Abstract
The conformational changes in the agonist binding domain of the glycine-binding GluN1 and glutamate-binding GluN2A subunits of the N-methyl d-aspartic acid receptor upon binding agonists of varying efficacy have been investigated by luminescence resonance energy transfer (LRET) measurements. The LRET-based distances indicate a cleft closure conformational change at the GluN1 subunit upon binding agonists; however, no significant changes in the cleft closure are observed between partial and full agonists. This is consistent with the previously reported crystal structures for the isolated agonist binding domain of this receptor. Additionally, the LRET-based distances show that the agonist binding domain of the glutamate-binding GluN2A subunit exhibits a graded cleft closure with the extent of cleft closure being proportional to the extent of activation, indicating that the mechanism of activation in this subunit is similar to that of the glutamate binding α-amino-5-methyl-3-hydroxy-4-isoxazole propionate and kainate subtypes of the ionotropic glutamate receptors.
Keywords: Fluorescence Resonance Energy Transfer (FRET); Glutamate Receptors Ionotropic (AMPA, NMDA); Ion Channels; Membrane Trafficking; Neurotransmitter Receptors
Introduction
Glutamate receptors are the main mediators of excitatory signal transmission in the mammalian central nervous system. Glutamate binding to an extracellular domain in the receptor triggers the formation of a cation-permeable transmembrane channel in the receptor. The structures of the isolated agonist binding domain of the three subtypes of glutamate receptors have provided the first insight into the conformational changes and mechanism by which the agonist could control the activation and desensitization of the channel (1–10). However, most of the structures are of the α-amino-5-methyl-3-hydroxy-4-isoxazole propionate (AMPA) subtype, for which currently there are >60 structures in various ligated states (1, 8, 11). There is also significant insight into the dynamic state of the agonist binding domain for the AMPA receptors and on the role of specific agonist-protein interactions (9, 12–26). These studies show that for a number of cases the agonist binding domains (ABDs)3 of the AMPA receptors show a graded cleft closure with the extent of cleft closure correlating to the extent of activation (efficacy) of the agonist. Similar investigations on the kainate receptors also show a similar mechanism of activation. The NMDA subtype of the glutamate receptor, on the other hand, is the least studied in terms of structures (2, 3, 27). These receptors are different from the other subtypes as they are obligate heteromers of the glycine-binding GluN1 or GluN3 subunits and the glutamate-binding GluN2 (28–30), whereas non-NMDA receptors such as AMPA and kainate receptors can form functional homotetrameric channels activated solely by glutamate. For the NMDA receptors, the structures of the agonist binding domain of the GluN1 subunit bound to full agonist glycine and partial agonists such as d-cycloserine showed no significant differences (2, 27). Based on these results, it has been suggested that the NMDA receptors could follow a two-state model, where the cleft exists in either an open or closed state, and that the different coupling efficiencies between the ABD and the activation of the channel is due a shift in the equilibrium between these two states. However, this conclusion is based on a limited number of structures. Additionally, it is important to note that the structures of all of the partial agonists showed similar closed cleft conformations and no open state structures, contrary to what would be expected for a two-state model. If there is a shift in the equilibrium, one would expect to see at least some of these partial agonist-bound structures in the open form (assuming that the crystallographic constraints are not playing a role). Apart from the glycine-binding GluN1 subunit there is currently only one structure of the GluN2 subunit, the glutamate-bound form (3). Hence, it is still unknown whether the GluN2 subunit exhibits a graded cleft closure similar to that observed to the homologous glutamate-binding AMPA and kainate receptors. Here, we have used LRET-based measurements to study the extent of cleft closure in the GluN2A and GluN1 subunits of the NMDA receptors with agonists of varying efficacy.
EXPERIMENTAL PROCEDURES
Modifications Introduced in the NMDA Receptor
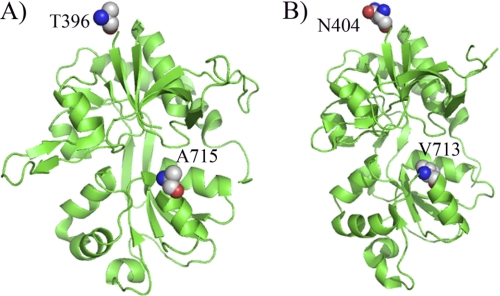
For studying the mechanism of cleft closure in the NMDA receptor, we modified GluN1 and GluN2A subunits of the NMDA receptor to eliminate the single non–disulfide-bonded accessible cysteines, Cys2 and Cys459, in GluN1 and Cys204, Cys399, and Cys460 in GluN2A by mutating them to a serine residue. These modified subunits, referred to as GluN1* and GluN2A*, were used for LRET investigations by introducing cysteines that can then be attached to donor and acceptor fluorophores to measure the extent of cleft closure in GluN1* and GluN2A* subunits of NMDA receptor, respectively. Mutations were introduced into the plasmids using the Stratagene QuikChange Site-directed Mutagenesis kit, and the integrity of the plasmid after mutagenesis was verified by sequencing. The sites tagged for LRET measurements are displayed in the GluN1-GluN2A crystal structure (Fig. 1).
FIGURE 1.
Sites that were tagged in the (A) GluN1* and (B) GluN2A* subunit to probe for cleft closure conformational changes using LRET measurements are highlighted in the crystal structure of the ABD of GluN1 bound to glycine and GluN2A bound to glutamate.
Receptor Expression and Tagging
Isolation and maintenance of oocytes were performed as described previously in Mg2+-free storage solution (31). The GluN1*:GluN2A* mutant receptors were expressed in Xenopus laevis oocytes by injecting the RNA (mMessage mMachine T7 kit; Ambion) in a 1:2 ratio, respectively. Injection, preblocking of oocytes and membrane preparations were performed as previously detailed (31). To measure background LRET signal, 1 unit of thrombin (Calbiochem) was added to the membrane preparation and allowed to digest for 2–3 h at 4 °C. LRET lifetimes were recorded after digestion with thrombin and subtracted from the nondigested signal to obtain the final specific LRET lifetime for the modified NMDA receptor protein.
Fluorophores
ATTO 465 maleimide was purchased from Sigma-Aldrich, the maleimide derivative of terbium chelate was purchased from Invitrogen, and the fluorescein maleimide was purchased from Biotium.
Fluorescence Spectroscopy
A cuvette-based fluorescence lifetime spectrometer QuantaMaster model QM3-SS (Photon Technology International) was used for performing fluorescence measurements. The excitation source was a high power pulsed xenon lamp. Additionally, the collected emitted light was passed through a monochromator onto a detector. Using a Peltier TE temperature controller, the sample was held at a constant 15 °C. Fluorescan software was used to collect data (Photon Technology International), and Origin 4.0 software was used to analyze the data (OriginLab Corp.). Data from three to four sets of data were averaged and fitted to obtain the lifetimes shown in Table 1. Each individual dataset was examined to ensure that similar trends were maintained. The donor only lifetimes were collected at 545 nm, and the sensitized emissions of acceptor were measured at 510 nm for ATTO 465 and 515 nm for fluorescein to obtain the LRET lifetimes. The fluorescence decay was fitted to a function that is a sum of discrete exponentials, and the goodness of the exponential fit was determined from the random residual distribution with a chi-square value being close to unity.
TABLE 1.
Fluorescence lifetimes and distances for GluN1*:GluN2A* receptors
| Protein | Donor:Acceptor fluorophore pair | Ligated state | Donor lifetime | Sensitized emission lifetime | Distance |
|---|---|---|---|---|---|
| μs | μs | Å | |||
| GluN1*:GluN2A*N404C-Th, V713C | Terbium chelate:fluorescein | Apo | 1680 ± 40 | 520 ± 44 | 39.4 ± 0.6 |
| GluN1*:GluN2A*N404C-Th, V713C | Terbium chelate:fluorescein | Antagonist (DLAPV) | 1694 ± 21 | 498 ± 26 | 38.9 ± 0.4 |
| GluN1*:GluN2A* N404C-Th, V713C | Terbium chelate:fluorescein | HQ | 1628 ± 12 | 390 ± 25 | 37.1 ± 0.4 |
| GluN1*:GluN2A* N404C-Th, V713C | Terbium chelate:fluorescein | Glutamate | 1608 ± 8 | 295 ± 41 | 35.1 ± 0.8 |
| GluN1*T396 C-Th, A715C:GluN2A* | Terbium chelate:ATTO 465 | Glycine | 1681 ± 10 | 788 ± 22 | 35.0 ± 0.1 |
| GluN1*T396 C-Th, A715C:GluN2A* | Terbium chelate:ATTO 465 | ACPC | 1754 ± 5 | 766 ± 22 | 34.6 ± 0.2 |
| GluN1*T396 C-Th, A715C:GluN2A* | Terbium chelate:ATTO 465 | DCS | 1704 ± 15 | 720 ± 12 | 34.8 ± 0.1 |
R0 values were determined for each pair of donor and acceptor fluorophores as described previously (9). The R0 values measured for the following donor:acceptor pairs are as follows: terbium chelate:fluorescein is 45 Å, and terbium chelate:ATTO 465 is 36 Å. The time constants of donor fluorescence decay in the absence of acceptor and sensitized acceptor fluorescence in the presence of the acceptor, along with the R0 values were inserted into Förster's equation to calculate the distance between the donor and acceptor fluorophore. Two-tailed t test was performed for statistical analysis of differences in the measured LRET distances (Table 1), and a p value ≤0.05 was considered significant in our experiments.
Two-electrode Voltage Clamp
Two-electrode voltage clamp measurements were used to test the functionality of all of the constructs used in the LRET measurements. The recordings were performed as detailed previously (31). The dose-response curves for the wild type and mutants used in LRET measurements are shown in supplemental Fig. 1.
RESULTS AND DISCUSSION
Conformational Changes in the ABD of the GluN2A Subunit of NMDA Receptors
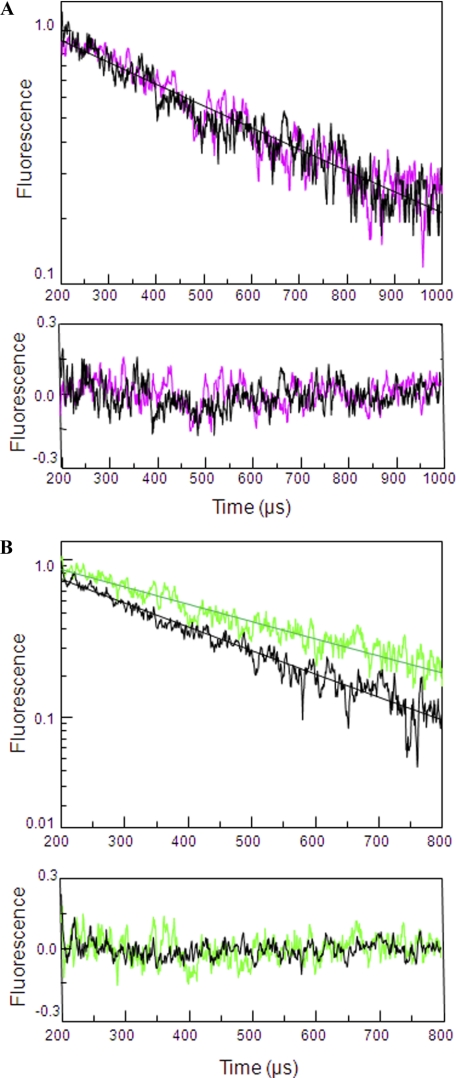
To study the mechanism of cleft closure at the ABD on the GluN2A subunit of NMDA receptors we used a wide range of agonists with varying extent of activation. LRET lifetimes were used to determine distances between cysteine mutants introduced at residue 404 present at the N terminus of the ABDs of GluN2A in domain 1 and residue 713 present in domain 2 (Fig. 1). The residues were chosen based on the subunit arrangement of the receptors (32, 33) such that a readout of cleft closure conformational change within the dimer is obtained. To minimize background LRET signal and hence determine the LRET specific to the GluN2A subunit of NMDA receptors, a thrombin cleavage site was introduced following the cysteine at site 404 in the GluN2A* subunit (N404C-Th). This allowed for selectively cleaving the donor or acceptor fluorophore tagged to the site. The LRET lifetime obtained after digestion with thrombin represents the background signal, which was subtracted from the original signal to obtain the final, specific LRET signal. The GluN1*:GluN2A*N404C-Th, V713C was tagged with maleimide derivatives of terbium chelate and fluorescein that served as donor and acceptor fluorophores, respectively. The tagged receptor was used to measure the distance between residues 404 and 713 within a GluN2A subunit (Fig. 2) under various ligated states. The LRET distances were measured under apo-, antagonist (DLAPV)-, partial agonist (homoquinolinic acid (HQ))-, and full agonist (glutamate)-bound conditions. Saturating concentrations of agonists/antagonist were used for the ligated state conditions. The saturating concentration of agonist was determined based on the dose-response curves generated under varying concentrations of agonists for the full agonist glutamate, whereas for other ligands data from previous literature were used (2, 27, 34). The LRET lifetime and distances are shown in Table 1. The LRET lifetimes obtained after subtracting the thrombin-digested background signal could be well represented by a single lifetime for the various ligated states. The LRET lifetime and distance for GluN1*:GluN2A*N404C-Th, V713C receptor decreased from 39.4 ± 0.6 Å for apo state to 35.1 ± 0.8 Å for the glutamate-bound state, indicating a statistically significant 4 Å decrease in cleft closure (two-tailed test, p = 0.0017). The lifetime and distance for the partial agonist (HQ) was at an intermediate distance (37.1 ± 0.4 Å) indicative of a partially closed cleft with a statistically significant difference of 2 Å between apo and HQ-bound form (two-tailed test, p = 0.0052). It is also to be noted that the antagonist (DLAPV)-stabilized conformation was similar to the open cleft seen for the apo form. The lifetime and distance for the antagonist-stabilized state (DLAPV) was 38.9 ± 0.4, similar in distance to the apo form. Thus, based on the lifetime and distance measurements it can be concluded that there is a negative correlation between the cleft closure distance measured between residues 404 and 713 on the GluN2A subunit and extent of activation with full agonists exhibiting shorter distance and partial agonist longer distances. These results support a cleft closure-based mechanism for partial agonism where the extent of cleft closure acts as the coupling mechanism by which agonists mediate extent of activation in GluN2A subunit of NMDA receptors.
FIGURE 2.
Cleft closure in the ABD of GluN2A* subunit. A, apo (black)- and antagonist (DLAPV in magenta)-bound LRET lifetimes for GluN1*:GluN2A*N404C-Th, V713C labeled with terbium chelate:fluorescein as measured by the sensitized emission of acceptor at 515 nm. The residuals for the above lifetime fits are shown below each measurement, and the y-axis is in linear scale. B, LRET lifetimes for GluN1*:GluN2A*N404C-Th, V713C labeled with terbium chelate:fluorescein as measured by the sensitized emission of acceptor at 515 nm under saturating concentrations of full agonist (glutamate in black) and partial agonist (HQ in green). The residuals for the above lifetime fits are shown below each measurement the y-axis in linear scale.
Conformational Changes in the ABD of the GluN1 Subunit of NMDA Receptors
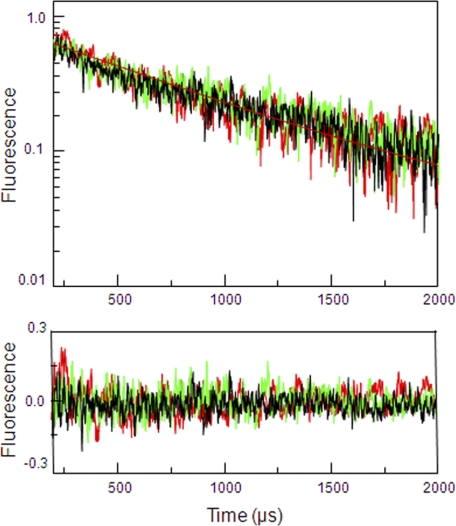
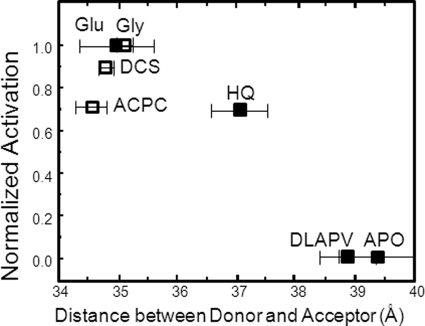
To draw a complete understanding of the conformational changes associated with agonist binding in NMDA receptors it is important that we measure the structural changes associated with the GluN1 subunit of NMDA receptors. Crystal structures of GluN1 ABD in complex with agonists of varying efficacies have been previously reported (27). These structures show no changes in the extent of cleft closure between agonists of different activation at the GluN1 ABD of NMDA receptors (27). This proved to be an anomaly to the widely held relationship between cleft closure and activation across the different members of the glutamate receptor family. Here, we have used LRET to determine the conformational changes in the ABD of GluN1 subunit in the full-length NMDA receptors. Similar to the LRET strategy described in the previous section for the GluN2A subunit, two sites were chosen in the modified GluN1* subunit to serve as donor and acceptor sites as a means of determining the extent of cleft closure. Residue 396 at the N terminus of the ABDs of GluN1 in domain 1 and residue 715 present in domain 2 were mutated to cysteines. This construct GluN1*T396C-Th, A715C:GluN2A* was expressed in Xenopus oocytes, and maleimide derivatives of terbium chelate:ATTO 465 were employed as donor and acceptor fluorophores. The extent of cleft closure was measured in the presence of saturating concentrations of full agonist (glycine) and partial agonists (d-cycloserine (DCS) and 1-aminocyclopropane-1-carboxylic acid (ACPC)). The final LRET lifetimes obtained upon subtracting the thrombin-digested background signal and distances are given in Fig. 3 and Table 1. The ensemble LRET distances obtained were in good agreement with the distances from the GluN1 ABD crystal structures (Table 2) and indicate that on average there is no change in the extent of cleft closure between agonist of varying efficacies (Fig. 4). The two-tailed test p value for the difference in LRET distances measured between the full agonist (glycine)- and partial agonists (DCS/ACPC)-bound forms is >0.05 and hence considered to be not statistically significant.
FIGURE 3.
Cleft closure in the ABD of GluN1* subunit. LRET lifetimes for GluN1*T396C-Th, A715C:GluN2A* labeled with terbium chelate:ATTO 465 were measured by the sensitized emission of acceptor at 510 nm under saturating concentrations of full agonist (glycine in black) and partial agonists (DCS in green, ACPC in red). The residuals for the above lifetime fits are shown below each measurement, and the y-axis is in linear scale.
TABLE 2.
Comparison of distances for GluN1*:GluN2A* receptors under various ligated state with respective available crystal structures of ABD
| Protein | Ligated state | Distance based on LRET | X-ray distance (Cα-Cα) |
|---|---|---|---|
| GluN1 396–715 | Glycine | 35.0 ± 0.1 | 34.0 |
| (1PB7) | |||
| GluN1 396–715 | ACPC | 34.6 ± 0.2 | 30.5 |
| (397–715, 1Y20) | |||
| GluN1 396–715 | DCS | 34.8 ± 0.1 | 33.9 |
| (1PB9) | |||
| GluN2A 404–713 | Glutamate | 35.1 ± 0.8 | 35.7 |
| (2A5T) |
FIGURE 4.
Dependence of cleft closure versus extent of activation for the ABD of GluN1* (open squares) and GluN2A* (filled squares) subunit.
It has been previously proposed that the GluN1 subunit could follow a two-state model where there is a shift in the equilibrium between the closed cleft and open cleft states. The LRET-based lifetimes do not support this mechanism as the lifetimes can be fit by a single exponential and do not require two exponentials. However, it should be noted that because the two states are not significantly different, it is likely that there is a large overlap between the states that the protein probes in the apo and ligated states, thus not allowing for a clear differentiation in the states and LRET lifetimes. Additionally, even in the closely related AMPA receptors there are several mutants and agonists that deviate from the cleft closure mechanism. Structural and spectroscopic investigations such as NMR (22, 35, 36, 38) and single molecule FRET investigations (37) of these mutants do not support a shift to a two-state model but indicate the role of other mechanisms such as the dynamics of the protein and differences at the level of specific interactions such as hydrogen bonds that contribute to differences in activation by different ligands. Also, it should also be noted that although there is a correlation between the average cleft closure and activation for the GluN2 subunit, the dynamics and specific interactions at the level of side chains may also play a role in activation for this subunit. Future NMR investigations and single molecule structural methods such as single molecule FRET should be able to shed light on the role of these in mediating the mechanism of agonism both at the GluN1 and GluN2 subunit of the NMDA receptors (37).
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant R01GM073102.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- ABD
- agonist binding domain
- ACPC
- 1-aminocyclopropane-1-carboxylic acid
- DCS
- d-cycloserine
- DLAPV
- DL-2-amino-5-phosphonopentanoic acid
- GluN1
- NMDA receptor subunit 1
- GluN2A
- NMDA receptor subunit 2A
- GluN3
- NMDA receptor subunit 3
- HQ
- homoquinolinic acid
- LRET
- luminescence resonance energy transfer
- Th
- thrombin.
REFERENCES
- 1. Armstrong N., Gouaux E. (2000) Neuron 28, 165–181 [DOI] [PubMed] [Google Scholar]
- 2. Furukawa H., Gouaux E. (2003) EMBO J. 22, 2873–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Furukawa H., Singh S. K., Mancusso R., Gouaux E. (2005) Nature 438, 185–192 [DOI] [PubMed] [Google Scholar]
- 4. Armstrong N., Jasti J., Beich-Frandsen M., Gouaux E. (2006) Cell 127, 85–97 [DOI] [PubMed] [Google Scholar]
- 5. Sun Y., Olson R., Horning M., Armstrong N., Mayer M., Gouaux E. (2002) Nature 417, 245–253 [DOI] [PubMed] [Google Scholar]
- 6. Mayer M. L. (2005) Neuron 45, 539–552 [DOI] [PubMed] [Google Scholar]
- 7. Nanao M. H., Green T., Stern-Bach Y., Heinemann S. F., Choe S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 1708–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mayer M. L., Armstrong N. (2004) Annu. Rev. Physiol. 66, 161–181 [DOI] [PubMed] [Google Scholar]
- 9. Ramanoudjame G., Du M., Mankiewicz K. A., Jayaraman V. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 10473–10478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Naur P., Vestergaard B., Skov L. K., Egebjerg J., Gajhede M., Kastrup J. S. (2005) FEBS Lett. 579, 1154–1160 [DOI] [PubMed] [Google Scholar]
- 11. Mayer M. L., Ghosal A., Dolman N. P., Jane D. E. (2006) J. Neurosci. 26, 2852–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jayaraman V., Keesey R., Madden D. R. (2000) Biochemistry 39, 8693–8697 [DOI] [PubMed] [Google Scholar]
- 13. Madden D. R., Thiran S., Zimmermann H., Romm J., Jayaraman V. (2001) J. Biol. Chem. 276, 37821–37826 [DOI] [PubMed] [Google Scholar]
- 14. Cheng Q., Thiran S., Yernool D., Gouaux E., Jayaraman V. (2002) Biochemistry 41, 1602–1608 [DOI] [PubMed] [Google Scholar]
- 15. Deming D., Cheng Q., Jayaraman V. (2003) J. Biol. Chem. 278, 17589–17592 [DOI] [PubMed] [Google Scholar]
- 16. Madden D. R., Cheng Q., Thiran S., Rajan S., Rigo F., Keinänen K., Reinelt S., Zimmermann H., Jayaraman V. (2004) Biochemistry 43, 15838–15844 [DOI] [PubMed] [Google Scholar]
- 17. Jayaraman V. (2004) Methods Enzymol. 380, 170–187 [DOI] [PubMed] [Google Scholar]
- 18. Cheng Q., Jayaraman V. (2004) J. Biol. Chem. 279, 26346–26350 [DOI] [PubMed] [Google Scholar]
- 19. McFeeters R. L., Oswald R. E. (2004) FASEB J. 18, 428–438 [DOI] [PubMed] [Google Scholar]
- 20. Cheng Q., Du M., Ramanoudjame G., Jayaraman V. (2005) Nat. Chem. Biol. 1, 329–332 [DOI] [PubMed] [Google Scholar]
- 21. Du M., Reid S. A., Jayaraman V. (2005) J. Biol. Chem. 280, 8633–8636 [DOI] [PubMed] [Google Scholar]
- 22. Ahmed A. H., Loh A. P., Jane D. E., Oswald R. E. (2007) J. Biol. Chem. 282, 12773–12784 [DOI] [PubMed] [Google Scholar]
- 23. Mankiewicz K. A., Jayaraman V. (2007) Braz. J. Med. Biol. Res. 40, 1419–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mankiewicz K. A., Rambhadran A., Du M., Ramanoudjame G., Jayaraman V. (2007) Biochemistry 46, 1343–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mankiewicz K. A., Rambhadran A., Wathen L., Jayaraman V. (2008) Biochemistry 47, 398–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fenwick M. K., Oswald R. E. (2008) J. Mol. Biol. 378, 673–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Inanobe A., Furukawa H., Gouaux E. (2005) Neuron 47, 71–84 [DOI] [PubMed] [Google Scholar]
- 28. Johnson J. W., Ascher P. (1987) Nature 325, 529–531 [DOI] [PubMed] [Google Scholar]
- 29. Benveniste M., Mayer M. L. (1991) Biophys. J. 59, 560–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Clements J. D., Westbrook G. L. (1991) Neuron 7, 605–613 [DOI] [PubMed] [Google Scholar]
- 31. Gonzalez J., Rambhadran A., Du M., Jayaraman V. (2008) Biochemistry 47, 10027–10032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rambhadran A., Gonzalez J., Jayaraman V. (2010) J. Biol. Chem. 285, 15296–15301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sobolevsky A. I., Rosconi M. P., Gouaux E. (2009) Nature 462, 745–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Erreger K., Geballe M. T., Kristensen A., Chen P. E., Hansen K. B., Lee C. J., Yuan H., Le P., Lyuboslavsky P. N., Micale N., Jørgensen L., Clausen R. P., Wyllie D. J., Snyder J. P., Traynelis S. F. (2007) Mol. Pharmacol. 72, 907–920 [DOI] [PubMed] [Google Scholar]
- 35. McFeeters R. L., Oswald R. E. (2002) Biochemistry 41, 10472–10481 [DOI] [PubMed] [Google Scholar]
- 36. Maltsev A. S., Ahmed A. H., Fenwick M. K., Jane D. E., Oswald R. E. (2008) Biochemistry 47, 10600–10610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Landes C. F., Rambhadran A., Taylor J. N., Salatan F., Jayaraman V. (2011) Nat. Chem. Biol. 7, 168–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Maltsev A. S., Oswald R. E. (2010) J. Biol. Chem. 285, 10154–10162 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.