Abstract
The intermediate conductance Ca2+-activated K+ channel (IKCa channel) encoded by KCa3.1 is responsible for the control of proliferation and differentiation in various types of cells. We identified novel spliced variants of KCa3.1 (human (h) KCa3.1b) from the human thymus, which were lacking the N-terminal domains of the original hKCa3.1a as a result of alternative splicing events. hKCa3.1b was significantly expressed in human lymphoid tissues. Western blot analysis showed that hKCa3.1a proteins were mainly expressed in the plasma membrane fraction, whereas hKCa3.1b was in the cytoplasmic fraction. We also identified a similar N terminus lacking KCa3.1 variants from mice and rat lymphoid tissues (mKCa3.1b and rKCa3.1b). In the HEK293 heterologous expression system, the cellular distribution of cyan fluorescent protein-tagged hKCa3.1a and/or YFP-tagged hKCa3.1b isoforms showed that hKCa3.1b suppressed the localization of hKCa3.1a to the plasma membrane. In the Xenopus oocyte translation system, co-expression of hKCa3.1b with hKCa3.1a suppressed IKCa channel activity of hKCa3.1a in a dominant-negative manner. In addition, this study indicated that up-regulation of mKCa3.1b in mouse thymocytes differentiated CD4(+)CD8(+) phenotype thymocytes into CD4(−)CD8(−) ones and suppressed concanavalin-A-stimulated thymocyte growth by down-regulation of mIL-2 transcripts. Anti-proliferative effects and down-regulation of mIL-2 transcripts were also observed in mKCa3.1b-overexpressing mouse thymocytes. These suggest that the N-terminal domain of KCa3.1 is critical for channel trafficking to the plasma membrane and that the fine-tuning of IKCa channel activity modulated through alternative splicing events may be related to the control in physiological and pathophysiological conditions in T-lymphocytes.
Keywords: Calcium, Confocal Microscopy, Membrane Trafficking, Potassium Channels, RNA Splicing, Ca2+-activated K+ Channel, Dominant-negative, Lymphoid Organ, Thymocyte
Introduction
The Ca2+-activated K+ (KCa) channel is a family of the K+ channel superfamily that can directly communicate with Ca2+ signal pathways and is responsible for various cellular processes by controlling membrane potential. In nonexcitable cells, hyperpolarization of the membrane potential, which is induced by activation of the KCa channel, increases the driving force for Ca2+ entry. Activation of the KCa channel is therefore strongly associated with sustained Ca2+ influx and the Ca2+-dependent signaling pathway (1, 2). The intermediate conductance KCa channel (IKCa)2 is present in a variety of non-neuronal peripheral tissues, e.g. spleen, thymus, peripheral blood leukocytes, T-lymphocytes, and undifferentiated vascular smooth muscles, and it participates in the control of cell proliferation and migration (3, 4). Physiological roles of KCa3.1 in T-lymphocytes are as follows: 1) maintenance of the intracellular Ca2+ levels required for mitogen activation by membrane potential hyperpolarization (5, 6), and 2) the control of apoptotic cell volume decrease, the initiation step in Ca2+-induced apoptosis (7). Recent studies have shown that IKCa channels are possible molecular targets for pharmacological intervention in various diseases, e.g. autoimmune diseases, restenosis, urinary incontinence, and cystic fibrosis (8–11).
Four subtypes of small/intermediate conductance Ca2+-activated K+ channels (KCa2.1, KCa2.3, and KCa3.1) have been identified by molecular cloning studies and are activated via a mechanism involving Ca2+ binding to the EF hands of calmodulin (4, 12, 13). KCa2.x and KCa3.1 can make heteromeric complexes of four pore-forming α subunits with different pharmacological profiles and cell surface expression from homotetramers (14, 15). Additionally, alternative splicing is a common mechanism for generating K+ channel diversity (16). Of interest, two alternatively spliced variants of KCa2.3, KCa2.3–1B and KCa2.3–1C, which have a different first exon from the original KCa2.3 and thereby lack transmembrane segment 1 (S1), have caused dominant-negative suppression of KCa2.x and KCa3.1 by trapping these channel proteins intracellularly (14, 15, 17). For example, KCa2.3–1B, which is implicated in schizophrenia (15, 18), suppresses KCa2.1 channel activity in human Jurkat T cells, and KCa2.3–1C suppresses KCa2.x and KCa3.1 channel activities almost completely and KCa3.1 channel activity partially. Kolski-Andreaco et al. (17) found that the dominant-inhibitory segment (DIS) in the C terminus plays an essential role in KCa2/3 subunit interaction and reduces functional KCa2/3 channel expression on the cell surface. Several reports have shown that the cell surface trafficking of KCa3.1 is modulated by calmodulin and C-terminal leucine zipper (19, 20); however, the dominant-negative KCa3.1 isoforms have not been identified yet.
In this study, we identified the alternatively spliced variants of KCa3.1 (KCa3.1-ΔN) in lymphoid tissues of humans, mice, and rats and examined their tissue distribution using real time PCR and Western blot analyses. Also, the dominant-negative regulation of the original KCa3.1a by KCa3.1-ΔN was determined by fluorescence imaging and electrophysiological techniques. Furthermore, to clarify the physiological significance of KCa3.1-ΔN in immune function, we examined the functional impacts of increased KCa3.1-ΔN on mouse thymocyte proliferation and CD4/CD8 thymocyte subsets using cell proliferation assay and flow cytometry analysis, respectively. The role of KCa3.1b in mouse thymocytes was also examined by generating mKCa3.1b-overexpressing thymocytes.
EXPERIMENTAL PROCEDURES
RNA Extraction, RT-PCR, and Real Time PCR
Total RNA extraction and RT-PCR from mice and rats were performed as reported previously (21). All experiments were carried out in accordance with the guiding principles for the care and use of laboratory animals (Science and International Affairs Bureau of the Japanese Ministry of Education, Science, Sports and Culture) and also with the approval of the ethics committee at Nagoya City University. Human RNA was purchased from Invitrogen. The resulting cDNA product was amplified with gene-specific primers (21), and primers are designated using Primer ExpressTM software (version 1.5; Applied Biosystems, Foster City, CA) (supplemental Table S1). The amplification profile was follows: a 15-s denaturation step at 96 °C and a 1–2-min primer extension step at 60 °C. The amplified products were separated on 1.0–2.5% agarose gels and visualized by ethidium bromide staining. Obtained full-length KCa3.1 clones were ligated into pBluescript SK(+) (Stratagene, La Jolla, CA) and pECFP-C1/pEYFP-C1 (BD Biosciences, Palo Alto, CA). To confirm the nucleotide sequences, amplified PCR products and plasmid constructs were sequenced with an ABI PRIZM 3100 genetic analyzer (Applied Biosystems).
Real time quantitative PCR was performed using SYBR Green chemistry on an ABI 7000 sequence detector system (Applied Biosystems) as reported previously (21). Regression analysis of the mean values of four multiplex RT-PCRs for log10 diluted cDNA was used to generate standard curves. Unknown quantities relative to the standard curve for a particular set of primers were calculated, yielding transcriptional quantification of the gene products for KCa3.1 spliced variants relative to the endogenous standard, glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
Western Blot, Immunoprecipitation, and Immunocytochemistry
Protein fractions of the plasma membrane and cytoplasmic region were prepared from mouse and rat tissues as reported previously (21), and those from human tissues were purchased from the BioChain Institute (Hayward, CA). Protein fractions were solubilized with 2× sample buffer with 1% SDS and subjected to SDS-PAGE (10%). The blots were incubated with anti-KCa3.1 antibody specific to the C-terminal region (anti-KCa3.1-C; Alomone Labs, Jerusalem, Israel) and then incubated with anti-rabbit horseradish peroxidase-conjugated IgG (Chemicon International, Inc., Temecula, CA). An enhanced chemiluminescence detection system (Amersham Biosciences) was used for the detection of the bound antibody. Resulting images were analyzed by a LAS-1000 device (Fujifilm, Tokyo, Japan).
For immunoprecipitation, cells and tissue homogenates from the mouse thymus were incubated with anti-KCa3.1 antibody specific to the N-terminal region (anti-KCa3.1-N, ab65985; Abcam, Cambridge, MA) overnight at 4 °C, and immunoprecipitation was performed using the Seize Classic mammalian immunoprecipitation kit according to the experimental manual supplied by Pierce. After the obtained fractions were subjected to SDS-PAGE, the blots were incubated with anti-KCa3.1-C (Alomone Labs) antibody, and the images were detected as described above. Immunocytochemical staining of KCa3.1 in mouse thymocytes was performed as shown in our previous report (21), and Alexa Fluor® 488 goat anti-rabbit IgG (Invitrogen) was used as a secondary antibody.
Subcellular Localization of KCa3.1 Spliced Variants
To visualize the intracellular distribution of human and mouse KCa3.1 spliced variants by laser scanning confocal microscopy (LSM 510; Carl Zeiss, Oberkochen, Germany), the full-length of hKCa3.1a–d and mKCa3.1a and -b clones was subcloned into pECFP-C1 and pEYFP-C1, and each plasmid construct was referred to as pECFP-hKCa3.1a, pEYFP-hKCa3.1b, pEYFP-hKCa3.1c, pEYFP-hKCa3.1d, pECFP-mKCa3.1a, and pEYFP-mKCa3.1b. HEK293 cell line (Health Science Research Resources Bank (HSRRB), Tokyo, Japan) was maintained as reported previously (21). Cells were transiently transfected with plasmid constructs using Lipofectamine 2000 reagent (Invitrogen). Cells were used in the experiments 48–72 h after transfection. Control cells were transfected with plasmid vector alone. Spectral line profiles of the fluorescence and quantification of fluorescent signals were performed using Image-Pro Plus software (version 5.1; Silver Spring, MD) and Image J software (version 1.42, National Institutes of Health). The fluorescence intensity in and below the plasma membrane within 1.0 μm was calculated. Confocal parameters were set such that pixel saturation was less than 10% and that signal intensities were in the linear range of photomultiplier sensitivity.
RNA Injection to Oocytes and Electrophysiology
Adult female Xenopus laevis frogs were purchased from Hamamatsu Seibutsu Kyozai (Shizuoka, Japan) and maintained in the laboratory until used. The specific cRNA for hSK4a–d was synthesized in vitro from the linearized plasmids using the mMESSAGE mMACHINETM in vitro transcription kit (Ambion, Austin, TX). Defolliculated stage V and VI X. laevis oocytes were harvested, and 50 nl of cRNA per oocyte was injected at a concentration of 0.5–1.0 μg/μl using a microinjector. Oocytes were incubated at 20 °C in standard ND96 solution (in mm: 96 NaCl, 2 KCl, 1.8 CaCl2, 1 MgCl2, and 5 HEPES, pH 7.4, adjusted with NaOH) supplemented with antibiotics to permit channel expression. One or 2 days after the injection of cRNA, oocytes were voltage-clamped using two microelectrodes (22). Membrane currents were recorded using an amplifier (CEZ-1250; Nihon Koden, Tokyo, Japan) and a pen recorder (FBR-252A; DKK-TOA, Tokyo, Japan). All experiments were performed at room temperature (23 ± 1 °C). Ionomycin, DC-EBIO, and charybdotoxin were purchased from Sigma, Tocris Bioscience (Ellisville, MO), and Peptide Institute Inc. (Osaka, Japan), respectively.
Preparation of Concanavalin-A-stimulated Mouse Thymocytes and Excess Expression of EYFP-fused mKCa3.1b in Primary Cultured Thymocytes
Mouse thymocytes were prepared by pressing the thymus with frosted slide glasses in the medium containing RPMI 1640 (Invitrogen) supplemented with 10% heat-inactivated fetal calf serum (Invitrogen) and antibiotics (penicillin and streptomycin) in the presence and absence of concanavalin-A (Con-A, 5 μg/ml) (Con-A(+) and Con-A(−), respectively). After 60 min, Con-A(−) thymocytes were cultivated with Con-A-supplemented medium for 0–2 days. Primary cultured Con-A(+) thymocytes were transiently transfected with pEYFP-C1 (control vector alone) or pEYFP-mKCa3.1b using Lipofectamine LTX with Plus reagent (Invitrogen). Cells were used in the experiments 2 days after transfection. Determination of maximal efficacy was performed according to the manufacturer's instructions.
Measurement of Mouse Thymocyte Viability by MTT Assay
MTT (Sigma) cell proliferation assay was performed to assess the viability of mouse thymocytes. Briefly, 4 × 105 cells/ml were cultured in duplicate in 96-well plates for 0–2 days. After addition of MTT reagent into each well, lysing solution was added, and the absorbance was measured in a multiple well plate reader, Multiscan JX (Thermo Electron Corp., Waltham, MA) at a test wavelength of 540 nm and a reference wavelength of 630 nm. TRAM-34 was purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Flow Cytometric Analysis
Cell surface markers were analyzed with a FACScan flow cytometer (BD LSR, BD Biosciences) acquiring at least 10,000 events. Data were analyzed using CellQuest software (BD Biosciences). The lymphocyte gate was established by analysis of forward angle versus right angle light scatter. The percentage of positive-staining cells was determined by comparing test histograms with those obtained using fluorescein isothiocyanate (FITC)-labeled CD4 and phycoerythrin-labeled CD8 antibodies (BD Biosciences).
Apoptosis Measurement by Annexin V Staining
Apoptotic cells were analyzed with annexin V-EGFP or -phycoerythrin apoptosis detection kit (Medical and Biological Laboratories (MBL), Nagoya, Japan) by confocal laser microscopy (A1R, Nikon, Tokyo, Japan). Annexin V staining was performed according to the manufacturer's instructions.
Statistical Analysis
Statistical significance between and among multiple groups was evaluated using Student's t test, Welch's t test, or Tukey's test after F test or analysis of variance. Data are presented as the means ± S.E.
RESULTS
Molecular Cloning of Alternatively Spliced Variants of KCa3.1 in Human, Murine, and Rat Thymus
We first examined KCa3.1 protein expression in the mouse thymus and spleen by Western blotting using anti-KCa3.1C antibody. Unexpectedly, band signal with a smaller molecular weight (around 30 kDa) than predicted from mouse KCa3.1 (around 50 kDa) was detected, and disappeared by preincubation of primary anti-KCa3.1C antibody with excess antigen (supplemental Fig. S1). We then designed specific PCR primers to isolate full-length KCa3.1 from human thymus (see supplemental Table S1), and two different fragments were detected (data not shown). As in the results from DNA sequencing, we identified novel KCa3.1 spliced variants, named hKCa3.1b (Fig. 1A, amino acid sequences, and supplemental Fig. S2A, nucleotide sequences). In addition, from much less abundant PCR fragments, we also identified two additional KCa3.1 spliced variants, hKCa3.1c and hKCa3.1d (Fig. 1A and supplemental Fig. S2A). Ghanshani et al. (25) have shown that human KCa3.1 is composed of nine exons on chromosome 19q13.2. On the basis of genomic sequence information of human KCa3.1 (GenBankTM accession number, NT_011109), hKCa3.1b lacked partial sequences of exon 2 and/or exon 3 compared with original KCa3.1 (hKCa3.1a) (supplemental Fig. S2D), resulting in the truncation of N-terminal transmembrane segments from S1 to S3. We also isolated rodent KCa3.1 spliced variants, named mKCa3.1b (Fig. 1B and supplemental Fig. S2B) and rKCa3.1b (Fig. 1C and supplemental Fig. S2C) from mouse and rat thymus. mKCa3.1b lacked exon 3 but gained a novel exon 4, exon 4a, compared with the original mKCa3.1, mKCa3.1a, resulting in the truncation of N-terminal sequences (S1–S5) (supplemental Fig. S2Db). rKCa3.1b lacked exon 2 and partial sequences of exon 1 and exon 3, also resulting in the truncation of N-terminal sequences (S1–S4). We also found an additional rKCa3.1 variant, rKCa3.1c, with different C-terminal sequences from original rKCa3.1a, lacking exon 7 and exon 8; however, rKCa3.1c was a much less abundant isoform than rKCa3.1a as a result of competitive PCR assay (data not shown).
FIGURE 1.
Alternatively spliced variants of KCa3.1. The alignment of the amino acid sequences of human (A), mouse (B), and rat (C) KCa3.1 (hKCa3.1, mKCa3.1, and rKCa3.1) spliced variants. Residues identical to the original KCa3.1, KCa3.1a, are shown as asterisks, and dashes indicate gaps introduced into the sequence to improve alignment. S1–S6 and P indicate the transmembrane segment 1–6 and pore-forming domain, respectively.
Transcriptional Expression of KCa3.1 Spliced Variants in Human, Mouse, and Rat Tissues
Human KCa3.1 transcripts are expressed in a variety of peripheral tissues, e.g. thymus, spleen, placenta, prostate, lung, stomach, urinary bladder, and colon (12, 13). To identify the relative expression levels of KCa3.1 spliced variants in human, mouse, and rat tissues, we performed real time PCR analyses using specific primers for the following: (i) hKCa3.1a/b; (ii) hKCa3.1a; (iii) mKCa3.1a/b; (iv) mKCa3.1a; (v) mKCa3.1b; (vi) rKCa3.1a/b; and (vii) rKCa3.1a (see supplemental Table S1). In human tissues, expressions of KCa3.1a and KCa3.1b isoforms (KCa3.1a/b) relative to an internal standard, GAPDH, were 0.045, 0.014, 0.049, 0.053, 0.027, and 0.011 in the thymus, spleen, lymph node, placenta, colon, and urinary bladder (bladder), whereas hKCa3.1a expressions were 0.035, 0.008, 0.006, 0.017, 0.010, and 0.008 (mean values in triplicate) (Fig. 2A). Taken together, hKCa3.1b transcripts were expressed in human tissues examined at various levels, and its predominant expression was observed in the lymph nodes and placenta. Additionally, in mouse tissues, mKCa3.1a expressions were 0.026 ± 0.005, 0.028 ± 0.005, 0.041 ± 0.008, 0.063 ± 0.005, and 0.037 ± 0.005 in the thymus, spleen, lymph node (mesenteric), colon, and urinary bladder, respectively (n = 5 for each), whereas mKCa3.1b expression was 0.023 ± 0.002, 0.017 ± 0.004, 0.009 ± 0.002, 0.003 ± 0.000, and 0.003 ± 0.000 (n = 5 for each) (Fig. 2B). Similar results were obtained in rat lymphoid tissues (supplemental Fig. S3A). Each PCR product was confirmed by DNA sequencing. Taken together, the N terminus lacking the KCa3.1 isoform, KCa3.1b, was also expressed at a significant level in the thymus, spleen, and lymph nodes of rodents.
FIGURE 2.
Quantitative real time PCR for KCa3.1 spliced variant expressions in humans (A) and mice (B) tissues. Values are shown for steady state transcripts relative to GAPDH in the same preparation. A, results are expressed as the means of triplicate analyses. B, results are expressed as the means ± S.E. (n = 5).
Protein Expression of KCa3.1 Spliced Variants in Human, Mouse, and Rat Lymphoid Tissues
We calculated the deduced molecular weight of human KCa3.1 spliced variants using GENETYX-Win software version 4.0 (Genetyx Corp., Tokyo, Japan). The deduced molecular masses in hKCa3.1a–d were 36.4, 47.6, 33.2, and 32.7 kDa, respectively. Because anti-KCa3.1 antibody obtained from Alomone Labs was raised against a peptide mapped at the C terminus of KCa3.1, anti-KCa3.1 antibody can recognize not only KCa3.1a but also newly identified KCa3.1 spliced variants. Western blot analyses showed two bands specific for anti-KCa3.1 antibody with molecular masses of 48 and 36 kDa in the plasma membrane and cytosolic fraction of human thymus, spleen, and lymph node (Fig. 3A). In particular, a strong 36-kDa band, which was almost identical to the predicted molecular mass of hKCa3.1b, was observed in the cytosolic fraction of human lymph nodes. Similarly, we calculated the deduced molecular mass of mouse and rat KCa3.1 spliced variants, and the calculated molecular masses in mKCa3.1a, mKCa3.1b, rKCa3.1a, and rKCa3.1b were 50.6, 25.0, 47.8, and 30.9 kDa, respectively. In mouse lymphoid tissues (thymus, spleen, and mesenteric lymph node), two bands specific for anti-KCa3.1 antibody with molecular masses of ∼50 and 25 kDa were observed in the plasma membrane and cytosolic fractions, respectively (Fig. 3B). Similarly, in rat lymphoid tissues, two bands specific for anti-KCa3.1 antibody with molecular masses of ∼50 and 30 kDa were observed in the plasma membrane and cytosolic fractions, respectively (supplemental Fig. S3B). These signals disappeared by preincubation with the excess antigen, as shown in supplemental Fig. S1 (data not shown), indicating that the N terminus lacking KCa3.1 isoforms was mainly distributed in the cytosolic region but not the plasma membrane in the lymphoid tissues examined. Similar results were obtained from at least three separate experiments.
FIGURE 3.
KCa3.1 protein expressions in thymus, spleen, and lymph node (mesenteric) of humans (A) and mice (B). Protein fractions from cytosol (C) and plasma membrane (M) were probed by immunoblotting with anti-KCa3.1-C antibody. Molecular mass standards are shown in kilodaltons on the left side of the panels. C, co-immunoprecipitation of KCa3.1b with KCa3.1a. C, panel a, plasma membrane protein fraction of mouse thymus was probed by immunoblotting (IB) with anti-KCa3.1-C antibody, which recognized the N-terminal region of KCa3.1 (anti-KCa3.1-N) (left lane). Right lane shows the membrane blotted with KCa3.1-N with excess antigen. C, panel b, mouse thymus protein lysate was immunoprecipitated with anti-KCa3.1-N and probed by immunoblotting with anti-KCa3.1-C, which recognized the C terminus of KCa3.1. Arrowheads indicate the migrating positions of KCa3.1a (50 kDa) and KCa3.1b (25 kDa). Molecular mass standards are shown in kilodaltons on the left side of the panels.
To determine whether KCa3.1a is immunoprecipitated with these KCa3.1 isoforms, we performed immunoprecipitation using anti-KCa3.1 antibody raised against peptide mapping at the N terminus of KCa3.1, recognizing KCa3.1a but not the N terminus lacking KCa3.1 isoforms (anti-KCa3.1-N) as a primary antibody for immunoprecipitation. We first confirmed whether anti-KCa3.1-N recognizes KCa3.1a alone. As shown in Fig. 3C, panel a, when anti-KCa3.1-N was used as the primary antibody in Western blot analysis, no band signals with 25 kDa, predicted molecular mass for mKCa3.1b, were observed in membrane fraction proteins from mouse thymus. Signals of 50 kDa, the predicted molecular mass for mKCa3.1a, disappeared by preincubation with excess antigen. We then confirmed the interaction between mKCa3.1a and mKCa3.1b using an in vitro binding assay with whole tissue lysates of mouse thymus. When immunoprecipitated proteins with anti-KCa3.1-N antibody were stained with anti-KCa3.1-C antibody in Western blotting, a band of 25 kDa for mKCa3.1b was readily detected (Fig. 3C, panel b). These findings suggest that KCa3.1a strongly interacts with the N terminus lacking KCa3.1 isoforms with a DIS motif in mammalian native tissues.
Cellular Localization of KCa3.1 Spliced Variants in HEK293 Cells
Green fluorescence protein (GFP) relatives, CFP and YFP, have been used extensively to study the subcellular localization of KCa3.1 spliced variants by laser-scanning confocal fluorescence microscopy in living cells. To investigate whether KCa3.1 spliced variants function as trafficking defect isoforms, we examined the cellular localization of hKCa3.1 spliced variants in HEK293 cells, which was transfected with CFP-tagged hKCa3.1a and/or YFP-tagged hKCa3.1b/c/d. Both fluorescence proteins were tagged to the N termini of KCa3.1 spliced variants. To confirm that the fluorescence protein tag itself does not affect KCa3.1a channel activity, we performed electrophysiological recordings in transiently CFP-tagged KCa3.1a-expressed HEK293 cells. When whole-cell patch clamp recordings using 500-ms ramps from −120 to 40 mV were performed to assess the level of IKCa currents under symmetric K+ conditions, clotrimazole-sensitive IKCa currents were observed in hKCa3.1a-transfected HEK cells, whereas clotrimazole-sensitive currents were not detectable in mock-transfected HEK293 cells (data not shown). These results are consistent with the functional properties of hKCa3.1a without CFP tags.
As shown in Fig. 4A, in CFP-hKCa3.1a-expressed HEK293 cells, CFP signals were localized along the plasma membrane (Fig. 4A, panel a); however, in YFP-hKCa3.1b-expressed HEK293 cells, YFP signals were observed in the whole area of the cells (Fig. 4A, panel b). Similar distribution was observed in YFP-hKCa3.1c- and YFP-hKCa3.1d-expressed HEK293 cells, respectively (Fig. 4A, panels c and d). Of importance, CFP-hKCa3.1a (green) and YFP-hKCa3.1b signals (red) were well overlapped (yellow) when CFP-hKCa3.1a and YFP-hKCa3.1b were co-expressed in HEK293 cells (Fig. 4B). Fig. 4, C and D, showed the quantitative analyses of CFP/YFP fluorescence intensities, showing that membrane trafficking of hKCa3.1a was defected by co-expression with hKCa3.1b. Similarly, the co-expression of CFP-hKCa3.1a with YFP-hKCa3.1c and YFP-hKCa3.1d showed the trafficking-defective effects of hKCa3.1c and hKCa3.1d in HEK293 cells, respectively (supplemental Fig. S4, A and B). We also tested the possibility that the co-localization of CFP-hKCa3.1a with YFP-hKCa3.1b/c/d was an artifact of bleeding through the emission of light from one fluorophore into the detection range of the other. Cells labeled with only one fluorophore did not result in the detection of light when the filter was set for the other fluorophore (data not shown). KCa3.1 can function as a cell volume regulator (7); however, the diameter of the KCa3.1a-expressed transfectant was not significantly larger than the other spliced isoform-expressed transfectants and native HEK cells (data not shown).
FIGURE 4.
Cellular distribution of human KCa3.1 in HEK293 cells. A, CFP-tagged hKCa3.1a-transfected HEK293 cells, CFP image (upper panel) and the profile on the dotted line (lower panel) are shown in green (panel a). In YFP-tagged hKCa3.1b/c/d-transfected HEK293 cells, YFP image (upper panel) and the profile of relative intensity (lower panel) are shown in red. B, CFP-tagged hKCa3.1a and YFP-tagged hKCa3.1b-cotransfected HEK293 cells, CFP (upper, left) and YFP (upper, right) images are shown in green and red, respectively, and the merged image (lower, left) and the profiles on the dotted line (lower, right) of CFP and YFP as yellow. Bar, 20 μm. C, summarized data from A. D, summarized data from B and supplemental Fig. S4. Data are expressed as the means ± S.E. **, p < 0.01 versus plasma membrane fractions.
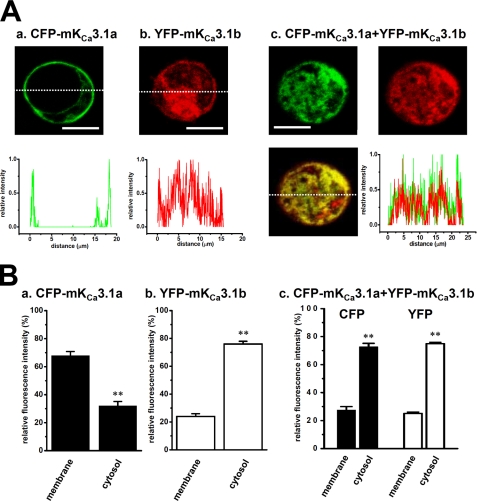
Similar to hKCa3.1b, mKCa3.1b had a DIS motif, but its molecular weight was smaller than hKCa3.1 (Fig. 1, A and B); therefore, we examined the trafficking-defective effects of mKCa3.1b on the cellular localization of mKCa3.1a in CFP-mKCa3.1a and YFP-mKCa3.1b-co-expressed HEK293 cells. Similar to the results in human KCa3.1 variants, the trafficking-deficient effects of mKCa3.1b were clearly observed in CFP-mKCa3.1a and YFP-mKCa3.1b-co-expressed HEK293 cells, as were the trafficking-deficient effects of mKCa3.1a by co-expression of mKCa3.1b (Fig. 5). These results strongly suggest that the N-terminal domain of KCa3.1 may include a critical membrane-trafficking motif and the N terminus lacking KCa3.1 isoforms may function as the dominant negatives of functional KCa3.1a. Additionally, we confirmed no imaging artifacts caused by reciprocal detection of light emitted by CFP or YFP.
FIGURE 5.
Cellular distribution of murine KCa3.1 in HEK293 cells. A, CFP-tagged mKCa3.1a-transfected HEK293 cells, CFP image (upper panel) and the profile on the dotted line (lower panel) are shown in green (panel a). In YFP-tagged mKCa3.1b-transfected HEK293 cells, YFP image (upper panel) and the profile on the dotted line (lower panel) are shown in red (panel b). In CFP-tagged mKCa3.1a and YFP-tagged mKCa3.1b-co-transfected HEK293 cells, CFP (upper, left) and YFP (upper, right) images are shown in green and red, respectively, and the merged image (lower, left) and the profiles on the dotted line (lower, right) of CFP and YFP in yellow (panel c). Bar, 20 μm. B, summarized data from A, panels a–c. Data are expressed as the means ± S.E. **, p < 0.01 versus plasma membrane fractions.
Dominant-negative Inhibition of KCa3.1a Currents by KCa3.1 Spliced Variants
To determine whether hKCa3.1b/c/d function as dominant-negative isoforms of functional KCa3.1a, electrophysiological experiments were performed using Xenopus oocyte expression systems. As shown in Fig. 6A, two electrode recordings using a ramp pulse from −120 to 40 mV were performed to assess the level of IKCa current under physiological conditions, and hKCa3.1a-expressing X. laevis oocytes showed ionomycin (0.5 μm) and a potential IKCa channel opener, DC-EBIO (10 μm)-activated and charybdotoxin (ChTX) (0.1 μm)-inhibited K+ currents (988.6 ± 112.3 nA at −30 mV, n = 5). ChTX-sensitive IKCa currents were inward at negative potentials, reversed at −80 mV, and showed little rectification. A voltage-gated ChTX-insensitive component was also present, which served as an internal control to demonstrate the specificity of hKCa3.1b/c/d dominant-inhibitory activity (data not shown). Ionomycin and DC-EBIO had no significant effect on the currents in mock- and water-injected oocytes (data not shown), suggesting that the currents observed in Fig. 6A are mostly caused by the expression of hKCa3.1a. When expressed with hKCa3.1b/c/d, they did not produce functional channels (n = 3 for each) (see Fig. 6, B–D, right columns). No ChTX-sensitive currents were observed in water-injected oocytes (data not shown).
FIGURE 6.
Dominant-negative inhibition of IKCa currents by hKCa3.1 spliced variants in the Xenopus oocyte expression system. Human KCa3.1 spliced variants were expressed in Xenopus oocytes by injection of the corresponding cRNA. A, representative traces from oocytes injected with the cRNA for hKCa3.1a. Ionomycin- and DC-EBIO-activated and ChTX-inhibited currents were studied by two-electrode voltage clamp. Current to voltage relations were derived by voltage ramp protocols under physiological conditions. B–D, co-expression of hKCa3.1b (B), hKCa3.1c (C), and hKCa3.1d (D) with hKCa3.1a largely reduced hKCa3.1a-induced IKCa currents. Current amplitudes were measured at −30 mV. Oocytes were injected with the cRNA for hKCa3.1a and hKCa3.1b/c/d at 1:0, 1:1, 1:10, and 0:1 ratios as indicated. Means ± S.E. of current amplitudes from 3 to 8 oocytes for each condition. *, **, p < 0.05, 0.01, significantly different as determined by Tukey's test.
We next ascertained whether hKCa3.1b/c/d exhibited the dominant-inhibitory activity of hKCa3.1a. Co-injection of hKCa3.1a and hKCa3.1b cRNAs in the ratios of 1:1 and 1:10 significantly decreased ChTX-sensitive currents at −30 mV in oocytes by 60.7% (n = 7) and 10.6% (n = 3), respectively (p < 0.05 and 0.01 versus hKCa3.1a alone, n = 8) (Fig. 6B). Similar to these results, co-injection of hKCa3.1a and hKCa3.1c cRNA in the ratios of 1:1 and 1:10 significantly decreased the currents by 15.7% (n = 7) and 0.3% (n = 3), respectively (p < 0.01 versus hKCa3.1a alone, n = 8) (Fig. 6C), and co-injection of hKCa3.1a and hKCa3.1d cRNA in the ratios of 1:1 and 1:10 significantly decreased the currents by 63.8% (n = 4) and 12.1% (n = 3), respectively (p < 0.05 and 0.01 versus hKCa3.1a alone, n = 5) (Fig. 6D). These clearly indicated that hKCa3.1b/c/d had dominant-negative effects on functional hKCa3.1a channel activity.
Up-regulation of mKCa3.1b in Mouse Thymocytes Isolated without Concanavalin-A Stimulation
To investigate the physiological role of the KCa3.1b spliced isoform in immune cell function, we examined whether the mKCa3.1 isoform expression was altered by the treatment with a T-lymphocyte-selective mitogen, Con-A. Mahaut-Smith and Mason (26) have demonstrated that the earliest events following mitogenic stimulation such as Con-A of thymocytes include an increase in intracellular Ca2+ and a concomitant activation of IKCa channel activity. Mouse thymocytes were isolated in the presence (Con-A(+)) and absence (Con-A(−)) of Con-A, and 60 min later, both cells were cultivated in the presence of Con-A. As shown in Fig. 7A, panel a, in Con-A(+) thymocytes, the expression levels of both mKCa3.1a and mKCa3.1b transcripts after 0, 1, and 2 day(s) (d0, d1, and d2) were almost similar to those in native thymus tissue (see Fig. 2B). Unexpectedly, in Con-A(−) thymocytes, the expression of mKCa3.1b transcripts (Fig. 7A, panel b, open columns) was up-regulated with 4–5-fold increase at d1 and d2, without change in mKCa3.1a transcript expression (Fig. 7A, panel b, solid columns) (n = 5 for each, p < 0.01 versus d0).
FIGURE 7.
Gene and protein expressions and cellular distribution of mKCa3.1 in Con-A(+) and Con-A(−) thymocytes. Quantitative real time PCR analysis of KCa3.1 spliced variant expressions in Con-A(+) (A, panel a) and Con-A(−) (A, panel b) thymocytes at day 0 (d0), day 1 (d1), and day 2 (d2). Values are shown for steady state transcripts relative to GAPDH in the same preparation. Results are expressed as the means ± S.E. (n = 5). **, p < 0.01 versus Con-A(+). B, KCa3.1 protein expressions in Con-A(−) thymocytes at d0 and d2. Protein fractions from plasma membrane (M) and cytosol (C) were probed by immunoblotting with anti-KCa3.1-C antibody. Summarized results were obtained as the optical density of each band signal relative to that of mKCa3.1a band signal in plasma membrane fraction at d0 (n = 5 for each). **, p < 0.01 versus d0. C, summarized data of cellular distribution of KCa3.1 in Con-A(+) and Con-(−) thymocytes. Data are expressed as the means ± S.E. (n = 16 for each). **, p < 0.01 versus Con-A(+). D, summarized data of the widths at half-maximum intensity in the plasma membrane of Con-A(+) and Con-A(−) thymocytes. Data are expressed as the means ± S.E. (n = 16 for each). **, p < 0.01 versus Con-A(+).
Similar to the results of Fig. 3B, Western blot assay showed two bands specific for anti-KCa3.1-C antibody with molecular masses of ∼50 and 25 kDa in the plasma membrane and cytosolic fraction of thymocytes at d0 (supplemental Fig. S5A, left panel). Different from thymocytes at d0, mKCa3.1b protein expression in plasma membrane fraction was clearly detected in Con-A(−) thymocytes at d2 (supplemental Fig. S5A, right panel). Densitometric analyses of anti-KCa3.1-C antibody responses showed significant increase in mKCa3.1b signal in plasma membrane fraction of Con-A(−) thymocytes at d2 (n = 5 for each, p < 0.01 versus d0) (Fig. 7B). Protein expression patterns of mKCa3.1a/b in Con-A(+) thymocytes at d2 were almost identical to those in thymocytes at d0 (data not shown). Positive signals for mKCa3.1a and mKCa3.1b disappeared by preincubation with the excess antigen (data not shown). Additionally, cellular distribution of mKCa3.1 was examined in Con-A(+) and Con-A(−) thymocytes by confocal laser microscope. Similar to Fig. 4C, the quantitative analyses of Alexa Fluor 488® fluorescence intensities were performed (Fig. 7C and supplemental Fig. S6). When anti-KCa3.1-C antibody was used as a primary antibody, relative fluorescence intensity in the plasma membrane of Con-A(+) thymocytes was significantly higher than in that of Con-A(−) thymocytes (Con-A(+), 62.7 ± 1.7%; Con-A(−), 53.6 ± 1.8%, n = 16 for each, p < 0.01) (Fig. 7C, solid columns, and supplemental Fig. S6, B and C). Moreover, the widths at half-maximum intensity below the plasma membrane of Con-A(+) and Con-A(−) thymocytes were calculated (supplemental Fig. S6, B and C). The width in Con-A(−) was significantly larger than that in Con-A(+): 0.67 ± 0.09 and 1.21 ± 0.14 mm in Con-A(+) and Con-A(−), respectively (n = 16 for each, p < 0.01) (Fig. 7D). The high level of KCa3.1 expression in plasma membrane were also observed in freshly isolated thymocytes (71.1 ± 2.3%, n = 10) (supplemental Fig. S6A). We performed similar experiments using anti-KCa3.1-N antibody, recognizing KCa3.1a alone; however, this antibody was not adequate for immunocytochemistry experiments (data not shown). Taken together, mKCa3.1b, which is up-regulated in Con-A(−) thymocytes, may interact with mKCa3.1a in plasma membrane and suppress functional expression of mKCa3.1a.
Effects of IKCa Channel Inhibition and Excess mKCa3.1b Expression on Cell Growth in Mouse Thymocytes
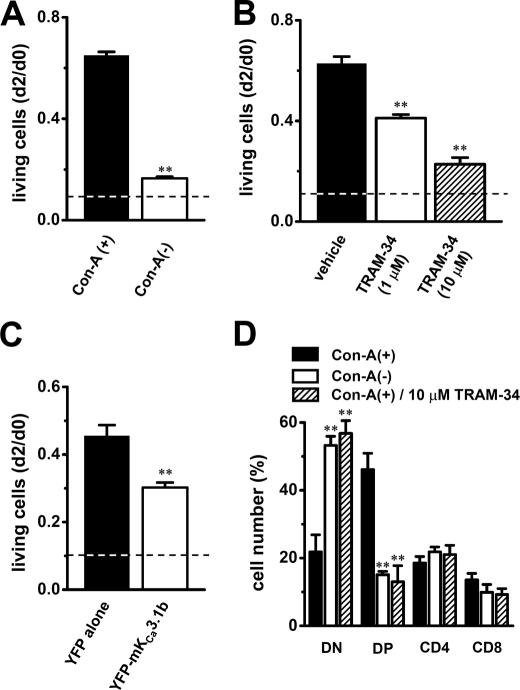
We employed an MTT proliferation assay to determine the effects of up-regulation of mKCa3.1b on Con-A-stimulated thymocyte growth. Values of living cells were expressed as the ratio of absorption spectra of the formed formazan at d2 to that at d0. When isolated thymocytes were cultivated in the absence of Con-A for 2 days, the value of living cells was 0.11 ± 0.01 (n = 6) (Fig. 8A, dashed line). Compared with Con-A(+) thymocytes (Fig. 8A, solid column, 0.65 ± 0.01, n = 6), Con-A(−) thymocytes exhibited significantly reduced levels of living cells at d2 (Fig. 8A, open column, 0.17 ± 0.01, n = 6, p < 0.01). Similarly, significant decrease in living cells was observed in Con-A(+) thymocytes by treatment with TRAM-34 in a concentration-dependent manner (1 μm, 0.41 ± 0.01; 10 μm, 0.23 ± 0.03) (n = 6 for each, p < 0.01 versus vehicle (0.1% DMSO, 0.63 ± 0.03)) (Fig. 8B). When cultivated in the absence of Con-A for 2 days (including 0.1% DMSO), the value of living cells was 0.14 ± 0.01 (n = 6) (Fig. 8B, dashed line). Several investigations have shown that selective pharmacological KCa3.1 blockade or KCa3.1 deficiency does not promote apoptosis in thymocytes and fibroblasts (5, 27). We quantified apoptotic cells by annexin-V labeling by confocal laser microscopy. In both Con-A(+) and Con-A(−) thymocytes, annexin V-positive cells were less than 1%, and no significant differences were observed. Similarly, application of TRAM-34 produced no increase in annexin V-positive cells. Cell morphology appeared well maintained in all groups examined as visualized by phase contrast microscopy.
FIGURE 8.
Effects of mKCa3.1b overexpression and IKCa current blockade on cell growth and CD4/CD8 phenotype expression in mouse thymocytes. Cell viability at d2 was evaluated by MTT cell proliferation assay. Value of living cells was expressed as the absorption spectra of the formed formazans and was calculated relative to that in thymocytes at d0. A, value of living cells in Con-A(−) thymocytes (open column) at d2 was compared with that in Con-A(+) thymocytes (solid column) (n = 6 for each). **, p < 0.01 versus Con-A(+). B, value of living cells in Con-A(+) thymocytes treated with 1 μm (open column) and 10 μm (hatched column) TRAM-34 at d2 was compared with that in Con-A(+) thymocytes treated with vehicle (0.1% DMSO) (n = 6 for each). **, p < 0.01 versus vehicle. C, value of living cells in Con-A(+) thymocytes transfected with YFP-mKCa3.1b (open column) at d2 was compared with that in Con-A(+) transfected with YFP alone (solid column) (n = 4 for each). **, p < 0.01 versus YFP alone. Dashed lines show value of living cells in thymocytes cultured in the absence of Con-A. D, reduced numbers of CD4(+)CD8(+) double-positive (DP) thymocyte populations in Con-A(−) and Con-A(+) treated with TRAM-34. Thymocytes were stained for CD4 and CD8 and examined by flow cytometry. The bar graph summarized each subset from separate (n = 5 for each) experiments as follows: CD4(−)CD8(−) double-negative (DN), CD4(+)CD8(+) double-positive (DP), CD4(+)CD8(−) single-positive (CD4), and CD4(−)CD8(+) single-positive (CD8). **, p < 0.01 versus Con-A(+).
We further employed an MTT proliferation assay using mKCa3.1b overexpressed thymocytes. Under the condition with maximal transfection efficacy, gene and protein expressions of YFP-mKCa3.1b were determined by real time PCR and Western blot examinations, respectively. As shown in supplemental Fig. S5, B and C, expressions of both mKCa3.1 transcripts and YFP-mKCa3.1b (∼45 kDa) proteins were significantly increased by mKCa3.1b overexpression. mKCa3.1-C primer pairs were used for real time PCR (see supplemental Table S1). Only specific band signals in supplemental Fig. S5C had disappeared by preincubation of primary antibody with control peptide (data not shown). As shown in Fig. 8C, YFP-mKCa3.1b-expressed Con-A(+) thymocytes (open column) exhibited significant reduction of living cells at d2 (YFP alone, 0.45 ± 0.03; YFP-mKCa3.1b, 0.30 ± 0.01, n = 4 for each, p < 0.01). Approximately 15% of YFP-positive cells were observed by flow cytometric analysis. Interestingly, excess expression of human KCa3.1b, hKCa3.1b with around 90% (28:31) sequence similarity to mKCa3.1b in DIS segment, did not produce a significant decrease in living cells (data not shown). Overexpression of mKCa3.1b produced less than 1% of annexin V-positive cells, and cell morphology appeared well maintained in all groups examined as visualized by phase contrast microscopy.
Flow Cytometric Analysis of Mice Thymocyte Populations
We next examined whether up-regulation of mKCa3.1b and the inhibition of IKCa channel activity affected thymocyte phonotypes in Con-A(−) and Con-A(+) treated with TRAM-34 using anti-CD4 and anti-CD8 antibodies by flow cytometry. The percentages of thymocytes showing CD4(−)CD8(−) double-negative (DN), CD4(+)CD8(+) double-positive (DP), CD4(+)CD8(−) single-positive (CD4), and CD4(−)CD8(+) single-positive (CD8) were summarized in Fig. 8D. In freshly isolated thymocytes, ∼75% of cells showed DP phenotypes (75.7 ± 5.8%, n = 3) (supplemental Fig. S7A). In Con-A(+) thymocytes, the majority of DP phenotype cells was observed (46.1 ± 4.8%, n = 5) (Fig. 8D, supplemental Fig. S7B); however, in Con-A(−) thymocytes, the differentiation into DN phenotype cells was observed, which represent a minor population in the freshly isolated and Con-A(+) thymocytes (DN: 53.2 ± 2.7%, n = 5, p < 0.01 versus Con-A(+)) (Fig. 8D and supplemental Fig. S7C). Similarly, the significant decrease in number of DP phenotype cells was observed in Con-A(+) treated with 10 μm TRAM-34 (13.0 ± 4.7%, n = 4, p < 0.01). No significant differences were observed in CD4 and CD8 phenotype cells between Con-A(+) and Con-A(−) thymocytes (Fig. 8D). In thymocytes cultivated in the absence of Con-A for 2 days, over 80% DN phenotype cells were observed (data not shown). These results suggest that up-regulation of mKCa3.1b may be responsible for the differentiation of DP phenotype cells into DN ones, resulting in the cell growth arrest.
Inhibition of Interleukin-2 Transcription in Thymocytes by Up-regulation of mKCa3.1b
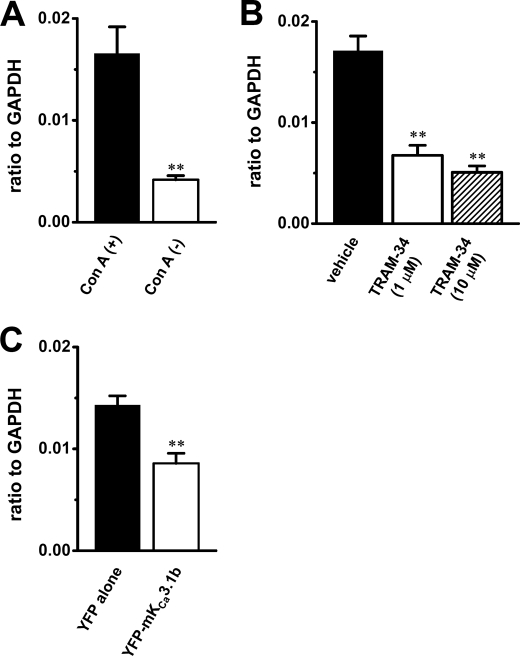
Interleukin-2 (IL-2) is essential for T cell activation and proliferation. We examined the effects of up-regulation of mKCa3.1b and inhibition of IKCa channel activity on the transcription of mice IL-2 (mIL-2) by real time PCR assay. As shown in Fig. 9A, ∼75% decrease in mIL-2 transcripts was observed in Con-A(−) thymocytes (n = 4, p < 0.01 versus Con-A(+)). Additionally, inhibition of IKCa channel activity by TRAM-34 (1 and 10 μm) induced the significant decrease in mIL-2 transcription in a concentration-dependent manner (∼60 and 70% inhibition, respectively) (n = 4 for each, p < 0.01 versus vehicle) (Fig. 9B). In Con-A(+) thymocytes overexpressing YFP-mKCa3.1b, a significant decrease in mIL-2 transcripts was also observed (∼40% inhibition) (n = 4, p < 0.01 versus YFP alone) (Fig. 9C). These suggest that the decrease in mIL-2 transcription may be at least in part involved in the cell growth arrest in thymocytes overexpressing mKCa3.1b.
FIGURE 9.
Quantitative real time PCR for interleukin-2 expression in mouse thymocytes. A, closed and open columns show interleukin-2 expression in Con-A(+) and Con-A(−) thymocytes, respectively. Values are shown for steady state transcripts relative to GAPDH in the same preparation (n = 4 for each, p < 0.01). B, closed, open, and hatched columns show interleukin-2 expression in Con-A(+) thymocytes treated with vehicle (0.1% DMSO), 1 μm TRAM-34, and 10 μm TRAM-34, respectively (n = 4 for each, p < 0.01 versus vehicle). C, closed and open columns show interleukin-2 expression in Con-A(+) thymocytes transfected with pEYFP-C (YFP alone) and pEYFP-mKCa3.1b (YFP-mKCa3.1b), respectively (n = 4 for each, p < 0.01).
DISCUSSION
In this study, we identified the N terminus lacking spliced variants of the intermediate conductance Ca2+-activated K+ channel, KCa3.1 (KCa3.1-ΔN), from lymphoid tissues of human and rodents. KCa3.1-ΔN isoforms have dominant-negative effects on membrane trafficking and the channel activity of functional KCa3.1a, suggesting that, in addition to the up-regulation of KCa3.1a, down-regulation of KCa3.1-ΔN may also be associated with the mechanisms underlying the enhancement of IKCa channel activity in activated T-lymphocytes. Additionally, we indicated that up-regulation of mKCa3.1b in mouse thymocytes differentiated CD4(+)CD8(+) DP phenotype thymocytes into CD4(−)CD8(−) DN ones, which represent a minor population in the native thymus, and suppressed Con-A-stimulated thymocyte growth by down-regulation of mIL-2 transcripts. The role of KCa3.1b in mouse thymocytes was also confirmed by generating mKCa3.1b-overexpressing thymocytes.
N Terminus of KCa Critical for Channel Trafficking to Plasma Membrane
In voltage-gated K+ (Kv) channels, it is well known that the cytoplasmic N- and C-terminal domains modify channel gating, membrane trafficking, and protein degradation (28–30). Several investigations have shown that the N-terminal domains of KCa3.1 and KCa2.x with a similar intron-exon structure to KCa3.1 play an important role for channel assembly and membrane trafficking using the N terminus lacking recombinant mutants and N-terminal polypeptides (6, 14, 19, 20, 31–33); however, KCa3.1 alternative splicing has not been identified in native tissues. This is the first identification of KCa3.1-ΔN variants significantly expressed in human and rodent lymphoid organs (Figs. 1–3, andsupplemental Fig. S3). Alternative splicing patterns differed among species in various tissues (supplemental Fig. S2); however, the most dominant was commonly KCa3.1b in human and rodent lymphoid tissues. KCa3.1-ΔN exerted the membrane-trafficking inhibition of KCa3.1a proteins (Figs. 4 and 5 and supplemental Fig. S4) and the dominant-negative effects of its activity (Fig. 6) by the formation of interaction partners with KCa3.1a (Fig. 3C). This inhibitory effect of alternative splicing on IKCa channel activity may be involved in the abnormalities in T cell function and its related diseases by KCa3.1 dysfunction. Indeed, disruption of KCa3.1 channel function by up-regulation of mKCa3.1b and pharmacological blockade of IKCa channel activity was associated with the cell growth arrest and the cell differentiation into CD4/CD8 subsets in mouse thymocytes (Fig. 8).
Different from the cellular distribution of mKCa3.1b proteins in mKCa3.1a/b-co-expressed HEK293 cells (Fig. 5, A, panel c, and B, panel c), mKCa3.1b proteins in Con-A(−) thymocytes distributed to plasma membrane fraction at a relatively abundant level (Fig. 7, B and C, and supplemental Fig. S5A). We identified the interacting proteins with the C-terminal domain of KCa3.1 using the yeast two-hybrid system, and we isolated a cytoskeletal protein, spectrin α, nonerythrocytic 1 (SPTAN1) and a targeting protein to plasma membrane, FYN tyrosine kinase.3 In lymphocytes, spectrin is aggregated to a discrete region of plasma membrane (34). Therefore, this discrepancy of KCa3.1b distribution can be explained by the interaction of the C terminus of KCa3.1b with cytoskeletal and membrane-targeting proteins in native thymocytes. Indeed, KCa3.1 proteins in Con-A(−) thymocytes distributed in a wider area below the plasma membrane, in comparison with those in Con-A(+) thymocytes (Fig. 7D).
Possible Pathophysiological Significances of KCa3.1 Alternative Splicing
Alternative splicing can result in significant changes in the original function in various disease pathogeneses; for example, the N-terminal truncated isoform of KCa2.3, KCa2.3–1B, is implicated in schizophrenia as a potent dominant-negative regulator of KCa2.3 (18). In a chronic inflammatory disease, multiple sclerosis and experimental autoimmune encephalomyelitis animal model with similarities to multiple sclerosis in pathogenicity, alternative splicing of several genes has been reported (35–37). Functional regulation of immune response-related molecules by alternative splicing during cell proliferation and differentiation is therefore expected to provide new therapeutic strategies for the treatment of immune diseases (38). KCa3.1 is involved in the proliferation and differentiation of naive and central memory T-lymphocytes (39–41), and also relatively higher levels of IKCa channel activity are detected in Th1 cells compared with Th2 cells (42). Cell type-specific and disease-associated alternative splicing of KCa3.1 has not been found yet; however, the increase in IKCa channel activity by the down-regulation of KCa3.1-ΔN may have potential in the pathogenesis of Th1-mediated autoimmune diseases, such as multiple sclerosis and delayed-type hypersensitivity.
In addition, KCa3.1 gene expression is positively regulated by activator protein-1 (AP-1, Fos/Jun complex) and Ikaros-2 and negatively regulated by repressor element 1-silencing transcription factor (REST) (40, 43). Recent studies have reported that N-terminal spliced variants of Ca2+- and Na+-activated K+ channels (Slo/KCa1.1 and Slack/KCa4.1) are regulated by alternative promoters (44, 45). As shown in Figs. 2 and 3, the expression pattern of KCa3.1a and KCa3.1b was quite different between the thymus and mesenteric lymph node, suggesting that the alternative transcriptional regulators of KCa3.1 described above may be responsible for the tissue-specific and cell type-specific expressions of KCa3.1 alternative transcripts. A recent study has shown that alternative splicing of KCa1.1 is post-transcriptionally regulated by microRNA (46). Future investigations will provide new insight into post-transcriptional regulation of KCa3.1 splicing by microRNA.
Membrane depolarization induced by IKCa channel blockade suppresses Ca2+ influx through a Ca2+ release-activated Ca2+ channel in T-lymphocytes (47, 48). Membrane depolarization also causes up-regulation of cyclin-dependent kinase inhibitor p27Kip1 and a subsequent cell cycle arrest in phase G0/G1 in several types of cells, including T-lymphocytes (49–51). These are strongly associated with the suppression of IL-2 transcription/production, and thereby T cell proliferation and differentiation are inhibited (50–53). In this study, we indicated that up-regulation of mKCa3.1b and pharmacological blockade of IKCa channel activity suppressed cell growth, IL-2 transcription, and maintenance of CD4(+)CD8(+) DP phenotype in mouse thymocytes (Figs. 8 and 9). In Con-A(−) and TRAM-34-treated Con-A(+) thymocytes, expression of p27Kip1 transcripts was ∼2-fold up-regulated, compared with that in Con-A(+) thymocytes (supplemental Fig. S8). These results suggest that KCa3.1b may be responsible for the fine-tuning of such signaling pathways in T-lymphocytes and may play a key regulatory role in governing the termination of immune response in T-lymphocytes.
A balance between the translation and degradation of ion channels is also of importance to regulate its activity. It has been reported that the N termini of KCa2.3 and KCa3.1 play an important role in shuttling them to the proteasome for degradation with a different mechanism (54, 55). Gao et al. (54) have clarified that S3/S4 domains are essential for the degradation of KCa3.1 proteins. As shown in Fig. 1, KCa3.1b isolated from human and rodent thymus lacked S3 and/or S4 domains; therefore, the lack of these domains in KCa3.1b could relate to a discrepancy between the results of real time PCR and Western blotting examinations, particularly observed in mouse lymph nodes (Figs. 2B and 3B). In addition to the post-transcriptional regulation of KCa3.1, the differences in protein degradation between KCa3.1a and KCa3.1b may be an important mechanism underlying the physiological and pathophysiological regulation of IKCa channel activity.
In conclusion, this is the first time that novel alternative splicing of the KCa3.1 gene in the N terminus has been identified in the lymphoid tissues of humans and rodents with their evident trafficking-defective and functionally dominant-negative effects on the original KCa3.1a. The putative molecular mechanism underlying the generation of alternative splicing of KCa3.1 remains to be determined; however, the novel regulation of the alternative splicing of KCa3.1 may play important roles in physiological and pathophysiological settings. This study strongly suggests that the altered combinatorial splicing profiles of KCa3.1 transcripts may provide a different and new perspective in understanding their role in generating autoimmune disorders, cancer, and restenosis, and the dominant negatives may be new targets and markers for diseases generated by KCa3.1 dysfunction.
Supplementary Material
Acknowledgment
We thank Dr. Shimada (Osaka University, Japan) for the gift of the plasmid vector for Xenopus oocyte expression.
This work was supported by a Grant-in-aid for Scientific Research (C) 21590098 from the Ministry of Education, Culture, Sports, Science and Technology, Japan (to S. O.), the Salt Science Research Foundation Grant 10C6 (to S. O.), Takeda Science Foundation (to S. O.), a grant-in-aid for scientific research (B) from the Ministry of Education, Culture, Sports, Science and Technology (to Y. I.), and Grant-in-aid for Scientific Research on Priority Areas 20056027 from the Ministry of Education, Culture, Sports, Science and Technology (to Y. I.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S8 and Table S1.
The nucleotide sequence(s) reported in this paper has been submitted to the DDBJ/GenBankTM/EBI Data Bank with accession number(s) AB128983, AB128984, AB128985, AB128930, AB292802, and AB292803.
S. Ohya, unpublished data.
- IKCa
- intermediate conductance KCa channel
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- CFP
- cyan fluorescent protein
- h
- human
- m
- murine
- r
- rat
- DIS
- dominant-inhibitory segment
- ChTX
- charybdotoxin
- DN
- double-negative
- DP
- double-positive
- Con-A
- concanavalin-A
- DC-EBIO
- 5,6-dichloro-1-ethyl-1,3-dihydro-2H-benzimidazol-2-one.
REFERENCES
- 1. Ghatta S., Nimmagadda D., Xu X., O'Rourke S. T. (2006) Pharmacol. Ther. 110, 103–116 [DOI] [PubMed] [Google Scholar]
- 2. Stocker M. (2004) Nat. Rev. Neurosci. 5, 758–770 [DOI] [PubMed] [Google Scholar]
- 3. Logsdon N. J., Kang J., Togo J. A., Christian E. P., Aiyar J. (1997) J. Biol. Chem. 272, 32723–32726 [DOI] [PubMed] [Google Scholar]
- 4. Köhler M., Hirschberg B., Bond C. T., Kinzie J. M., Marrion N. V., Maylie J., Adelman J. P. (1996) Science 273, 1709–1714 [DOI] [PubMed] [Google Scholar]
- 5. Begenisich T., Nakamoto T., Ovitt C. E., Nehrke K., Brugnara C., Alper S. L., Melvin J. E. (2004) J. Biol. Chem. 279, 47681–47687 [DOI] [PubMed] [Google Scholar]
- 6. Fanger C. M., Rauer H., Neben A. L., Miller M. J., Rauer H., Wulff H., Rosa J. C., Ganellin C. R., Chandy K. G., Cahalan M. D. (2001) J. Biol. Chem. 276, 12249–12256 [DOI] [PubMed] [Google Scholar]
- 7. Elliott J. I., Higgins C. F. (2003) EMBO Rep. 4, 189–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jensen B. S., Strøbaek D., Olesen S. P., Christophersen P. (2001) Curr. Drug Targets 2, 401–422 [DOI] [PubMed] [Google Scholar]
- 9. Köhler R., Wulff H., Eichler I., Kneifel M., Neumann D., Knorr A., Grgic I., Kämpfe D., Si H., Wibawa J., Real R., Borner K., Brakemeier S., Orzechowski H. D., Reusch H. P., Paul M., Chandy K. G., Hoyer J. (2003) Circulation 108, 1119–1125 [DOI] [PubMed] [Google Scholar]
- 10. Chandy K. G., Wulff H., Beeton C., Pennington M., Gutman G. A., Cahalan M. D. (2004) Trends Pharmacol. Sci. 25, 280–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wulff H., Pennington M. (2007) Curr. Opin. Drug Discov. Devel. 10, 438–445 [PubMed] [Google Scholar]
- 12. Ishii T. M., Silvia C., Hirschberg B., Bond C. T., Adelman J. P., Maylie J. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 11651–11656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Joiner W. J., Wang L. Y., Tang M. D., Kaczmarek L. K. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 11013–11018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Miller M. J., Rauer H., Tomita H., Rauer H., Gargus J. J., Gutman G. A., Cahalan M. D., Chandy K. G. (2001) J. Biol. Chem. 276, 27753–27756 [DOI] [PubMed] [Google Scholar]
- 15. Monaghan A. S., Benton D. C., Bahia P. K., Hosseini R., Shah Y. A., Haylett D. G., Moss G. W. (2004) J. Biol. Chem. 279, 1003–1009 [DOI] [PubMed] [Google Scholar]
- 16. Snyders D. J. (1999) Cardiovasc. Res. 42, 377–390 [DOI] [PubMed] [Google Scholar]
- 17. Kolski-Andreaco A., Tomita H., Shakkottai V. G., Gutman G. A., Cahalan M. D., Gargus J. J., Chandy K. G. (2004) J. Biol. Chem. 279, 6893–6904 [DOI] [PubMed] [Google Scholar]
- 18. Tomita H., Shakkottai V. G., Gutman G. A., Sun G., Bunney W. E., Cahalan M. D., Chandy K. G., Gargus J. J. (2003) Mol. Psychiatry 8, 524–535 [DOI] [PubMed] [Google Scholar]
- 19. Joiner W. J., Khanna R., Schlichter L. C., Kaczmarek L. K. (2001) J. Biol. Chem. 276, 37980–37985 [DOI] [PubMed] [Google Scholar]
- 20. Syme C. A., Hamilton K. L., Jones H. M., Gerlach A. C., Giltinan L., Papworth G. D., Watkins S. C., Bradbury N. A., Devor D. C. (2003) J. Biol. Chem. 278, 8476–8486 [DOI] [PubMed] [Google Scholar]
- 21. Ohya S., Kuwata Y., Sakamoto K., Muraki K., Imaizumi Y. (2005) Am. J. Physiol. Heart Circ. Physiol. 289, H1635–H1642 [DOI] [PubMed] [Google Scholar]
- 22. Ohya S., Takii T., Yamazaki H. F., Matsumori M., Onozaki K., Watanabe M., Imaizumi Y. (1997) FEBS Lett. 401, 252–258 [DOI] [PubMed] [Google Scholar]
- 23. Deleted in proof.
- 24. Deleted in proof.
- 25. Ghanshani S., Coleman M., Gustavsson P., Wu A. C., Gargus J. J., Gutman G. A., Dahl N., Mohrenweiser H., Chandy K. G. (1998) Genomics 51, 160–161 [DOI] [PubMed] [Google Scholar]
- 26. Mahaut-Smith M. P., Mason M. J. (1991) J. Physiol. 439, 513–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Grgic I., Kiss E., Kaistha B. P., Busch C., Kloss M., Sautter J., Müller A., Kaistha A., Schmidt C., Raman G., Wulff H., Strutz F., Gröne H. J., Köhler R., Hoyer J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 14518–14523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hopkins W. F., Demas V., Tempel B. L. (1994) J. Neurosci. 14, 1385–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Levitan E. S., Takimoto K. (2000) Trends Cardiovasc. Med. 10, 317–320 [DOI] [PubMed] [Google Scholar]
- 30. Strang C., Cushman S. J., DeRubeis D., Peterson D., Pfaffinger P. J. (2001) J. Biol. Chem. 276, 28493–28502 [DOI] [PubMed] [Google Scholar]
- 31. Roncarati R., Decimo I., Fumagalli G. (2005) Mol. Cell. Neurosci. 28, 314–325 [DOI] [PubMed] [Google Scholar]
- 32. Decimo I., Roncarati R., Grasso S., Clemens M., Chiamulera C., Fumagalli G. (2006) Biosci. Rep. 26, 399–412 [DOI] [PubMed] [Google Scholar]
- 33. Srivastava S., Choudhury P., Li Z., Liu G., Nadkarni V., Ko K., Coetzee W. A., Skolnik E. Y. (2006) Mol. Biol. Cell 17, 146–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pauly J. L., Bankert R. B., Repasky E. A. (1986) J. Immunol. 136, 246–253 [PubMed] [Google Scholar]
- 35. Lundmark F., Duvefelt K., Iacobaeus E., Kockum I., Wallström E., Khademi M., Oturai A., Ryder L. P., Saarela J., Harbo H. F., Celius E. G., Salter H., Olsson T., Hillert J. (2007) Nat. Genet. 39, 1108–1113 [DOI] [PubMed] [Google Scholar]
- 36. Gillett A., Maratou K., Fewings C., Harris R. A., Jagodic M., Aitman T., Olsson T. (2009) PLoS ONE 4, e7773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hu X., Barnum S. R., Wohler J. E., Schoeb T. R., Bullard D. C. (2010) Mol. Immunol. 47, 1692–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mourich D. V., Iversen P. L. (2009) Curr. Opin. Mol. Ther. 11, 124–132 [PubMed] [Google Scholar]
- 39. Wulff H., Beeton C., Chandy K. G. (2003) Curr. Opin. Drug Discov. Devel. 6, 640–647 [PubMed] [Google Scholar]
- 40. Ghanshani S., Wulff H., Miller M. J., Rohm H., Neben A., Gutman G. A., Cahalan M. D., Chandy K. G. (2000) J. Biol. Chem. 275, 37137–37149 [DOI] [PubMed] [Google Scholar]
- 41. Cahalan M. D., Chandy K. G. (2009) Immunol. Rev. 231, 59–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fanger C. M., Neben A. L., Cahalan M. D. (2000) J. Immunol. 164, 1153–1160 [DOI] [PubMed] [Google Scholar]
- 43. Cheong A., Bingham A. J., Li J., Kumar B., Sukumar P., Munsch C., Buckley N. J., Neylon C. B., Porter K. E., Beech D. J., Wood I. C. (2005) Mol. Cell 20, 45–52 [DOI] [PubMed] [Google Scholar]
- 44. Brenner R., Thomas T. O., Becker M. N., Atkinson N. S. (1996) J. Neurosci. 16, 1827–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Brown M. R., Kronengold J., Gazula V. R., Spilianakis C. G., Flavell R. A., von Hehn C. A., Bhattacharjee A., Kaczmarek L. K. (2008) J. Physiol. 586, 5161–5179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pietrzykowski A. Z., Friesen R. M., Martin G. E., Puig S. I., Nowak C. L., Wynne P. M., Siegelmann H. T., Treistman S. N. (2008) Neuron 59, 274–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Deleted in proof.
- 48. Hogan P. G., Lewis R. S., Rao A. (2010) Annu. Rev. Immunol. 38, 491–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Boussiotis V. A., Freeman G. J., Taylor P. A., Berezovskaya A., Grass I., Blazar B. R., Nadler L. M. (2000) Nat. Med. 6, 290–297 [DOI] [PubMed] [Google Scholar]
- 50. Ghiani C. A., Yuan X., Eisen A. M., Knutson P. L., DePinho R. A., McBain C. J., Gallo V. (1999) J. Neurosci. 19, 5380–5392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dohda T., Maljukova A., Liu L., Heyman M., Grandér D., Brodin D., Sangfelt O., Lendahl U. (2007) Exp. Cell Res. 313, 3141–3152 [DOI] [PubMed] [Google Scholar]
- 52. Wonderlin W. F., Strobl J. S. (1996) J. Membr. Biol. 154, 91–107 [DOI] [PubMed] [Google Scholar]
- 53. Di L., Srivastava S., Zhdanova O., Ding Y., Li Z., Wulff H., Lafaille M., Skolnik E. Y. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 1541–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gao Y., Chotoo C. K., Balut C. M., Sun F., Bailey M. A., Devor D. C. (2008) J. Biol. Chem. 283, 9049–9059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gao Y., Balut C. M., Bailey M. A., Patino-Lopez G., Shaw S., Devor D. C. (2010) J. Biol. Chem. 285, 17938–17953 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.