm). Data points were acquired every 2 s. Typically, immediately before the assay, an end point fluorescent reading with excitation and emission wavelengths of 485 nm and 525 nm, respectively, was done, in order to ensure uniform dye loading of all of the sample wells. Background fluorescence was measured for 20 s prior to the addition of stimulant, and results are presented as a ratio of sample fluorescence at any given time point divided by background fluorescence. All experimental conditions were in triplicate and were assayed simultaneously.
Membrane Potential (Vm) Measurement
Changes in HUVEC membrane potential in response to VEGF were determined as described previously (8), with the use of the slow response bis-oxonol dye indicator DiBaC4(3) (Invitrogen) (30). HUVECs were seeded at a density of 1 × 104 cells/well in a sterile flat clear-bottomed black-walled 96-well microtiter plate (Corning Glass). Cells were allowed to attach for 24 h in a humidified incubator (5% CO2, in air, 37 °C). The following day, the medium was removed; cells were washed twice with PBS and serum-starved in S/F medium for 45 min at 37 °C. Concurrently, DiBaC4(3) was dissolved in S/F medium. Following the incubation period, 50 μl of S/F medium containing the appropriate concentrations of inhibitors or vehicle and 100 nm DiBaC4(3) was added, and the cells were incubated for a further 15 min at 37 °C. The assay plate was then transferred into the assay chamber of a FLIPR plate reader (Molecular Devices), and HUVECs were challenged with 50 ng/ml VEGF in S/F medium for 400 s at 37 °C. The changes in Vm were monitored immediately after the addition of stimulant, as a measure of changes in fluorescence intensity at 37 °C on a FLIPR plate reader (Molecular Devices) with excitation of 485 nm and emission wavelength of 525 nm. Data points were acquired every 2 s as before. Samples were in triplicate, and results are presented as the ratio of sample fluorescence at any given time point divided by background fluorescence, as described for the [Ca2+]i experiments. Uniform dye application was ensured by an end point fluorescent reading as described above for the Ca2+ measurement.
Data Analysis
The data were expressed as means ± S.E. Where applicable, statistical significance was determined by Student's paired, one-tailed t test. Values of p < 0.05 were deemed statistically significant unless otherwise stated.
RESULTS
VGSCα and VGSCβ Expression in HUVECs
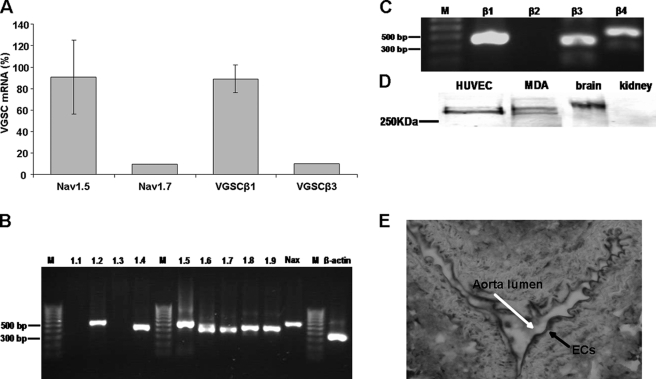
RT-PCR using the SYBR Green method (see “Experimental Procedures”) showed that the major VGSCα isoforms in HUVECs were Nav1.5, Nav1.6, and Nav1.7, representing 56, 38, and 6% of total VGSCα mRNA, respectively (Fig. 1, A and B). RT-PCR also revealed that the VGSCα subtypes Nav1.2, Nav1.4, Nav1.8, Nav1.9, and NaX were also present, but these constituted less than 0.2% of the total VGSCα mRNA. Nav1.1 and Nav1.3 were not detected by this method. Resolving the products of the PCRs by agarose electrophoresis confirmed the specificity of the primers (Fig. 1B). Moreover, Nav1.6 mRNA was present as two bands, consistent with expression of neonatal and mistranscribed/exon-skipped truncated isoforms, neither of which would yield functional channels (15). We therefore concluded that potentially the most significant VGSCα isoforms were Nav1.5 and Nav1.7, comprising ∼91 and 9% of the total VGSCα mRNAs in HUVECs, respectively (Fig. 1A). For the β subunits, VGSCβ1 was the predominant isoform, representing 89% of total mRNA. VGSCβ3 mRNA constituted ∼10% of the total, whereas VGSCβ4 mRNA was ≪1% and VGSCβ2 was undetectable (Fig. 1, A and C). A pan-VGSCα antibody revealed a protein of ∼250 kDa in HUVECs, and the overall level was comparable with that in MDA-MB-231 breast cancer cells (Fig. 1D). Adult mouse brain and kidney were used as positive and negative controls, respectively. The expression of ion channels in ECs in vitro is known to vary depending on the isolation method and the culture conditions (22). In order to confirm the physiological relevance of our in vitro data, we stained sections of mouse aorta using a pan-VGSC antibody and confirmed the presence of VGSCs in vivo (Fig. 1E).
FIGURE 1.
PCR and Western blots for VGSCα/β expression in HUVECs. A, semiquantified relative (%) levels of mRNA of the major VGSCα and VGSCβ isoforms. Data are presented as mean ± S.E. The expression level of the predominant VGSC subunit is expressed as a percentage of the total respective VGSCα/β mRNA levels. Error bars represent the propagated errors (2−ΔΔCT analysis). Shown is mRNA expression of VGSCα isoforms (B) and VGSCβ isoforms (C). D, Western blot showing VGSCα protein. E, immunohistochemical staining with a pan-VGSC antibody of mouse aorta. Black arrow, endothelial cells; white arrow, the aortic lumen.
VGSCs Are Active in HUVECs
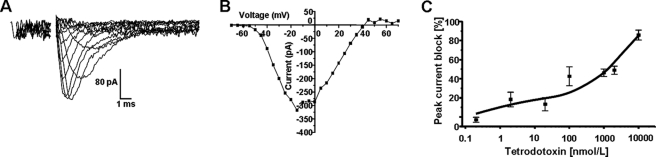
Next, we investigated whether the VGSC protein detected is functional. Whole-cell patch clamp recordings showed HUVECs to have a resting potential of −42.9 ± 3.9 mV (n = 22) and a whole-cell capacitance of 44.9 ± 3.2 picofarads (n = 43). About 40% (18 of 45) of HUVECs tested expressed depolarization-activated inward currents (Fig. 2, A and B). These currents activated at −41.1 ± 1.3 mV (n = 4) showed a peak current density of 2.9 ± 0.7 pA/picofarads (n = 18) and were abolished in Na+-free medium (not shown). The inward currents were suppressed by the highly specific VGSC blocker TTX in a dose-dependent manner. Dose-response relationships had two components with IC50 values of 1–10 and >200 nmol/liter, implying that both TTX-S and TTX-R VGSC activities contributed to the Na+ current, in a ratio of 1:5 on average (Fig. 2C). Thus, the electrophysiology was consistent with the RT-PCR regarding the subtypes of VGSC expressed.
FIGURE 2.
VGSC activity in HUVECs. A, electrophysiological whole-cell recordings. Traces show activation of an inward current by applying 5-mV steps, from −70 mV to +70 mV, from a holding potential of −100 mV, with an interpulse interval of 2 s. Alternate traces only are shown for clarity. B, a typical current-voltage relationship for the inward currents recorded as in A. C, TTX dose-response curve. Data points denote means ± S.E. (error bars) (n ≥ 4). Cells were pulsed from a holding potential of −100 mV to −10 mV for 30 ms with a repeat interval of 10 s. The effect of TTX was recorded on the fifth pulse. The intracellular pipette solution contained Cs+ to block outward (K+) currents in all recordings shown.
TTX Inhibits HUVEC Angiogenic Activities
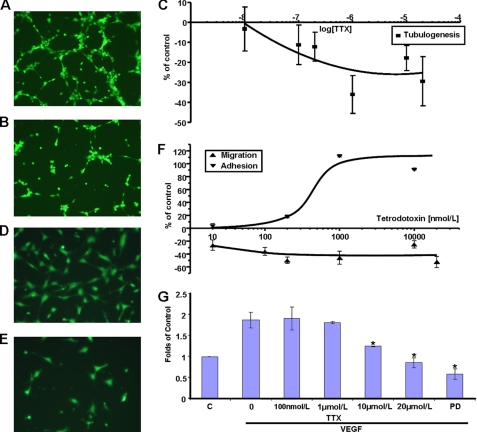
HUVEC tubular differentiation on MatrigelTM, a surrogate assay used to model angiogenesis in vitro (25), was significantly inhibited by exposure to TTX (Fig. 3, A–C). TTX concentrations up to 200 nmol/liter had no effect, whereas increasing the concentration to 1 μmol/liter reduced tubule length to 64 ± 9% of controls, with no further effect of TTX up to 10 μmol/liter (Fig. 3C). We concluded that tubulogenesis was controlled mainly by TTX-R VGSC, presumably Nav1.5, activity. EC migration toward proangiogenic cytokines constitutes a crucial part of the angiogenesis cascade (19, 31). Furthermore, VGSC activity is known to promote migration of cancer cells (17). In Transwell assays, TTX reduced HUVEC chemotaxis toward VEGF in a dose-dependent manner (Fig. 3, D–F). A maximum of 50 ± 5% inhibition was seen at 200 nmol/liter; increasing the TTX concentration to 10 μmol/liter had no further effect. These data implied control mainly by a TTX-S VGSC, presumably Nav1.7. As shown previously for cancer cells (26), TTX increased HUVEC adhesion in a dose-dependent manner. There was little effect on HUVEC adhesion at TTX concentrations up to 200 nmol/liter; the effect peaked at 1 μmol/liter with a 112 ± 1.8% increase, and 10 μmol/liter produced no further significant change (Fig. 3F). These data were consistent with HUVEC adhesion also being controlled by TTX-R VGSC activity. Finally, we investigated the effect of TTX on VEGF-induced proliferation. VEGF increased the number of viable HUVECs by 1.87 ± 0.18-fold over 72 h, in agreement with previous work (32) (Fig. 3G). Nanomolar concentrations of TTX had no apparent effect; increasing the concentration of TTX to 10 μmol/liter significantly reduced the number of viable cells to 1.25 ± 0.01-fold of unstimulated controls, and 20 μmol/liter TTX reduced their number further (0.86 ± 0.12-fold). The specific MEK inhibitor PD98059 also suppressed VEGF-induced HUVEC proliferation (0.59 ± 0.13-fold), confirming that the underlying signaling was via ERK1/2 (33).
FIGURE 3.
Effects of TTX on HUVEC functions. Representative photomicrographs of HUVECs plated onto MatrigelTM. A, control; B, 1 μmol/liter TTX. C, quantification of tubule length at 4 h relative to controls. Representative photomicrographs of HUVECs migrated through Transwell filters toward VEGF. D, control. E, 200 nmol/liter TTX. F, migration (▴) was measured at 16 h, and adhesion (▾) was measured at 24 h. Data points denote mean ± S.E. (n = 3–5). G, HUVEC proliferation assay. Cells were exposed to VEGF for 72 h in the presence of TTX or PD98059, as indicated. C, unstimulated controls; absorbance set at 1. Bars, represent mean ± S.E. (error bars) (n = 3 in triplicate). *, p < 0.05 versus controls.
siRNA Knockdown of Nav1.7 but Not Nav1.5 Reduces HUVEC Chemotaxis toward VEGF
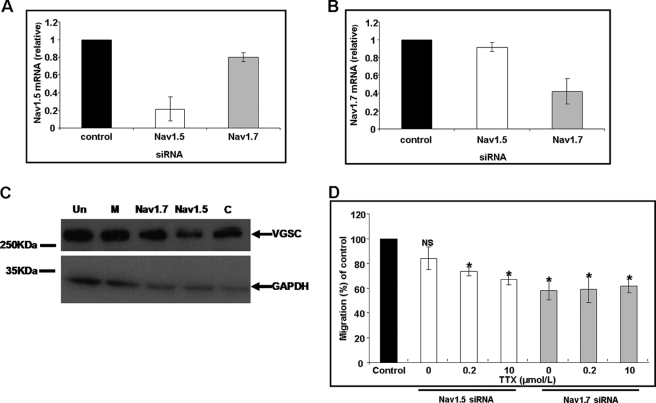
Pooled siRNA duplexes targeting Nav1.7 or Nav1.5 gave specific down-regulation of VGSC subtypes 72 h post-transfection. Nav1.7 mRNA levels were reduced to 0.42 ± 0.14-fold, and Nav1.5 mRNA levels were reduced to 0.22 ± 0.13 fold, compared with control siRNA. In both cases, the expression level of the non-targeted alternative isoform was essentially unaffected, and control siRNA transfections had no effect on expression (Fig. 4, A and B). Specific knockdown was also achieved at the protein level with no significant reduction in mock-transfected or control siRNA-treated HUVECs (Fig. 4C). Nav1.7 down-regulation resulted in a 42 ± 7.6% reduction in migration toward VEGF compared with control siRNA-treated HUVECs, and applying TTX (200 nmol/liter to 10 μmol/liter) to Nav1.7-depleted cells produced no further decrease (Fig. 4D). Knockdown of Nav1.5 expression did not alter HUVEC migration toward VEGF, whereas TTX (200 nmol/liter to 10 μmol/liter) still reduced migration to levels comparable with those observed in the Nav1.7-knockdown cells (33.3 ± 4% reduction). These results also were consistent with the pharmacology (Fig. 3F).
FIGURE 4.
Effect of VGSC (Nav1.5 and Nav1.7) siRNAs on chemotaxis. mRNA measurements following treatments of HUVECs with Nav1.5 (A) and Nav1.7 (B) siRNA. Normalized levels of mRNA at 72 h relative to cells transfected with non-targeting siRNA are shown. White bars, Nav1.5 siRNA-transfected HUVECs. Gray bars, Nav1.7 siRNA-transfected HUVECs. Bars, mean ± S.E. from 2−ΔΔCT analysis (n = 3). C, representative Western blots of proteins from siRNA-treated HUVECs using a pan-VGSC primary antibody and GAPDH. Un, untreated; M, mock-transfected; other lane labels are as in A. D, Transwell chemotaxis of HUVECs treated with Nav1.5 or Nav1.7 siRNA. Data are shown as percentage controls. Samples are as in A but also showing the effect of TTX applied post-transfection. Each bar denotes mean ± S.E. (error bars) (n = 3). *, p < 0.05.
VGSC Activity Is Required for ERK1/2 Activation by VEGF
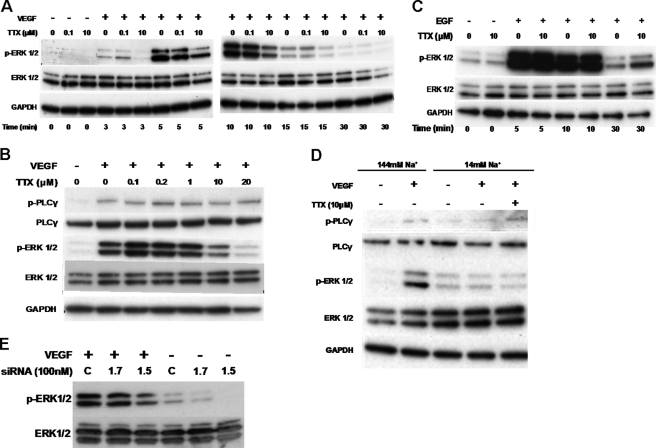
VGSC activity potentiated HUVEC tubulogenesis (Fig. 3C) and VEGF-stimulated proliferation (Fig. 3G). Because both of these functions depend on ERK1/2 activation (32–34), we studied the effects of TTX on ERK1/2 phosphorylation. Stimulation of HUVECs with VEGF resulted in a marked increase in the levels of phospho-ERK1/2Thr-202/Tyr-204. Preincubation with nanomolar concentrations of TTX had no effect, whereas micromolar concentrations of TTX markedly reduced VEGF-induced ERK1/2 phosphorylation (Fig. 5, A and B). In contrast, TTX had no effect on the levels of phospho-PLCγ (Fig. 5B), which can be used as an indication of VEGFR-2 activity (6) and as an internal control to show that cells were equally stimulated. Selectivity of inhibition of VEGFR-ERK1/2 signaling was demonstrated by the fact that 10 μmol/liter TTX had no apparent effect on the activation of ERK1/2 by epidermal growth factor (EGF), which signals via RAS-RAF and MEK (Fig. 5C) (35, 36). As an alternative way of reducing VGSC activity, we lowered the extracellular Na+ concentration ([Na+]o) 10-fold. This significantly decreased the phospho-ERK1/2 level in VEGF-stimulated HUVECs. Treating the cells with 10 μmol/liter TTX in the low Na+ medium produced no further effect (Fig. 5D). Furthermore, the levels of phospho-PLCγ were similar under control or low [Na+]o conditions, indicating that the cells were similarly stimulated and that low [Na+]o does not alter VEGFR2 activity (6, 31). The pharmacological data were complemented by an RNAi approach. By employing the same siRNA duplexes as in Fig. 4, we targeted Nav1.5 and Nav1.7 expression in HUVECs. Knockdown of Nav1.7 had no obvious effect on ERK1/2 phosphorylation in comparison with the control. On the other hand, in HUVECs treated with Nav1.5 siRNA, phospho-ERK1/2 levels in response to VEGF were attenuated (Fig. 5E).
FIGURE 5.
TTX inhibits ERK1/2 activation upon VEGF stimulation of HUVEC. HUVECs were incubated with VEGF in the presence of TTX for the indicated times. A, Western blots of phospho-ERK1/2, total ERK1/2, and GAPDH (n = 4). B, phospho-PLCγ1, total PLCγ, p-ERK/12, and total ERK levels (n = 3). C, HUVECs were incubated with EGF in the presence of TTX as indicated, and blots were probed as in A (n = 3). D, HUVECs were stimulated with VEGF in a “low Na+” medium; ERK1/2 and PLCγ1 activation were analyzed as before (n = 3). E, representative Western blot of phosphorylated and total ERK1/2 from HUVECs transfected with siRNA (100 nm), serum-starved and challenged with VEGF (50 ng/ml for 10 min). Un, untreated; M, mock-transfected; 1.7, Nav1.7 siRNA; 1.5, Nav1.5 siRNA.
Possible Role of PKC as the Link between VGSC Activity and ERK1/2 Phosphorylation
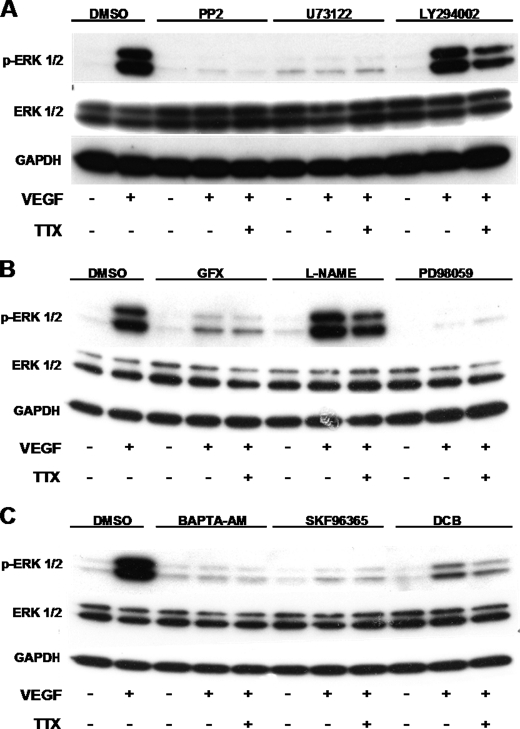
Next, we investigated the relationship of VGSC to the established VEGFR2-ERK1/2 signaling pathway, which is reportedly via Src-PLCγ-PKC-Raf and independent of PI3K (6, 7, 28, 35). We showed that VGSC influences VEGF-induced ERK1/2 activation independently of PI3K and also NOS (Fig. 6, A and B). It was previously reported (37) that VEGFR-ERK1/2 signaling requires intracellular Ca2+; indeed, BAPTA-AM abolished ERK1/2 phosphorylation, and TTX had no further effect (Fig. 6C). Additionally, inhibition of cation entry with SKF-96365 completely suppressed the activation of ERK1/2 by VEGF, highlighting the importance of extracellular cations in this process (Fig. 6C). Based on these data, we hypothesized that the Ca2+-sensitive candidate linking VGSC activity with the VEGF-induced ERK1/2 activation could be PKC.
FIGURE 6.
VEGF activates ERK1/2 through a pathway that involves Src, PLCγ, PKC, and Ca2+. HUVECs were treated with 1 μm Src inhibitor PP2, 1 μm PLC inhibitor, 30 μm PI3K inhibitor LY294002 (A); 1 μm PKC inhibitor GF109203X, 1 mm general NOS inhibitor l-NAME, 30 μm MEK inhibitor PD98059 (B); 10 μm cell-permeable Ca2+ chelator BAPTA-AM, 50 μm inhibitor of non-selective cation channels SKF-96365, or 30 μm NCX inhibitor DCB (C) for 30 min before exposure to 50 ng/ml VEGF. 10 μm TTX was added to the indicated samples for 10 min before stimulation with VEGF for 10 min. VEGF-induced ERK1/2 activation in cell lysates was determined by Western blot as described in the legend to Fig. 5 (n = 3).
VGSC Activity Is Required for VEGF Activation of B-Raf and PKD/PKCμ via PKCα
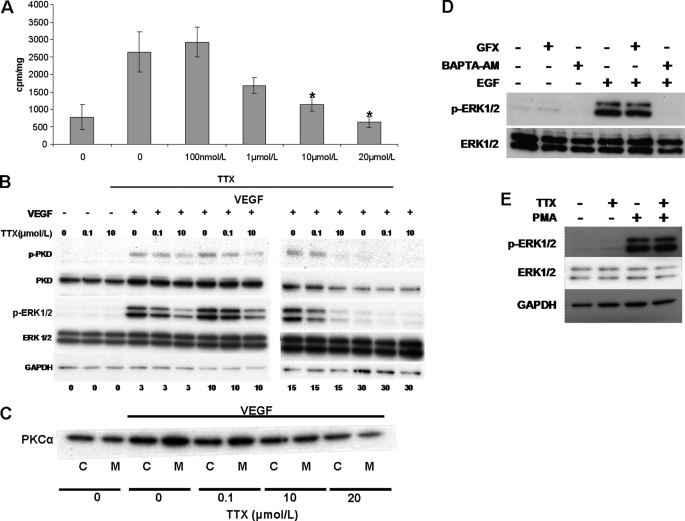
We used TTX to investigate the role of VGSC activity in VEGF-induced B-Raf activation, a process crucial for ERK1/2 phosphorylation and dependent on PKC activation in ECs (35, 36, 38). VEGF stimulation of HUVECs resulted in a ∼3–4-fold increase in B-Raf activity compared with controls (2,634 ± 581 versus 767 ± 362 cpm/mg; Fig. 7A). Micromolar TTX reduced the VEGF-induced B-Raf activity to the level of controls (627 ± 166 cpm/mg for 20 μmol/liter TTX), in agreement with our previous pathway activation data (Fig. 5); 100 nmol/liter TTX had no significant effect on B-Raf activity (2,919 ± 436 cpm/mg; Fig. 7A). PKCμ/PKD requires PKC-dependent phosphorylation of Ser-744/Ser-748 for catalytic activity (38). The PKCα isoform was found to activate PKCμ/PKD upstream of ERK1/2 in VEGF-stimulated HUVECs (38). The level of PKDSer-744/748 was monitored as an indication of PKCα activity in intact HUVECs. PKD phosphorylation was readily detected after VEGF stimulation. Pretreatment with 100 nmol/liter TTX had no detectable effects; however, 10 μmol/liter TTX substantially reduced PKD phosphorylation at all time points (Fig. 7B). Phospho-ERK1/2 levels in the same samples were also significantly reduced, as shown earlier (Fig. 5A). Next, we investigated whether TTX would inhibit VEGF-induced PKCα translocation to the plasma membrane, a process that correlates with PKC activity (28). Crude membrane and cytosolic fractions were isolated, and PKCα was detected by Western blot. VEGF stimulation of HUVECs resulted in the translocation of PKCα to the membrane fraction. Preincubating the cells with 100 nmol/liter TTX had no detectable effect; however, 10 μmol/liter TTX reduced the amount of translocated PKCα, and 20 μmol/liter TTX had a more pronounced effect (Fig. 7C), in agreement with our previous results (Fig. 5B). In addition, the PKC inhibitor GFX109293X (1 μm) failed to suppress EGF-induced ERK1/2 phosphorylation (Fig. 7D), a process independent of VGSC activity (Fig. 5C). Furthermore, TTX (10 μmol/liter) did not diminish the ERK1/2 phosphorylation elicited by the PKC activator phorbol 12-myristate 13-acetate (PMA; Fig. 7E). Thus, these findings support our hypothesis that VGSC activity is essential for activation of PKC by VEGF and subsequent ERK1/2 phosphorylation.
FIGURE 7.
Micromolar concentrations of TTX inhibit VEGF-induced B-Raf activation, PKCμ/PKD phosphorylation, and PKCα translocation to the membrane fraction. HUVECs were stimulated with VEGF with or without TTX. A, B-Raf was immunoprecipitated from cell extracts, and activity was assayed using a B-Raf kinase assay kit (Upstate). Bars, mean ± S.E. cpm/mg protein (n = 4). *, p < 0.05 versus VEGF-stimulated control. B, blots show phospho-PKD, total PKD, phospho-ERK1/2, total ERK, and GAPDH (n = 3). C, PKCα distribution to cytosolic (C) or membrane (M) fractions obtained by ultracentrifugation of HUVEC cell lysates. (n = 3). D, HUVECs were treated with the PKC inhibitor GFX109203X or the cell-permeable Ca2+ chelator BAPTA-AM as described in Fig. 6. EGF-induced ERK1/2 phosphorylation was determined by Western blot as in Fig. 5 (n = 3). E, serum-starved HUVECs were stimulated with phorbol 12-myristate 13-acetate (PMA) (1 μm for 10 min) in the presence or absence of TTX (10 μm)), and phospho-ERK1/2 was determined as described in the legend to Fig. 5.
No Detectable Physical Association between VGSC and NCX1
One of the potential mechanisms by which VGSC activity could influence PKCα and downstream VEGF signaling is by altering the transmembrane Na+/Ca2+ gradient (e.g. by reversing or slowing down the activity of Na+/Ca2+ exchanger (NCX)). In HUVECs, Na+ loading with monensin would lead to Ca2+ entry through reverse-mode NCX (Ca2+ in, Na+ out) and increased NO production by the Ca2+-sensitive endothelial nitric-oxide synthase (39). Indeed, we found that inhibiting the activity of the NCX resulted in a marked decrease in the VEGF-induced phosphorylation of ERK1/2, and preincubation with TTX consistently resulted in a further decrease (Fig. 6C). The possibility that NCX1, the main NCX isoform present in HUVECs (40), and VGSC were physically associated was investigated by reciprocal immunoprecipitation. Lysates from HUVECs grown in control culture conditions or stimulated with VEGF were immunoprecipitated with a pan-VGSC or a NCX1 antibody. However, no interaction between NCX1 and VGSC could be detected in Western blots (supplemental Figs. S1 (A and B) and S2 (A and B)). VGSC and NCX do not need to be physically associated, however, in order for the channel to influence the exchanger's function. Na+ could diffuse and increase the transmembrane [Na+] gradient, thus potentially reversing the NCX, if in close proximity. By immunocytochemistry, diffuse NCX1 immunoreactivity was observed in the cytosol of HUVECs cultured in standard medium, whereas the pan-anti-VGSC antibody stained the plasma membrane and the cytosol. Under these conditions, we did not detect an obvious co-localization of NCX1 and VGSC (supplemental Fig. S2, C–H).
VGSC Activity Enhances VEGF-induced Intracellular Ca2+ Transients and Suppresses Membrane Depolarization
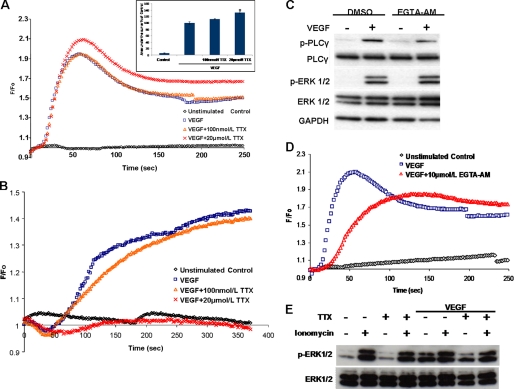
Because VEGF-induced ERK1/2 activation in ECs is a Ca2+-sensitive process (Fig. 6C) (37), we investigated whether VGSC activity would influence the well documented VEGF-induced increase in bulk [Ca2+]i (8). VEGF stimulation of HUVECs, loaded with the Ca2+ indicator Fluo-4NW, resulted in a rapid increase in [Ca2+]i. Preincubation of the cells with 100 nmol/liter TTX had minimal effect on the VEGF-induced Ca2+ response. Surprisingly, 20 μmol/liter TTX (the concentration most effective at inhibiting VEGF-induced ERK1/2 activation) augmented the VEGF-induced rise in [Ca2+]i, quantified as the area under the curve of the Ca2+ traces (132 ± 2.9% versus 100 ± 3.5%; Fig. 8A). Membrane potential can also modulate changes in [Ca2+]i in various cell types, including ECs (41). We also determined, therefore, whether VEGF would affect the Vm of HUVECs. VEGF transiently hyperpolarized the membrane, followed by a sustained depolarization (Fig. 8B), in agreement with a previous study (8). Preincubation of HUVECs with 100 nmol/liter TTX prior to VEGF application had no effect on the Vm changes. However, 20 μmol/liter TTX increased the duration of the VEGF-induced initial hyperpolarization and completely abolished the later depolarization (Fig. 8B). Additionally, voltage-gated Ca2+ channel (VGCC) activity has been reported in ECs (22), so VGSC-dependent membrane depolarization could lead to Ca2+ influx through VGCC activity. However, preincubation of HUVECs with nifedipine, a potent blocker of VGCCs, had no apparent effect on the VEGF-induced ERK1/2 activation (supplemental Fig. S3). In mast cells, ERK1/2 activation in response to antigen involves intracellular Ca2+ microdomains (42), so in order to investigate the possible contribution of such microdomains to the VEGF-induced ERK1/2 activation, we used differential [Ca2+]i chelation strategies. The “slow” membrane-permeable Ca2+ chelator EGTA-AM, which due to its slow Ca2+-binding kinetics does not affect local Ca2+ signals (43), showed no obvious inhibition of the VEGF-induced ERK1/2 activation (Fig. 8C) despite altering the kinetics of the Ca2+ response (Fig. 8D). On the other hand, the “fast” Ca2+ chelator BAPTA, which would interfere with Ca2+ microdomains (43), inhibited the VEGF-induced ERK1/2 activation (Fig. 6C). Finally, we investigated whether the application of the Ca2+ ionophore ionomycin rescues ERK1/2 phosphorylation in the presence of TTX (10 μmol/liter). Application of ionomycin (1 μmol/liter) for 11 min resulted in a marked increase of phospho-ERK1/2, and preincubation of HUVECs with TTX had no apparent effect (Fig. 8E). On the other hand, TTX attenuated, as expected, VEGF-induced ERK1/2 phosphorylation (50 ng/ml for 10 min). Conversely, the effect of TTX was alleviated when ionomycin was added 1 min prior to VEGF application (Fig. 8E). Thus, VGSC activity modulates ERK1/2 phosphorylation by influencing transmembrane Ca2+ fluxes, and the effect of VGSC inhibition on ERK1/2 is rescued if Ca2+ influx is initiated by the ionophore ionomycin.
FIGURE 8.
Micromolar TTX enhances VEGF-induced Ca2+ transients and abolishes VEGF-induced membrane depolarization. A, time courses of Ca2+-sensitive Fluo-4NW fluorescence during VEGF stimulation of HUVECs. HUVECs were stimulated with VEGF at time 0. Shown are unstimulated control (black), VEGF-stimulated control (blue), and 100 nmol/liter or 20 μmol/liter TTX plus VEGF (orange and red, respectively) (n = 3, in triplicate). The area under the curve of the Ca2+ trace was calculated for each condition. The area under the curve of the VEGF-stimulated control was set to 100%. Bars, mean ± S.E. (n = 3 in triplicate). *, p < 0.05 versus VEGF-stimulated control. B, representative traces of membrane potential-sensitive DiBAC4(3) fluorescence recorded during VEGF stimulation of HUVECs (details and key are as in A) (n = 3, in triplicate). C, HUVECs were incubated with EGTA-AM (10 μm, 20 min) prior to VEGF stimulation. Phospho-ERK1/2 and phospho-PLCγ levels were analyzed as in Fig. 5. D, representative time courses of Ca2+-sensitive Fluo-4NW fluorescence recorded during VEGF stimulation of HUVECs loaded with EGTA-AM as in C. Traces of unstimulated (black), VEGF-stimulated (blue), and EGTA-AM-loaded plus VEGF (red) HUVECs were obtained as in A (n = 3, in triplicate). E, serum-starved HUVECs were stimulated with ionomycin (1 μm), VEGF (50 ng/ml), or both in the presence or absence of TTX (10 μm), and ERK1/2 activation was determined with Western blot as described in the legend to Fig. 5.
DISCUSSION
Although VGSCs were reported earlier to be expressed in some ECs (11–14), the exact subtype(s) present, their functional significance, and their mode(s) of action have not been fully investigated. Here, using the HUVEC model of ECs, we have elucidated quantitatively, for the first time, the VGSC subtypes expressed, determined their roles in different aspects of angiogenesis in vitro, and provided evidence that short term VGSC activity significantly influences VEGF-stimulated signaling. Specifically, we conclude the following 1) TTX-S (Nav1.7) and TTX-R (Nav1.5) VGSCs are expressed in HUVECs, as determined consistently by both PCR and TTX sensitivity in electrophysiological recordings. 2) VGSC proteins are also expressed in HUVECs and mammalian endothelia in vivo. 3) VGSC activity enhances VEGF-stimulated cellular proliferation, chemotaxis, and tubulogenesis but reduces basal adhesion. Both TTX-S and TTX-R VGSCs differentially contribute to these angiogenic processes. 4) VGSC activity potentiates VEGF-induced ERK1/2 activation by attenuating membrane depolarization, altering [Ca2+]i kinetics and PKCα activity.
VGSC Expression Profile
We determined that the major subtypes of VGSCs expressed in HUVECs are Nav1.7 (TTX-S) and Nav1.5 (TTX-R), representing ∼9 and 91% of the total “functional” VGSCα mRNAs, respectively. The VGSC mRNA expression profile is consistent with our observed electrophysiology (TTX sensitivity) as well as a previous report of TTX-R currents in HUVECs (11) and cardiac microvascular ECs (12). Furthermore, in agreement with previous in vitro studies (11, 14), we demonstrated by immunohistochemistry of intact mouse aortic endothelium that VGSC protein is also expressed in ECs in vivo. We report here a peak VGSC current density of 3 pA/picofarads. This is lower than the values reported for human neuroblastoma (18) or breast cancer cell lines (17) but similar to a metastatic colon cancer cell line (18). Nonetheless, as in the case of the cancer cells, the VGSC activity was sufficient to exert significant control on HUVEC signaling and angiogenesis in vitro (Figs. 3 and 5).
Role of VGSC in Angiogenic Functional Assays
The angiogenic cascade can be dissected into a number of elementary cellular activities, including proliferation, chemotaxis, substrate adhesion, and tubular differentiation in response to angiogenic factors (25). Interestingly, there was a difference in concentrations of TTX required to block these complementary aspects of angiogenesis, implying differential control by TTX-S and TTX-R VGSCs. VEGF-induced chemotaxis was inhibited by nmol/liter TTX, consistent with TTX-S (Nav1.7) activity, whereas substrate adhesion, tubulogenesis, and proliferation were modulated by μmol/liter TTX, suggesting involvement of TTX-R (Nav1.5) VGSCs. Indeed, chemotaxis was inhibited by knockdown of Nav1.7 (but not Nav1.5), and TTX had no further effect (Fig. 4). Thus, a “minor” VGSC in terms of mRNA expression (Nav1.7) might still play a major functional role (i.e. VEGF-induced chemotaxis). A similar phenomenon was described in the heart, where TTX-S VGSCs, although minor relative to the main cardiac Nav1.5 expression, nevertheless contributed significantly to control of heart rate (44). On the other hand, tubulogenesis was controlled by the dominant Nav1.5 activity, as in the case of breast cancer cell invasiveness (17).
Studies of Intracellular Signaling Pathways
We investigated the impact of VGSC activity on VEGF-induced ERK1/2 activation because this pathway is implicated in both EC proliferation (38) and tubular differentiation (34), which we showed to be influenced by TTX-R/Nav1.5 activity. Indeed, micromolar TTX and Nav1.5 RNAi decreased ERK1/2 activation, consistent with a role for Nav1.5 in VEGF-induced ERK1/2 phosphorylation (Fig. 5). The classical PKCα isoform is central to VEGF-induced ERK1/2 activation, EC proliferation (38), migration, and tubular differentiation (45), making it a plausible target of VGSC activity. Indeed, TTX inhibited two PKCα-dependent processes: VEGF-induced PKCμ/PKD phosphorylation at Ser-744/748 and B-Raf activity (35, 38). Also, micromolar TTX decreased the translocation of PKCα to cell membranes. On the other hand, a PKC inhibitor failed to suppress EGF-induced ERK1/2 phosphorylation (Fig. 7D), a process not influenced by VGSC activity (Fig. 5C). Moreover, ERK1/2 activation by phorbol 12-myristate 13-acetate, an activator of PKCs, was not inhibited by TTX (Fig. 7E). Taken together, these findings provide evidence that VGSC influences PKC activity.
Studies of [Ca2+]i and Vm
Because classical PKC isoforms require Ca2+ in order to translocate to the plasma membrane and to become fully activated (46), we investigated the impact of VGSC activity on the intracellular Ca2+ level in HUVECs. 20 μmol/liter TTX significantly enhanced VEGF-induced Ca2+ traces (Fig. 8A). This was unexpected because μmol/liter TTX inhibited VEGF-induced ERK1/2 and PKCα activation (Figs. 5 and 7), both Ca2+-sensitive processes. The effect of VGSC activity on [Ca2+]i transients can be explained in conjunction with the results of the Vm measurements (Fig. 8B). Membrane hyperpolarization would increase the driving force for Ca2+ influx, whereas depolarization would have the opposite effect (41). As in the previous study investigating VEGF and membrane potential (8), an initial transient hyperpolarization followed by a sustained membrane depolarization was seen (Fig. 8B). The VEGF-induced hyperpolarization was attributed to the activation of Ca2+-activated K+ channels (8). 100 nmol/liter TTX, presumably inhibiting mainly Nav1.7, had no obvious effect on the Ca2+ or Vm transients. On the other hand, 20 μmol/liter TTX (blocking Nav1.5 activity) increased both the phasic and the plateau phases of the Ca2+ response while prolonging the initial membrane hyperpolarization and abolishing the subsequent tonic depolarization (Fig. 8, A and B). The Vm of ECs is proposed to be determined by the competitive effect of hyperpolarizing K+ channels and depolarizing Cl− channels (22). VEGF-induced HUVEC depolarization is reportedly controlled by Ca2+-sensitive and volume-regulated Cl− channels (8). However, the compounds used to block the chloride channels (clomiphene at 10 μmol/liter and tamoxifen at 10 μmol/liter) (8) could also inhibit Nav1.5 activity in heart (47) and TTX-S VGSCs in glial cells (48). Therefore, we could postulate that the reported effects (8) may be due, at least in part, to inhibition of Nav1.5 in agreement with our findings. Additionally, Na+ fluxes were recently described as a crucial regulator of HUVEC membrane potential, although the ion channel(s) involved was not determined (49). The finding that the Ca2+ ionophore ionomycin alleviated the effect of TTX on VEGF-induced ERK1/2 phosphorylation (Fig. 8E) further supports our conclusion that VGSC activity modulates Ca2+ influx from the extracellular milieu, a process crucial for ERK1/2 phosphorylation.
Ca2+ Microenvironments and VEGF Signaling
Although the effect of VGSC inhibition on Vm would satisfactorily explain the observed increase in bulk [Ca2+]i, the question remains as to why VEGF-induced ERK1/2 activation, a Ca2+-sensitive process, was attenuated. We postulated that local Ca2+ gradients (microdomains) rather than global [Ca2+]i might regulate this response. Indeed, EGTA, a slow Ca2+ chelator that would not affect Ca2+-sensitive processes within ∼100 nm of the Ca2+ source (43), did not suppress the VEGF-induced ERK1/2 activation despite altering the shape of the Ca2+ transient (Fig. 8, C and D), whereas the same concentration of BAPTA completely abolished the response (Fig. 6C). Hence, we can argue that although Nav1.5 inhibition increases bulk [Ca2+]i in response to VEGF, local [Ca2+] in close proximity to the plasma membrane may be reduced. This phenomenon could be of more general significance because Ca2+ microdomains have also been implicated in IgE-induced ERK1/2 activation via PKC in mast cells (42).
Source(s) of Ca2+ and Role of NCX
Finally, we investigated some of the possible mechanisms that could differentially regulate submembrane [Ca2+]i in conjunction with VGSCs. Ca2+ influx through Ca2+ release-activated Ca2+ (CRAC) channels is required for PKC and subsequent ERK1/2 activation in mast cells (42) and has been implicated in EC proliferation (52). The inhibitor of non-selective cation channels, SKF96365, that suppressed the VEGF-induced ERK1/2 phosphorylation (Fig. 6C), would inhibit CRAC-like currents, among other ionic activities (53). However, at present, there is no evidence implicating the membrane potential or VGSCs in regulation of CRAC currents. Another possibility is that VGSC-induced depolarization could affect the activity of VGCCs in HUVECs (21), but this was not supported by pharmacological inhibition (supplemental Fig. S3). In mouse brain, VGSC mediates Ca2+ entry through reverse-mode NCX (Na+ out, Ca2+ in) presumably through localized increase of [Na+]i (50). Although we found some evidence for NCX involvement in VEGF-induced ERK1/2 activation (Fig. 6C), a mechanism depending on VGSC-mediated [Na+] increase would theoretically require co-localization of the VGSC and NCX (50). However, such a “physical” association could not be demonstrated under the conditions employed in the present study (supplemental Fig. S1). On the other hand, NCX is electrogenic and can reverse its direction of activity, depending on the membrane potential (51). Membrane depolarization would favor reverse-mode NCX (Na+ out, Ca2+ in) in HUVECs (49). Moreover, recently, in cardiomyocytes, voltage-sensitive Na+ currents were shown to augment Ca2+ transients by activating reverse Na+-Ca2+ exchange. This increase in [Ca2+]i was attributed to a combination of localized rise in [Na+]o and plasma membrane depolarization and was not observed in cardiomyocytes derived from NCX cardiac specific knock-out mice (54). Consequently, VGSC activity could modulate transmembrane Ca2+ by influencing the membrane potential and thus the activity of NCX. Such a role for NCX in the VEGF-induced ERK1/2 activation and EC angiogenesis remains to be established and is currently under investigation.
Reciprocal Signaling between PKC and/or ERK1/2 and VGSCs in HUVECs?
PKC phosphorylates and suppresses Nav1.5 activity (55). Interestingly, activated ERK1/2 was reported to co-localize with multiple neuronal VGSC isoforms in neuromas (56), and more recently, ERK1/2 was shown to directly phosphorylate and alter the gating properties of Nav1.7 in dorsal root ganglion neurons (57). However, phosphorylation of Nav1.5 by ERK1/2 has not been reported yet, and the regulation of VGSC activity in the endothelium by post-translational modifications has not been investigated. Our results suggest that a reciprocal feedback loop could exist between VGSC, PKC, and ERK1/2 activities, and further studies are needed in order to investigate this intriguing possibility.
Conclusions
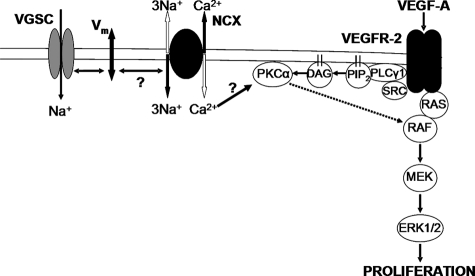
A limited number of studies have investigated VGSC signaling in non-excitable cells. Reported activities include enhancement of extracellular proteolysis (58), decreased protein kinase A activity in metastatic cancer cells (59), and Ca2+ release from the mitochondria of mast cells (60). Our study adds a novel dimension to this emerging field: a key role for VGSC activity and membrane potential in the regulation of VEGF-induced EC proliferation via PKCα and ERK1/2 (Fig. 9). To our knowledge, this is the first evidence for the control of Vm in non-excitable cells by VGSCs in response to physiological stimuli in vitro. Whether the VGSC contribution to agonist-induced ERK1/2 activation is specific to VEGF and ECs remains to be elucidated.
FIGURE 9.
A mechanistic model for the role of VGSC activity in VEGF HUVEC signaling. Heavy lines show direct interactions. Dashed lines show indirect interactions. Voltage-gated sodium channel (VGSC) activity is proposed to influence the [Ca2+]i close to the plasma membrane by modifying the membrane potential and subsequently reverse-mode NCX exchange (Ca2+ in, Na+ out). DAG, diacylglycerol; PIP2, phosphatidylinositol 4,5-bisphosphate.
Supplementary Material
Acknowledgments
We thank the members of the Tumour Biology and Metastasis Team, Cancer Research UK Cancer Therapeutics Unit at the Institute of Cancer Research, and the members of the Neuroscience Solutions to Cancer group at Imperial College London for many useful discussions and comments on the manuscript.
This work was supported by British Heart Foundation Studentship FS/06/22 (to P. A.). Additional support was provided by the Pro Cancer Research Fund. The NIHR Biomedical Research Centre was supported by the National Health Service.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. S1–S3.
- EC
- endothelial cell
- HUVEC
- human umbilical vein endothelial cell
- PLC
- phospholipase C
- TTX
- tetrodotoxin
- TTX-R and TTX-S
- TTX-resistant and -sensitive, respectively
- VGCC
- voltage-gated calcium channel
- VGSC
- voltage-gated sodium channels
- Vm
- membrane potential
- BAPTA
- 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid
- AM
- acetoxymethyl ester
- PKD
- protein kinase D
- NCX
- Na+/Ca2+ exchanger
- CRAC
- Ca2+ release-activated Ca2+
- l-NAME
- Nω-nitro-l-arginine methyl ester
- PMA
- phorbol 12-myristate 13-acetate
- DiBaC4(3)
- bis-(1,3-dibutylbarbituric acid) trimethine oxonol.
REFERENCES
- 1. Carmeliet P. (2005) Nature 438, 932–936 [DOI] [PubMed] [Google Scholar]
- 2. Chen C. H., Walterscheid J. P. (2006) Circ. Res. 99, 787–789 [DOI] [PubMed] [Google Scholar]
- 3. Isner J. M. (2002) Nature 415, 234–239 [DOI] [PubMed] [Google Scholar]
- 4. Semenza G. L. (2006) Circ. Res. 98, 1115–1116 [DOI] [PubMed] [Google Scholar]
- 5. Olsson A. K., Dimberg A., Kreuger J., Claesson-Welsh L. (2006) Nat. Rev. Mol. Cell Biol. 7, 359–371 [DOI] [PubMed] [Google Scholar]
- 6. Takahashi T., Yamaguchi S., Chida K., Shibuya M. (2001) EMBO J. 20, 2768–2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gerber H. P., McMurtrey A., Kowalski J., Yan M., Keyt B. A., Dixit V., Ferrara N. (1998) J. Biol. Chem. 273, 30336–30343 [DOI] [PubMed] [Google Scholar]
- 8. Dawson N. S., Zawieja D. C., Wu M. H., Granger H. J. (2006) FASEB J. 20, 991–993 [DOI] [PubMed] [Google Scholar]
- 9. Heath V. L., Bicknell R. (2009) Nat. Rev. Clin. Oncol. 6, 395–404 [DOI] [PubMed] [Google Scholar]
- 10. Catterall W. A. (2000) Neuron 26, 13–25 [DOI] [PubMed] [Google Scholar]
- 11. Gordienko D. V., Tsukahara H. (1994) Pflugers. Arch. 428, 91–93 [DOI] [PubMed] [Google Scholar]
- 12. Walsh K. B., Wolf M. B., Fan J. (1998) Am. J. Physiol. 274, H506–H512 [DOI] [PubMed] [Google Scholar]
- 13. Gosling M., Harley S. L., Turner R. J., Carey N., Powell J. T. (1998) J. Biol. Chem. 273, 21084–21090 [DOI] [PubMed] [Google Scholar]
- 14. Traub O., Ishida T., Ishida M., Tupper J. C., Berk B. C. (1999) J. Biol. Chem. 274, 20144–20150 [DOI] [PubMed] [Google Scholar]
- 15. Diss J. K., Archer S. N., Hirano J., Fraser S. P., Djamgoz M. B. (2001) Prostate 48, 165–178 [DOI] [PubMed] [Google Scholar]
- 16. Fraser S. P., Salvador V., Manning E. A., Mizal J., Altun S., Raza M., Berridge R. J., Djamgoz M. B. (2003) J. Cell. Physiol. 195, 479–487 [DOI] [PubMed] [Google Scholar]
- 17. Brackenbury W. J., Chioni A. M., Diss J. K., Djamgoz M. B. (2007) Breast Cancer Res. Treat. 101, 149–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. House C. D., Vaske C. J., Schwartz A. M., Obias V., Frank B., Luu T., Sarvazyan N., Irby R., Strausberg R. L., Hales T. G., Stuart J. M., Lee N. H. (2010) Cancer Res. 70, 6957–6967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Eccles S. A. (2004) Int. J. Dev. Biol. 48, 583–598 [DOI] [PubMed] [Google Scholar]
- 20. Larrivée B., Freitas C., Suchting S., Brunet I., Eichmann A. (2009) Circ. Res. 104, 428–441 [DOI] [PubMed] [Google Scholar]
- 21. Chioni A. M., Brackenbury W. J., Calhoun J. D., Isom L. L., Djamgoz M. B. (2009) Int. J. Biochem. Cell Biol. 41, 1216–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nilius B., Droogmans G. (2001) Physiol. Rev. 81, 1415–1459 [DOI] [PubMed] [Google Scholar]
- 23. Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 24. Jones N. P., Peak J., Brader S., Eccles S. A., Katan M. (2005) J. Cell Sci. 118, 2695–2706 [DOI] [PubMed] [Google Scholar]
- 25. Eccles S. A., Court W., Patterson L., Sanderson S. (2009) Methods. Mol. Biol. 467, 159–181 [DOI] [PubMed] [Google Scholar]
- 26. Palmer C. P., Mycielska M. E., Burcu H., Osman K., Collins T., Beckerman R., Perrett R., Johnson H., Aydar E., Djamgoz M. B. (2008) Eur. Biophys. J. 37, 359–368 [DOI] [PubMed] [Google Scholar]
- 27. Sanderson S., Valenti M., Gowan S., Patterson L., Ahmad Z., Workman P., Eccles S. A. (2006) Mol. Cancer Ther. 5, 522–532 [DOI] [PubMed] [Google Scholar]
- 28. Xia P., Aiello L. P., Ishii H., Jiang Z. Y., Park D. J., Robinson G. S., Takagi H., Newsome W. P., Jirousek M. R., King G. L. (1996) J. Clin. Invest. 98, 2018–2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wilsher N. E., Court W. J., Ruddle R., Newbatt Y. M., Aherne W., Sheldrake P. W., Jones N. P., Katan M., Eccles S. A., Raynaud F. I. (2007) Drug. Metab. Dispos. 35, 1017–1022 [DOI] [PubMed] [Google Scholar]
- 30. Epps D. E., Wolfe M. L., Groppi V. (1994) Chem. Phys. Lipids. 69, 137–150 [DOI] [PubMed] [Google Scholar]
- 31. Lamalice L., Le Boeuf F., Huot J. (2007) Circ. Res. 100, 782–794 [DOI] [PubMed] [Google Scholar]
- 32. Zeng H., Sanyal S., Mukhopadhyay D. (2001) J. Biol. Chem. 276, 32714–32719 [DOI] [PubMed] [Google Scholar]
- 33. Meadows K. N., Bryant P., Pumiglia K. (2001) J. Biol. Chem. 276, 49289–49298 [DOI] [PubMed] [Google Scholar]
- 34. Mavria G., Vercoulen Y., Yeo M., Paterson H., Karasarides M., Marais R., Bird D., Marshall C. J. (2006) Cancer Cell 9, 33–44 [DOI] [PubMed] [Google Scholar]
- 35. Takahashi T., Ueno H., Shibuya M. (1999) Oncogene 18, 2221–2230 [DOI] [PubMed] [Google Scholar]
- 36. Doanes A. M., Hegland D. D., Sethi R., Kovesdi I., Bruder J. T., Finkel T. (1999) Biochem. Biophys. Res. Commun. 255, 545–548 [DOI] [PubMed] [Google Scholar]
- 37. Gliki G., Abu-Ghazaleh R., Jezequel S., Wheeler-Jones C., Zachary I. (2001) Biochem. J. 353, 503–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wong C., Jin Z. G. (2005) J. Biol. Chem. 280, 33262–33269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Teubl M., Groschner K., Kohlwein S. D., Mayer B., Schmidt K. (1999) J. Biol. Chem. 274, 29529–29535 [DOI] [PubMed] [Google Scholar]
- 40. Szewczyk M. M., Davis K. A., Samson S. E., Simpson F., Rangachari P. K., Grover A. K. (2007) J. Cell Mol. Med. 11, 129–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. He P., Curry F. E. (1994) J. Appl. Physiol. 76, 2288–2297 [DOI] [PubMed] [Google Scholar]
- 42. Ng S. W., di Capite J., Singaravelu K., Parekh A. B. (2008) J. Biol. Chem. 283, 31348–31355 [DOI] [PubMed] [Google Scholar]
- 43. Parekh A. B. (2008) J. Physiol. 586, 3043–3054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Maier S. K., Westenbroek R. E., Yamanushi T. T., Dobrzynski H., Boyett M. R., Catterall W. A., Scheuer T. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 3507–3512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang A., Nomura M., Patan S., Ware J. A. (2002) Circ. Res. 90, 609–616 [DOI] [PubMed] [Google Scholar]
- 46. Oancea E., Meyer T. (1998) Cell 95, 307–318 [DOI] [PubMed] [Google Scholar]
- 47. Borg J. J., Yuill K. H., Hancox J. C., Spencer I. C., Kozlowski R. Z. (2002) J. Pharmacol. Exp. Ther. 303, 282–292 [DOI] [PubMed] [Google Scholar]
- 48. Smitherman K. A., Sontheimer H. (2001) J. Membr. Biol. 181, 125–135 [DOI] [PubMed] [Google Scholar]
- 49. Liang G. H., Kim M. Y., Park S., Kim J. A., Choi S., Suh S. H. (2008) Pflugers. Arch. 457, 67–75 [DOI] [PubMed] [Google Scholar]
- 50. Platel J. C., Boisseau S., Dupuis A., Brocard J., Poupard A., Savasta M., Villaz M., Albrieux M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 19174–19179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shigekawa M., Iwamoto T. (2001) Circ. Res. 88, 864–876 [DOI] [PubMed] [Google Scholar]
- 52. Abdullaev I. F., Bisaillon J. M., Potier M., Gonzalez J. C., Motiani R. K., Trebak M. (2008) Circ. Res. 103, 1289–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Putney J. W., Jr. (2001) Mol. Interv. 1, 84–94 [PubMed] [Google Scholar]
- 54. Larbig R., Torres N., Bridge J. H., Goldhaber J. I., Philipson K. D. (2010) J. Physiol. 588, 3267–3276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Qu Y., Rogers J. C., Tanada T. N., Catterall W. A., Scheuer T. (1996) J. Gen. Physiol. 108, 375–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Black J. A., Nikolajsen L., Kroner K., Jensen T. S., Waxman S. G. (2008) Ann. Neurol. 64, 644–653 [DOI] [PubMed] [Google Scholar]
- 57. Stamboulian S., Choi J. S., Ahn H. S., Chang Y. W., Tyrrell L., Black J. A., Waxman S. G., Dib-Hajj S. D. (2010) J. Neurosci. 30, 1637–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gillet L., Roger S., Besson P., Lecaille F., Gore J., Bougnoux P., Lalmanach G., Le Guennec J. Y. (2009) J. Biol. Chem. 284, 8680–8691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Brackenbury W. J., Djamgoz M. B. (2006) J. Physiol. 573, 343–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Carrithers M. D., Chatterjee G., Carrithers L. M., Offoha R., Iheagwara U., Rahner C., Graham M., Waxman S. G. (2009) J. Biol. Chem. 284, 8114–8126 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.