Abstract
The mammalian HRD1-SEL1L complex provides a scaffold for endoplasmic reticulum (ER)-associated degradation (ERAD), thereby connecting luminal substrates for ubiquitination at the cytoplasmic surface after their retrotranslocation through the endoplasmic reticulum membrane. In this study the stability of the mammalian HRD1-SEL1L complex was assessed by performing siRNA-mediated knockdown of each of its components. Although endogenous SEL1L is a long-lived protein, the half-life of SEL1L was greatly reduced when HRD1 is silenced. Conversely, transiently expressed SEL1L was rapidly degraded but was stabilized when HRD1 was coexpressed. This was in contrast to the yeast Hrd1p-Hrd3p, where Hrd1p is destabilized by the depletion of Hrd3p, the SEL1L homologue. Endogenous HRD1-SEL1L formed a large ERAD complex (Complex I) associating with numerous ERAD components including ERAD lectin OS-9, membrane-spanning Derlin-1/2, VIMP, and Herp, whereas transiently expressed HRD1-SEL1L formed a smaller complex (Complex II) that was associated with OS-9 but not with Derlin-1/2, VIMP, or Herp. Despite its lack of stable association with the latter components, Complex II supported the retrotranslocation and degradation of model ERAD substrates α1-antitrypsin null Hong-Kong (NHK) and its variant NHK-QQQ lacking the N-glycosylation sites. NHK-QQQ was rapidly degraded when SEL1L was transiently expressed, whereas the simultaneous transfection of HRD1 diminished that effect. SEL1L unassociated with HRD1 was degraded by the ubiquitin-proteasome pathway, which suggests the involvement of a ubiquitin-ligase other than HRD1 in the rapid degradation of both SEL1L and NHK-QQQ. These results indicate that the regulation of the stability and assembly of the HRD1-SEL1L complex is critical to optimize the degradation kinetics of ERAD substrates.
Keywords: E3 Ubiquitin Ligase, Endoplasmic Reticulum (ER), Mammal, Protein Degradation, Protein-Protein Interactions, ER Quality Control, ER-associated Degradation (ERAD), HRD1-SEL1L Ubiquitin Ligase Complex
Introduction
Misfolded or unassembled proteins that accumulate in the endoplasmic reticulum (ER)3 are degraded by cytosolic proteasomes through ER-associated degradation (ERAD) (1, 2). During the process of substrate retrotranslocation from the ER, proteins are ubiquitinated by ubiquitin ligases embedded in the ER membrane (3–5). Hrd1p/Der3p ubiquitin-ligase, which contains a cytosolic RING-H2 finger E3 domain, ubiquitinates ERAD substrates in Saccharomyces cerevisiae (6, 7).
Hrd1p is an unstable protein with six N-terminal transmembrane segments that is stabilized by formation of a stoichiometric complex with Hrd3p, a type-I transmembrane protein with a large luminal domain containing SEL1 repeats (8, 9). Mammals have two yeast Hrd1p homologues: HRD1/synoviolin (10, 11) and gp78/AMFR (12). SEL1L, the homologue of Hrd3p (13, 14), associates stoichiometrically with HRD1 (13, 15) but not with gp78 (16). HRD1-SEL1L ubiquitin-ligase forms a large complex in the ER membrane by physically interacting with Der1-like proteins 1 and 2 (Derlin-1/2), valosine-containing protein (VCP)/p97-interacting membrane protein (VIMP), p97/VCP, and Herp (15, 17).
Derlin is a human homologue of yeast Der1p containing four transmembrane regions. Mammals have three such homologues, Derlin-1, -2, and -3 (18, 19), and may be a part of the ER membrane retrotranslocation channel. The small membrane protein VIMP connects p97/VCP to Derlins (15, 19). p97/VCP is an AAA-ATPase involved in the extraction of ERAD clients (20). Herp exposes its N-terminal ubiquitin-like and C-terminal domains on the ER cytosolic surface (21) and interacts with HRD1 (17, 22).
Formation of a large complex containing Hrd1p-Hrd3p and Der1p has been reported in S. cerevisiae (23), as has the association of the ERAD lectin Yos9p and ER chaperone Kar2p on the luminal side of the complex (24–26). A recent study identified the analogous membrane ERAD complex and regulation mechanism of glycoprotein quality control in yeast and mammals (for review, see Refs. 27–30). Mammalian homologues of yeast Yos9p (OS-9 and XTP3-B) (16, 31) and the ubiquitin conjugating enzyme UBC6e (32) were shown to bind to the ERAD complex.
ERAD substrate degradation, which is well characterized in yeast, is mediated by distinct pathways containing different ubiquitin ligases (33, 34). ERAD-C substrates with misfolded cytosolic domains are degraded by Doa10p (35, 36), whereas ERAD-L proteins with misfolded ER-luminal domains require Hrd1p (37, 38). ERAD-M proteins with misfolded intramembrane domains also use Hrd1p for degradation, but the complex required is different, and Der1p and Usa1p are dispensable (26). In mammals, the required ubiquitin ligases and E3-containing complexes for each ERAD client are less clear. Mammalian ERAD-LS substrates, defined as soluble polypeptides with luminal lesions, were recently found to depend on HRD1-SEL1L and the ERAD lectins OS-9/XTP3-B (39). However, pathways for various ERAD clients remain to be elucidated.
To assess the functions of SEL1L and HRD1 in ERAD, each was silenced by RNA interference (RNAi). This revealed that SEL1L is destabilized when HRD1 is silenced. This mechanism is different from that of the yeast homologue Hrd1p-Hrd3p complex, where association with Hrd3p stabilizes the short-lived Hrd1p (8, 9). With the endogenously assembled large complex (Complex I), transiently expressed HRD1-SEL1L forms a smaller-sized complex (Complex II). While Complex II does not cosediment with the complex containing Derlin-1/2, Herp, and VIMP, Complex II does support the retrotranslocation and degradation of model ERAD substrates of misfolded glycoprotein NHK and non-glycosylated NHK-QQQ (16, 40, 41). While NHK-QQQ is degraded more rapidly when SEL1L is transiently expressed, the simultaneous transfection of HRD1 diminishes this effect. Accordingly, a model is proposed whereby formation and stabilization of HRD1-SEL1L ERAD complex regulates mammalian ERAD.
EXPERIMENTAL PROCEDURES
Cell Culture, Transfection, and Cell Extracts
HEK293 and HeLa cells were cultured in DMEM supplemented with 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin. Plasmids were transfected using FuGENE 6 (Roche Applied Science) or Lipofectamine 2000 transfection reagent (Invitrogen), according to the protocol supplemented by the manufacturer. The siRNA (30 nm) was transfected using Lipofectamine RNAiMAX (Invitrogen). Cells were harvested ∼24 h after plasmid transfection and 48 h after siRNA treatment. Cells were lysed in a buffer (150 mm NaCl, 50 mm Tris-HCl, pH 7.5) containing 1% Nonidet P-40 or 3% digitonin supplemented with protease inhibitors (0.2 mm 4-(2-aminoethyl)benzenesulfonyl fluoride, 2 mm N-ethylmaleimide, 1 μg/ml leupeptin, 1 μg/ml pepstatin). Digitonin was used to maintain HRD1-SEL1L complexes formed in the ER membrane (16, 32). After centrifugation at 12,000 × g for 20 min, the supernatant was used for further analysis.
Antibodies
A new lot of anti-SEL1L antibody was generated as described previously (16). Briefly, rabbits were immunized with the same peptide sequence conjugated to keyhole limpet hemocyanin as the antigen. This new lot was used for both Western blotting and immunoprecipitation. The mouse monoclonal antibody for SEL1L (LifeSpan Bioscience) was also used for Western blotting where indicated.
Mouse monoclonal anti-HRD1/synoviolin antibody was kindly provided by Dr. Toshiro Nakajima (St. Marianna University School of Medicine, Japan), and rabbit polyclonal anti-Herp antibody was a generous gift from Dr. Koichi Kokame (National Cerebral and Cardiovascular Center, Japan). The following antibodies were purchased from the companies indicated: rabbit polyclonal anti-OS-9 (Proteintec Group Inc.), rabbit anti-α1 antitrypsin (α1AT) (DAKO), rabbit anti-Derlin-1 and -2 (MBL), rabbit polyclonal anti-VIMP (Abcam), mouse monoclonal anti-p97 (Affinity Bioreagents), mouse anti-BiP (BD Transduction Laboratories), rabbit anti-c-myc (Santa Cruz Biotechnology), mouse anti-actin (Chemicon), horseradish peroxidase-conjugated anti-rabbit IgG (BTI), horseradish peroxidase-conjugated anti-mouse IgG (Zymed Laboratories Inc.), and alkaline phosphatase-conjugated anti-mouse IgG (Jackson ImmunoResearch Laboratories).
Plasmid Construction and siRNA Sequences
The plasmids used for transfection were described previously as follows: SEL1L (16), HRD1myc and the HRD1myc mutant lacking ubiquitin-ligase activity (42), NHK (43), and NHK-QQQ (41). StealthTM siRNAs (Invitrogen) were used to silence specific proteins. The targeting sequences to knockdown HRD1 were as follows: HRD1-2 (5′-3′, UGG AGG AGG CAG CAG CAA CAA CUG U) and HRD1-3 (5′-3′, UGU GGU GUC AGG GCA GUC UCU UGG C). The siRNAs used to silence SEL1L (16) and OS-9 (44) were described previously. StealthTM siRNA Negative Control Low GC and Medium GC were used as control nonspecific siRNAs.
Metabolic Labeling and Pulse-Chase Experiments
Cells were metabolically labeled with [35S]methionine/cysteine (Protein-labeling mixture, PerkinElmer Life Sciences) as previously described (44). For pulse-chase experiments, cells were labeled for 15 or 20 min as indicated and chased in normal growth medium for the indicated time. Cell extracts prepared as above were subjected to immunoprecipitation, and the specific signal was quantified after exposure on phospho-imaging plates. Proteasome inhibitor MG132 was added to the medium at a final concentration of 20 μm 4 h before pulse labeling and was present both during the pulse and chase periods.
Immunoprecipitation
Cell extracts prepared as above were mixed with appropriate antibodies and incubated overnight at 4 °C. Immune complexes were collected by Protein A- or Protein G-Sepharose beads and eluted by incubating in Laemmli buffer at 65 °C for 15 min. The eluates were separated by SDS-PAGE and quantified or used for Western blotting.
Western Blotting
Cell lysates separated by SDS-PAGE were blotted on a nitrocellulose or nylon membrane. The specific signals probed by the antibodies were visualized using an ECL kit (GE Healthcare) and were detected by exposure to x-ray films (Fuji Film) or by LAS4000 (GE Healthcare). Specific signals were visualized using alkaline phosphatase-conjugated 2nd antibodies and nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate solution (Sigma).
Sucrose Density Gradient Centrifugation
Cell extracts were applied to a 10–40% linear sucrose density gradient with a cushion of 60% sucrose or to a 10–40% sucrose density gradient generated by Gradient MasterTM (BioComp) without a cushion. Extracts were then centrifuged at 36,000 rpm for 16 h. Each 250-μl fraction collected from the top was adjusted to 1 × Laemmli buffer for separation by SDS-PAGE.
RT-PCR
The total RNA was prepared from cells using an RNA extraction kit (Qiagen). The cDNA was synthesized using 1 μg of total RNA and SuperScript III reverse transcriptase (Invitrogen). Specific DNA fragments were amplified using LA-Taq (Takara). The primers were SEL1L forward (5′-3′, CTC GCT AAC AGG AGG CTC AGT AG), SEL1L reverse (5′-3′, GGT GCC ACT GGC ATG CAT CTG AG), HRD1 forward (5′-3′, GCA TGG CAG TCC TGT ACA TCC), and HRD1 reverse (5′-3′, GCA CCA TCG TCA TCA GGA TGG). The primers for β-actin are as described (45).
RESULTS
siRNA-mediated HRD1 Knockdown Destabilizes SEL1L
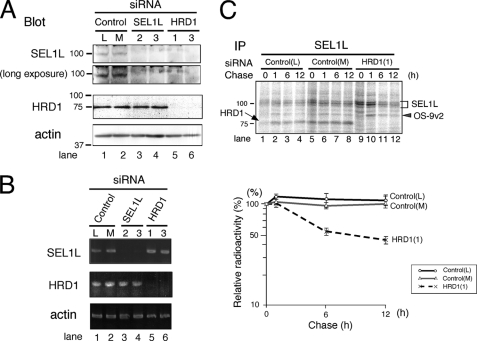
To analyze the function of the HRD1-SEL1L complex in ERAD, SEL1L or HRD1 was knocked down by small interfering RNA (siRNA)-mediated RNAi. Unexpectedly, in contrast to the yeast homologue Hrd1p-Hrd3p complex (8, 9), SEL1L disappeared not only by silencing SEL1L but also by the depletion of HRD1 in HEK 293 cells (Fig. 1A, SEL1L blot). The HRD1 expression level detected by Western blotting was unaffected when SEL1L was silenced (Fig. 1A, HRD1 blot). Similar results were obtained in HeLa cells (supplemental Fig. S1).
FIGURE 1.
SEL1L is destabilized by HRD1 knockdown. A, expression of SEL1L and HRD1 in HEK293 cells treated with siRNA targeted for SEL1L or HRD1 is shown. HEK293 cells were treated with 30 nm of StealthTM siRNA for 48 h. As a negative control, StealthTM siRNA Negative Control Low GC (L) and Medium GC (M) were used. Twenty μg of cell lysates were separated by 10% SDS-PAGE. The expression of each protein was analyzed by Western blotting. Mouse monoclonal anti-SEL1L antibody was used to detect SEL1L (upper panel). The positions of the molecular weight standards are shown on the left. B, detection of SEL1L and HRD1 transcripts by RT-PCR is shown. One mg of total RNA prepared from HEK293 cells treated with siRNA as in A was used. C, pulse-chase analysis of endogenous SEL1L is shown. HEK293 cells treated with siRNA (as in A) were labeled for 20 min and then chased for the time indicated. Cells were extracted in a buffer containing 3% digitonin. The positions of SEL1L, OS-9v2, and HRD1 are indicated by a bracket, gray arrowhead, and arrow, respectively. The positions of the molecular weight standards are indicated on the left. The relative radioactivity of SEL1L was quantified, and the signal intensity at the end of pulse-labeling period was set as 100% (lower panel). Data shown are the mean and corresponding S.E. of three independent experiments. IP, immunoprecipitate.
The specificity of each siRNA was analyzed by RT-PCR. Treatment of cells with two siRNAs targeted for SEL1L depleted the SEL1L mRNA, leaving the HRD1 transcript unaffected (Fig. 1B, lanes 3 and 4). Similarly, two siRNAs targeted for HRD1 down-regulated only the HRD1 transcript and did not influence the SEL1L or β-actin mRNA expression (Fig. 1B, lanes 5 and 6). The specificity of the siRNA for HRD1 was further assessed by Northern blotting analysis (supplemental Fig. S2). The results indicated that elimination of the SEL1L peptide by RNAi-mediated HRD1 silencing occurred post-translationally.
The intracellular half-life of endogenous SEL1L was assessed by pulse-chase experiments, and SEL1L was a stable protein with a half-life of more than 12 h (Fig. 1C, control siRNA). However, when HRD1 was silenced, approximately half of the SEL1L synthesized during the pulse period disappeared in 6 h (Fig. 1C, HRD1 siRNA). Collectively, these results suggest that endogenous SEL1L is a long-lived protein that is stabilized by HRD1.
Silencing SEL1L or HRD1 Alters the Sedimentation Rate of the HRD1-SEL1L-containing ERAD Complex
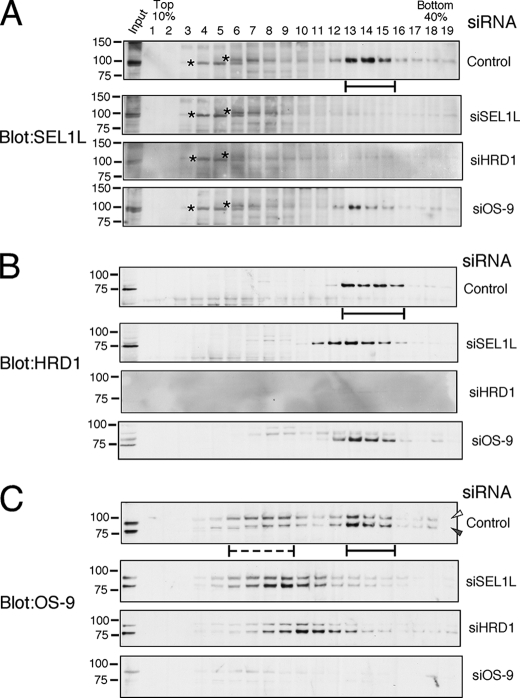
SEL1L and HRD1 are known to form a stoichiometric complex, and a large ERAD complex was generated by the association of various ERAD components including Derlin-1/2/3, Herp, VIMP, p97/VCP, OS-9, and so on (see Fig. 7, Complex I). Therefore, the sedimentation rate of each component of the large HRD1-SEL1L-containing ERAD complex was analyzed on a 10–40% linear sucrose density gradient when HRD1 or SEL1L was silenced. Endogenous SEL1L, HRD1, and OS-9 formed a large complex that was detected in fractions 13–16 (Fig. 2, solid bars). In cells treated with siRNA targeted for SEL1L or HRD1, SEL1L was not detected in any fraction (Fig. 2A). When SEL1L was silenced, HRD1 was fractionated in slightly smaller fractions (Fig. 2B, fractions 10–15). Endogenous OS-9 formed two separate complexes (Fig. 2C, fractions 6–9 (dotted line) and fractions 12–15 (solid line)) but converged in a single peak of smaller sedimentation rates by the depletion of SEL1L or HRD1 (Fig. 2C).
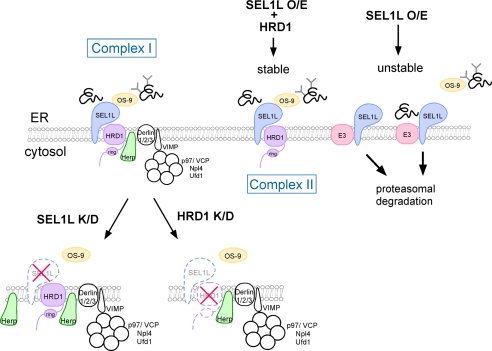
FIGURE 7.
Model for the formation of the ERAD complex containing HRD1-SEL1L in the ER membrane and ERAD regulation. The large complex containing HRD1-SEL1L (Complex I) regulates the ERAD of glycosylated NHK and non-glycosylated NHK-QQQ. By the transient expression of HRD1-SEL1L, a smaller-sized complex (Complex II) was formed in which OS-9 associates at the luminal side. Complex II was also capable of extraction and degradation of both NHK and NHK-QQQ. SEL1L that does not associate with HRD1 is rapidly degraded by an E3 other than HRD1. NHK-QQQ interacting with SEL1L was also rapidly degraded through the function of this E3. Destabilization of SEL1L by the silencing of HRD1 as well as depletion of SEL1L by siRNA treatment targeted for SEL1L resulted in the release of OS-9 from the membrane complex. Only the molecules analyzed in this study are shown. O/E, overexpression; K/D, knockdown.
FIGURE 2.
Analysis of HRD1-SEL1L complex by sucrose density gradient centrifugation. HEK293 cells were treated with 30 nm of StealthTM siRNA of negative control (Control) or targeted for SEL1L (siSEL1L), HRD1 (siHRD1), or OS-9 (siOS-9) for 48 h and were extracted by a buffer containing 3% digitonin. After fractionation by 10–40% sucrose density gradient centrifugation with a 60% sucrose cushion, SEL1L (A), HRD1 (B), and OS-9 (C) were detected by Western blotting. Solid bars indicate the fractions where the large ERAD complex containing HRD1-SEL1L and OS-9 sedimented. The dotted line shows the fractions of OS-9 with lower sedimentation rates. Asterisks indicate the nonspecific signals detected by the mouse monoclonal anti-SEL1L antibody. Open and gray arrowheads indicate the positions of OS-9v1 and v2, respectively.
Next, the sedimentation rates of ERAD components, including Herp, Derlin-1/2/3, VIMP, and p97/VCP, were analyzed when SEL1L or HRD1 was silenced. Fractions that included Herp, Derlin-1, Derlin-2, VIMP, and p97/VCP remained unchanged by the depletion of either SEL1L or HRD1 (supplemental Fig. S3). Thus, the membrane complex containing HRD1-SEL1L and OS-9 behaves differently than a complex comprised of Herp, Derlin-1/2, and VIMP.
The mass spectrometric analysis of the immunoprecipitates by the affinity-purified anti-SEL1L antibody revealed that HRD1 coimmunoprecipitated stoichiometrically as reported (15). In addition, OS-9 was found in the complex at a ratio of ∼1:3 (OS-9: HRD1-SEL1L) (supplemental Fig. S4A). The immunoprecipitation of metabolically labeled cells using specific antibodies against OS-9 and SEL1L confirmed the association of OS-9 with HRD1-SEL1L (supplemental Fig. S4B, lanes 1, 2, 7, and 8). These results suggest that the complex formation of HRD1-SEL1L with OS-9 is relatively stable, with an approximate ratio of 1:1:1/3. The ratio of OS-9 binding to HRD1-SEL1L was confirmed by assessing the radioactivity of co-immunoprecipitated proteins in radiolabeled cells after peptide N-glycosidase F (PNGase F) digestion (supplemental Fig. S4C). It should be noted that HRD1 remained in the SEL1L immunoprecipitates when OS-9 siRNA was used (supplemental Fig. S4B, lanes 9 and 10).
Transiently Expressed SEL1L Is Rapidly Degraded, but Co-expression of HRD1 Stabilizes SEL1L
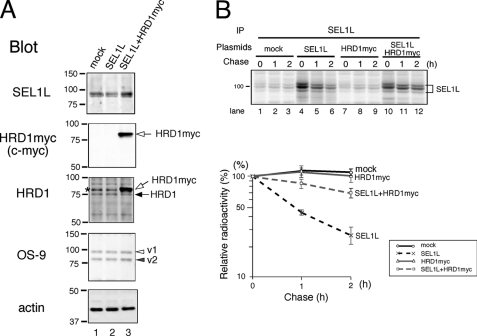
To further analyze the stability of SEL1L, SEL1L was transfected with or without HRD1. When only SEL1L was transfected, the total amount of SEL1L detected by Western blotting did not increase over the endogenous level (Fig. 3A, SEL1L blot, lanes 1 and 2). However, the SEL1L expression was increased mildly by the simultaneous transfection of SEL1L and HRD1. The total amount of SEL1L increased less than 2-fold over the endogenous level under our experimental conditions (Fig. 3A, SEL1L blot, lanes 1 and 3). Transfected myc-tagged HRD1 was expressed more than 5-fold over the endogenous HRD1 (Fig. 3A, HRD1 blot, lane 3). The overexpression of either SEL1L or HRD1-SEL1L did not change the amount of OS-9.
FIGURE 3.
Cotransfection of HRD1 stabilizes the transiently expressed SEL1L. A, HEK293 cells were transfected with SEL1L or SEL1L and myc-tagged HRD1. Equal amounts of cell lysates were separated by SDS-PAGE and analyzed by Western blotting. Open and closed arrows indicate the transfected HRD1myc and endogenous HRD1, respectively. Open and gray arrowheads are as in Fig. 2. The asterisk shows the nonspecific band detected by the anti-HRD1/synoviolin antibody. B, cells were transfected with SEL1L, HRD1myc, or SEL1L and HRD1. At 24 h after transfection, cells were pulse-labeled for 15 min. Cells were harvested and lysed in a buffer containing 1% Nonidet P-40 at the chase time indicated for immunoprecipitation (IP). The relative radioactivity of SEL1L is calculated as in Fig. 1C.
The half-life of SEL1L transfected with or without HRD1myc was examined by pulse-chase analysis. When SEL1L was overexpressed, newly synthesized SEL1L was rapidly degraded with a half-life of 1 h. SEL1L was markedly stabilized when HRD1 was cotransfected (Fig. 3B, quantified in the lower panel). These results are consistent with the destabilization of endogenous SEL1L when HRD1 is knocked down (Fig. 1).
Transient Expression of HRD1-SEL1L Forms a Smaller Complex (Complex II) in Addition to the Endogenously Assembled Complex (Complex I)
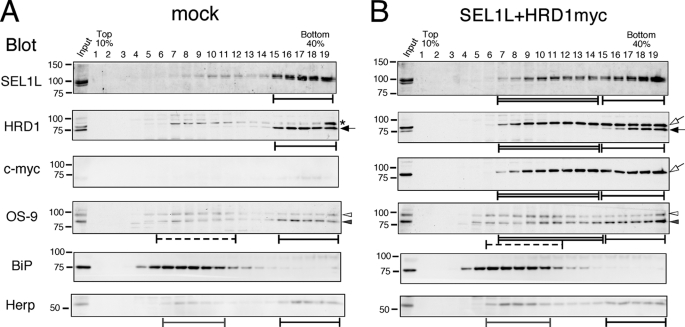
Next, the sedimentation rate of transfected HRD1-SEL1L was determined on a sucrose density gradient. SEl1L and HRD1myc were fractionated in broad fractions of 9–19 (Fig. 4B, Complex I and II, black and double-lined bars, respectively), although endogenous HRD1-SEL1L segregated in fractions close to the bottom (Fig. 4A, Complex I, black bars).
FIGURE 4.
Transient expression of SEL1L and HRD1 forms a smaller complex in addition to the large endogenous complex. Cell extracts transfected with mock (A) or SEL1L and HRD1 (B) were fractionated with a 10–40% sucrose density gradient generated using Gradient MasterTM without the 60% sucrose cushion. Black bars indicate the fractions where the large ERAD complex containing HRD1-SEL1L, OS-9, Herp, and Derlins was sedimented (Complex I). Double-lined bars show the smaller fractions detected by the transient expression of HRD1-SEL1L (Complex II). Dotted bars indicate the fractions containing OS-9 of a lower sedimentation rate. Fractions containing the second peak of Herp are indicated by the gray bars. The asterisk denotes the nonspecific signal observed by the anti-HRD1/synoviolin antibody. Arrows and arrowheads are as in Fig. 3.
OS-9 has four splice variants. In HEK 293 cells, OS-9v1 and v2 are abundantly expressed and detected by Western blotting (16, 31, 46). However, the functional differences between these two splice variants remains unclear. Interestingly, OS-9v2 was fractionated broadly when HRD1 and SEL1L were overexpressed, similar to the distribution of SEL1L and HRD1myc. However, the total amount of OS-9v2 detected by Western blotting did not increase (Fig. 3A). In contrast, the OS-9v1 distribution was almost unchanged, forming two separate peaks (Fig. 4, A and B, solid and dotted bars). These results suggest that OS-9v2, but not v1, preferentially associates with the HRD1-SEL1L complex.
Notably, the sedimentation fractions of Herp, Derlin-1/2, VIMP, and p97 remained unchanged even when HRD1-SEL1L was overexpressed (Fig. 4 and data not shown). These results suggest that HRD1-SEL1L is capable of forming two distinct complexes with different sedimentation rates, designated here as Complexes I and II. Endogenous HRD1-SEL1L forms a large Complex I associated with other ERAD components containing Herp, Derlin-1/2, and VIMP. However, when HRD1 and SEL1L were transiently expressed, we also observed the formation of Complex II associated with OS-9v2 but not Herp, Derlin-1/2, or VIMP.
HRD1-SEL1L-containing Complex II Is Functional in ERAD, and NHK-QQQ Degradation Is Enhanced by Transient SEL1L Expression
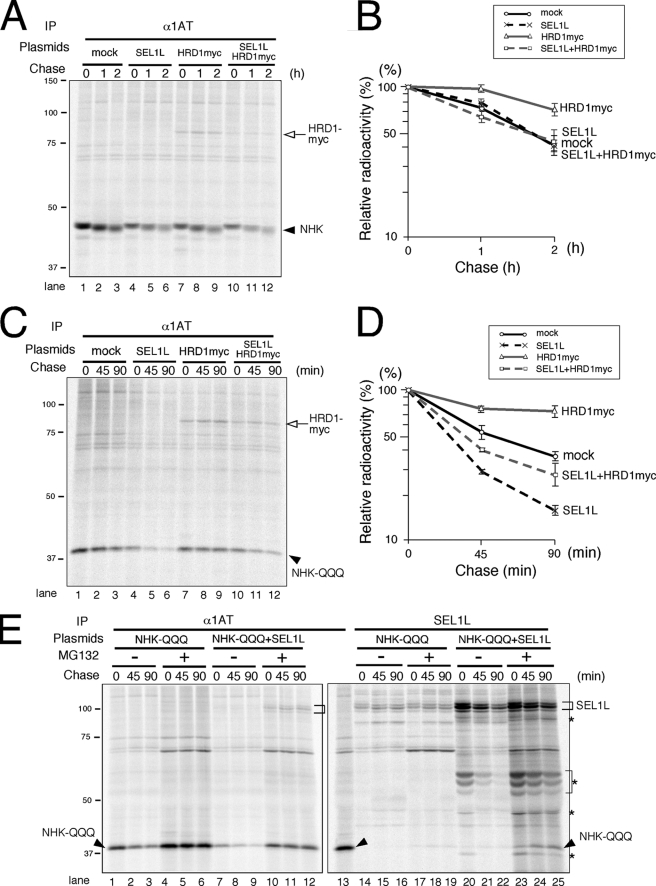
Silencing SEL1L abrogates the degradation of two model ERAD substrates, glycoprotein NHK and the non-glycosylated mutant NHK-QQQ (16, 31). HRD1 is also required for NHK disposal (31). The requirement of HRD1 for the degradation of NHK-QQQ was analyzed by RNAi (supplemental Fig. S5). However, because HRD1 silencing also eliminates the SEL1L peptide, knockdown experiments do not discriminate between the functions of SEL1L and HRD1 in ERAD. Therefore, we examined the effects of transiently expressing SEL1L and/or HRD1 on the degradations of NHK and NHK-QQQ. Transient expression of HRD1 moderately inhibited NHK degradation, whereas the transfection of SEL1L or both SEL1L and HRD1 did not affect the NHK degradation kinetics (Fig. 5A, quantified in B).
FIGURE 5.
Effect of transiently expressed HRD1-SEL1L on the degradation of NHK and NHK-QQQ. A, cells transfected with NHK, SEL1L, and/or HRD1 were pulse-labeled for 15 min and chased for the times indicated. Aliquots of cells extracted by 3% digitonin were immunoprecipitated (IP) with anti-α1 antitrypsin (α1AT) antibody. The arrowhead indicates the position of NHK. The open arrow shows coimmunoprecipitated HRD1myc. B, shown is quantification of NHK degradation in A. The relative radioactivity of NHK was calculated by setting the NHK intensity at the end of the pulse time as 100%. Results are the mean and S.E. (n = 3). C, cells transiently expressing NHK-QQQ, SEL1L, and/or HRD1 were metabolically labeled, and the degradation kinetics of NHK-QQQ was analyzed as in A. The arrowhead indicates the position of NHK-QQQ. D, radioactivity of NHK-QQQ in C was quantified as in B. E, cells were transfected with NHK-QQQ and SEL1L and treated with or without MG132 for 4 h before metabolic labeling. Arrowheads and brackets indicate the positions of NHK-QQQ and SEL1L, respectively. Asterisks are the degradation products generated from transiently expressed SEL1L.
When cells were lysed in a buffer containing 3% digitonin, SEL1L was found to be associated with OS-9v2, endogenous HRD1, and transfected HRD1myc (supplemental Fig. S6A). The transient expression of HRD1 inhibited NHK-QQQ degradation, similar to the effect on NHK. However, transiently expressed SEL1L markedly enhanced the disposal of NHK-QQQ (Fig. 5C, quantified in D). Cotransfection of SEL1L and HRD1 slightly accelerated the disappearance of NHK-QQQ, but the effect was relatively weak (Fig. 5C, quantified in D). As indicated by sucrose density gradient centrifugation, transient expression of HRD1-SEL1L forms a smaller Complex II in addition to the endogenous Complex I. If Complex II, which does not stably associate with Derlins, VIMP, or Herp, cannot support the retrotranslocation and degradation of ERAD substrates, then the degradation of client proteins should be inhibited. The observation of similar degradation kinetics for NHK and NHK-QQQ under conditions of HRD1-SEL1L overexpression thus suggests that the HRD1-SEL1L-containing Complex II is capable of sorting ERAD substrates for retrotranslocation and proteasomal degradation.
To confirm the interaction of the client proteins with the SEL1L complexes, we analyzed the immunoprecipitates of metabolically labeled cells. The NHK-QQQ band was hardly visible in the endogenous SEL1L immunoprecipitates (Fig. 5E, lanes 14–16, and supplemental Fig. S6B, lanes 14–16) but was detected when the proteasome inhibitor MG132 was added (Fig. 5E, lanes 17–19, and supplemental Fig. S7, lanes 5–7). In cells overexpressing SEL1L, coimmunoprecipitation of NHK-QQQ with SEL1L was easily observed even in the absence of MG132 (Fig. 5E, lanes 20–22, and supplemental Fig. S6B, lanes 17–19). Degradation of NHK-QQQ was markedly suppressed by the addition of MG132 (Fig. 5E, lanes 4–6). Notably, the enhanced disappearance of NHK-QQQ in the presence of overexpressed SEL1L was also strongly inhibited by MG132. This suggests that NHK-QQQ is degraded by the cytoplasmic proteasome even when SEL1L is overexpressed.
Transiently Expressed SEL1L Is Degraded by ERAD
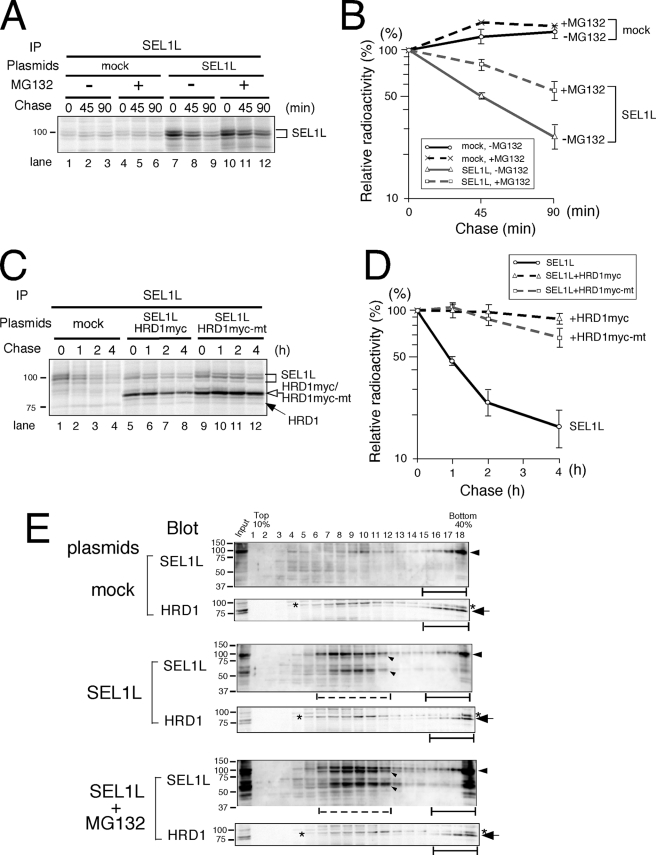
To determine the degradation pathway of transiently expressed SEL1L, the half-life of SEL1L was examined in the presence or absence of MG132. SEL1L degradation was inhibited by MG132 (Fig. 6A, quantified in B), suggesting that SEL1L is degraded by the proteasome.
FIGURE 6.
Transiently expressed SEL1L is degraded by the function of an E3 other than HRD1. A, cells transfected with mock or SEL1L were treated with or without MG132 for 4 h before metabolic labeling. Cells were labeled for 15 min and chased for the time indicated. SEL1L was immunoprecipitated (IP) using specific antibody. B, quantification of SEL1L immunoprecipitated in A. C, SEL1L was transfected with mock, HRD1myc, or mutant HRD1myc lacking ubiquitin-ligase activity. Cells were metabolically labeled and chased as in A. Cell aliquots extracted in 3% digitonin were subjected to immunoprecipitation. D, quantification of SEL1L analyzed in C. E, cells transfected with mock or SEL1L were treated with or without MG132 for 6 h, and cell extracts were fractionated with a 10–40% sucrose density gradient as in Fig. 4. Black bars indicate the fractions where the large molecular mass ERAD complex containing endogenous HRD1-SEL1L was detected, and dotted bars show the fractions that contained proteins with lower molecular mass where overexpressed SEL1L was detected. The arrowheads and arrows indicate SEL1L and HRD1, respectively. Small arrowheads indicate the degradation products of SEL1L. The asterisks denote the nonspecific signal observed with the anti-HRD1/synoviolin antibody.
Because the association with HRD1 stabilizes SEL1L, the E3 enzyme responsible for SEL1L ERAD must be an E3 other than HRD1. We further assessed the requirement of HRD1 ubiquitin-ligase activity for SEL1L stabilization. An HRD1 RING-H2 domain mutant lacking the enzyme activity stabilized SEL1L to a similar extent as wild-type HRD1 (Fig. 6C, lanes 9–12, quantified in D). This finding suggests that the E3 activity of HRD1 is irrelevant to SEL1L turnover.
Finally, the sedimentation rate of transiently expressed SEL1L in the presence or absence of the proteasome inhibitor MG132 was determined. SEL1L was detected in lower molecular mass fractions 6–11 (Fig. 6E, dotted line) as well as in high molecular mass fractions containing the endogenous HRD1-SEL1L complex. The low molecular mass fractions were smaller than Complex II formed by cotransfection of SEL1L and HRD1 (compare with Fig. 4B). When MG132 was added, the SEL1L degradation products accumulated, but the sedimentation rates of SEL1L were the same as in cells without the proteasome inhibitor (Fig. 6E). The distribution of endogenous HRD1 did not change in cells transiently expressing SEL1L even in the presence of the proteasome inhibitor, which confirms that SEL1L that is not associated with HRD1 is unstable.
DISCUSSION
In the present study we analyzed the effect of silencing SEL1L or HRD1 on the ERAD complex and found that SEL1L turnover is determined by HRD1. The half-life of endogenous SEL1L was longer than 12 h in our assay system, which coincides with the previously reported 15-h half-life of endogenous and transfected HRD1 in HeLa cells (10). When the relative amount of HRD1 was decreased, SEL1L was more rapidly degraded. Thus, the mode of HRD1-SEL1L ubiquitin-ligase complex regulation in the ER membrane is completely different from that of the yeast homologue Hrd1p-Hrd3p, in which unstable Hrd1p is stabilized by its association with Hrd3p (8, 9). The half-life of endogenous SEL1L is reported to be 3 h in human U373 glioblastoma-astrocytoma cells (13), which is inconsistent with our present results. Because we used human 293 cells, the discrepancy might depend upon the cell lines used. The amount of SEL1L and HRD1 synthesized in some cell lines may not be stoichiometric, which would result in a rapid degradation of the excess SEL1L that was not associated with HRD1. Additional studies are required to clarify this discrepancy.
Sedimentation analyses revealed that the ERAD lectin OS-9 dissociates from the large ERAD complex when either SEL1L or HRD1 is silenced (Fig. 7). OS-9 in 293 cells is composed of two splice variants, v1 and v2. Interestingly, OS-9v2 but not v1 preferentially associated with overexpressed HRD1-SEL1L. This finding suggests a functional difference between the two splice variants. Two models have been proposed for the function of OS-9. OS-9 may carry misfolded ERAD substrates to the membrane ERAD complex and/or may stably associate with the membrane complex to proofread the substrates (Ref. 31; for review, see Refs. 28 and 30). These two models are not mutually exclusive. Association of endogenous OS-9 with overexpressed HRD1-SEL1L suggests that at least a portion of OS-9 is a stable component of the complex. It is also unclear whether OS-9 regulates glycosylated substrates only as a lectin or if it also recognizes the misfolded polypeptides. Additional studies are required to clarify the function of OS-9. Moreover, near-stoichiometric association of OS-9 to the HRD1-SEL1L complex (∼1:3) suggests that HRD1-SEL1L has room for additional OS-9. Alternatively, the surface of SEL1L used for OS-9 binding can be occupied by other molecules, such as XTP3-B.
The sedimentation rates of Derlin-1/2, Herp, and VIMP did not change when either SEL1L or HRD1 was silenced. Both Herp (47) and Derlin-2 (48) are ER stress-responsive genes and are up-regulated by SEL1L depletion, as SEL1L deficiency causes ER stress (49). Although we also detected an increase in Herp and Derlin-2, the sedimentation profiles of these proteins remained unchanged (supplemental Fig. S3). Collectively, these results suggest that the large membrane ERAD complex is composed of the dynamic association of two sub-complexes; one containing Derlin-1/2, Herp, and VIMP and the other containing HRD1-SEL1L and OS-9.
In yeast, the Hrd1p-Hrd3p complex is required for both ERAD-C and ERAD-M pathways. However, Der1p and Usa1p are dispensable for the ERAD-M pathway (26), and the association of Der1p to the Hrd1p-Hrd3p complex is relatively weak (23). The mammalian homologues of Der1p and Usa1p are Derlin-1/2/3 and Herp, respectively. Therefore, the HRD1-SEL1L complex may be formed similar to the complex assembled in yeast ER, with non-inclusion of Derlin-1/2(/3) and Herp (Complex II).
Transiently expressed HRD1-SEL1L formed a smaller complex (Complex II) in addition to the large endogenous complex (Complex I) (Fig. 7). Complex II was associated with OS-9 but did not cosediment with Derlin-1/2, Herp, and VIMP. This is consistent with the two-subcomplex model suggested by the knockdown of either SEL1L or HRD1. NHK degradation depends on the Derlin2/3 complex (48). Because Complex II supports the extraction and degradation of ERAD substrates of both glycosylated NHK and non-glycosylated NHK-QQQ, Complex II likely interacts transiently or weakly with the Derlin-containing complex. Alternatively, the HRD1-SEL1L complex may be too saturated for cosedimentation with Derlins/Herp/VIMP. In the latter case, HRD1-SEL1L may associate with another complex for the retrotranslocation of ERAD clients when the Derlin-containing complex is unavailable.
Transiently expressed SEL1L was rapidly degraded by the proteasomal pathway with a half-life of 1 h, but coexpression of HRD1 markedly stabilized SEL1L. Conversely, when endogenous SEL1L was dissociated from HRD1 (i.e. by way of siRNA-mediated HRD1 knockdown), SEL1L became destabilized. These stabilization effects of HRD1 were independent of the ubiquitin-ligase activity. Thus, HRD1-unassociated SEL1L is degraded by another ubiquitin-ligase localized in the ER membrane (Fig. 7C, an E3, shown in pink).
NHK-QQQ was degraded more rapidly when SEL1L was overexpressed, decreasing the t½ of NHK-QQQ from 60 to 30 min. Although coimmunoprecipitation of NHK-QQQ with SEL1L was easily detected, it is unlikely that NHK-QQQ was degraded in its SEL1L-bound form, as the half-life of overexpressed SEL1L is 1 h. Rather, the faster degradation was likely because of the function of the E3 that recognized NHK-QQQ (Fig. 7, an E3, shown in pink). The requirement of the HRD1-SEL1L complex for ERAD likely prevents the rapid degradation of discrete substrates and affords time for proofreading. Various ERAD substrates require SEL1L or HRD1 for degradation (10, 13, 16, 22, 31, 39, 50, 51) but are still not completely identified now. The stabilization and assembly of the HRD1-SEL1L complex thus play a central role in mammalian ERAD regulation.
Supplementary Material
This work was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to K. N., I. W., and N. H.) and by the Hayashi Memorial Foundation for Female Natural Scientists (to N. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7.
- ER
- endoplasmic reticulum
- ERAD
- ER-associated degradation
- NHK
- α1-antitrypsin null (Hong Kong)
- VCP
- valosine-containing protein
- VIMP
- VCP/p97-interacting membrane protein.
REFERENCES
- 1. Ellgaard L., Helenius A. (2003) Nat. Rev. Mol. Cell Biol. 4, 181–191 [DOI] [PubMed] [Google Scholar]
- 2. Vembar S. S., Brodsky J. L. (2008) Nat. Rev. Mol. Cell Biol. 9, 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Plemper R. K., Wolf D. H. (1999) Trends Biochem. Sci. 24, 266–270 [DOI] [PubMed] [Google Scholar]
- 4. Tsai B., Ye Y., Rapoport T. A. (2002) Nat. Rev. Mol. Cell Biol. 3, 246–255 [DOI] [PubMed] [Google Scholar]
- 5. Hirsch C., Gauss R., Horn S. C., Neuber O., Sommer T. (2009) Nature 458, 453–460 [DOI] [PubMed] [Google Scholar]
- 6. Deak P. M., Wolf D. H. (2001) J. Biol. Chem. 276, 10663–10669 [DOI] [PubMed] [Google Scholar]
- 7. Bays N. W., Gardner R. G., Seelig L. P., Joazeiro C. A., Hampton R. Y. (2001) Nat. Cell Biol. 3, 24–29 [DOI] [PubMed] [Google Scholar]
- 8. Plemper R. K., Bordallo J., Deak P. M., Taxis C., Hitt R., Wolf D. H. (1999) J. Cell Sci. 112, 4123–4134 [DOI] [PubMed] [Google Scholar]
- 9. Gardner R. G., Swarbrick G. M., Bays N. W., Cronin S. R., Wilhovsky S., Seelig L., Kim C., Hampton R. Y. (2000) J. Cell Biol. 151, 69–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kikkert M., Doolman R., Dai M., Avner R., Hassink G., van Voorden S., Thanedar S., Roitelman J., Chau V., Wiertz E. (2004) J. Biol. Chem. 279, 3525–3534 [DOI] [PubMed] [Google Scholar]
- 11. Amano T., Yamasaki S., Yagishita N., Tsuchimochi K., Shin H., Kawahara K., Aratani S., Fujita H., Zhang L., Ikeda R., Fujii R., Miura N., Komiya S., Nishioka K., Maruyama I., Fukamizu A., Nakajima T. (2003) Genes Dev. 17, 2436–2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fang S., Ferrone M., Yang C., Jensen J. P., Tiwari S., Weissman A. M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 14422–14427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mueller B., Lilley B. N., Ploegh H. L. (2006) J. Cell Biol. 175, 261–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Biunno I., Cattaneo M., Orlandi R., Canton C., Biagiotti L., Ferrero S., Barberis M., Pupa S. M., Scarpa A., Ménard S. (2006) J. Cell. Physiol. 208, 23–38 [DOI] [PubMed] [Google Scholar]
- 15. Lilley B. N., Ploegh H. L. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 14296–14301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hosokawa N., Wada I., Nagasawa K., Moriyama T., Okawa K., Nagata K. (2008) J. Biol. Chem. 283, 20914–20924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schulze A., Standera S., Buerger E., Kikkert M., van Voorden S., Wiertz E., Koning F., Kloetzel P. M., Seeger M. (2005) J. Mol. Biol. 354, 1021–1027 [DOI] [PubMed] [Google Scholar]
- 18. Lilley B. N., Ploegh H. L. (2004) Nature 429, 834–840 [DOI] [PubMed] [Google Scholar]
- 19. Ye Y., Shibata Y., Yun C., Ron D., Rapoport T. A. (2004) Nature 429, 841–847 [DOI] [PubMed] [Google Scholar]
- 20. Ye Y., Meyer H. H., Rapoport T. A. (2001) Nature 414, 652–656 [DOI] [PubMed] [Google Scholar]
- 21. Kokame K., Agarwala K. L., Kato H., Miyata T. (2000) J. Biol. Chem. 275, 32846–32853 [DOI] [PubMed] [Google Scholar]
- 22. Okuda-Shimizu Y., Hendershot L. M. (2007) Mol. Cell 28, 544–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gauss R., Sommer T., Jarosch E. (2006) EMBO J. 25, 1827–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gauss R., Jarosch E., Sommer T., Hirsch C. (2006) Nat. Cell Biol. 8, 849–854 [DOI] [PubMed] [Google Scholar]
- 25. Denic V., Quan E. M., Weissman J. S. (2006) Cell 126, 349–359 [DOI] [PubMed] [Google Scholar]
- 26. Carvalho P., Goder V., Rapoport T. A. (2006) Cell 126, 361–373 [DOI] [PubMed] [Google Scholar]
- 27. Lederkremer G. Z. (2009) Curr. Opin. Struct. Biol. 19, 515–523 [DOI] [PubMed] [Google Scholar]
- 28. Tamura T., Cormier J. H., Hebert D. N. (2008) Trends Biochem. Sci. 33, 298–300 [DOI] [PubMed] [Google Scholar]
- 29. Aebi M., Bernasconi R., Clerc S., Molinari M. (2010) Trends Biochem. Sci. 35, 74–82 [DOI] [PubMed] [Google Scholar]
- 30. Hosokawa N., Kamiya Y., Kato K. (2010) Glycobiology 20, 651–660 [DOI] [PubMed] [Google Scholar]
- 31. Christianson J. C., Shaler T. A., Tyler R. E., Kopito R. R. (2008) Nat. Cell Biol. 10, 272–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mueller B., Klemm E. J., Spooner E., Claessen J. H., Ploegh H. L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 12325–12330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vashist S., Ng D. T. (2004) J. Cell Biol. 165, 41–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huyer G., Piluek W. F., Fansler Z., Kreft S. G., Hochstrasser M., Brodsky J. L., Michaelis S. (2004) J. Biol. Chem. 279, 38369–38378 [DOI] [PubMed] [Google Scholar]
- 35. Swanson R., Locher M., Hochstrasser M. (2001) Genes Dev. 15, 2660–2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ravid T., Kreft S. G., Hochstrasser M. (2006) EMBO J. 25, 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bordallo J., Plemper R. K., Finger A., Wolf D. H. (1998) Mol. Biol. Cell 9, 209–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hampton R. Y., Gardner R. G., Rine J. (1996) Mol. Biol. Cell 7, 2029–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bernasconi R., Galli C., Calanca V., Nakajima T., Molinari M. (2010) J. Cell Biol. 188, 223–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sifers R. N., Brashears-Macatee S., Kidd V. J., Muensch H., Woo S. L. (1988) J. Biol. Chem. 263, 7330–7335 [PubMed] [Google Scholar]
- 41. Hirao K., Natsuka Y., Tamura T., Wada I., Morito D., Natsuka S., Romero P., Sleno B., Tremblay L. O., Herscovics A., Nagata K., Hosokawa N. (2006) J. Biol. Chem. 281, 9650–9658 [DOI] [PubMed] [Google Scholar]
- 42. Morito D., Hirao K., Oda Y., Hosokawa N., Tokunaga F., Cyr D. M., Tanaka K., Iwai K., Nagata K. (2008) Mol. Biol. Cell 19, 1328–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hosokawa N., Wada I., Hasegawa K., Yorihuzi T., Tremblay L. O., Herscovics A., Nagata K. (2001) EMBO Rep. 2, 415–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hosokawa N., Kamiya Y., Kamiya D., Kato K., Nagata K. (2009) J. Biol. Chem. 284, 17061–17068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nagasawa K., Higashi T., Hosokawa N., Kaufman R. J., Nagata K. (2007) EMBO Rep. 8, 483–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bernasconi R., Pertel T., Luban J., Molinari M. (2008) J. Biol. Chem. 283, 16446–16454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kokame K., Kato H., Miyata T. (2001) J. Biol. Chem. 276, 9199–9205 [DOI] [PubMed] [Google Scholar]
- 48. Oda Y., Okada T., Yoshida H., Kaufman R. J., Nagata K., Mori K. (2006) J. Cell Biol. 172, 383–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Francisco A. B., Singh R., Li S., Vani A. K., Yang L., Munroe R. J., Diaferia G., Cardano M., Biunno I., Qi L., Schimenti J. C., Long Q. (2010) J. Biol. Chem. 285, 13694–13703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cattaneo M., Otsu M., Fagioli C., Martino S., Lotti L. V., Sitia R., Biunno I. (2008) J. Cell. Physiol. 215, 794–802 [DOI] [PubMed] [Google Scholar]
- 51. Omura T., Kaneko M., Okuma Y., Orba Y., Nagashima K., Takahashi R., Fujitani N., Matsumura S., Hata A., Kubota K., Murahashi K., Uehara T., Nomura Y. (2006) J. Neurochem. 99, 1456–1469 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.