Abstract
Sirt3 (silent mating type information regulation 2, homolog 3), a member of the sirtuin family of protein deacetylases with multiple actions on metabolism and gene expression is expressed in association with brown adipocyte differentiation. Using Sirt3-null brown adipocytes, we determined that Sirt3 is required for an appropriate responsiveness of cells to noradrenergic, cAMP-mediated activation of the expression of brown adipose tissue thermogenic genes. The transcriptional coactivator Pgc-1α (peroxisome proliferator-activated receptor-γ coactivator-1α) induced Sirt3 gene expression in white adipocytes and embryonic fibroblasts as part of its overall induction of a brown adipose tissue-specific pattern of gene expression. In cells lacking Sirt3, Pgc-1α failed to fully induce the expression of brown fat-specific thermogenic genes. Pgc-1α activates Sirt3 gene transcription through coactivation of the orphan nuclear receptor Err (estrogen-related receptor)-α, which bound the proximal Sirt3 gene promoter region. Errα knockdown assays indicated that Errα is required for full induction of Sirt3 gene expression in response to Pgc-1α. The present results indicate that Pgc-1α controls Sirt3 gene expression and this action is an essential component of the overall mechanisms by which Pgc-1α induces the full acquisition of a brown adipocyte differentiated phenotype.
Keywords: Adipocyte, Adipose Tissue, Adipose Tissue Metabolism, Sirtuins, Transcription Coactivators, Brown Adipose Tissue, Thermogenesis, Uncoupling Protein
Introduction
Brown adipose tissue plays a major role in the control of energy expenditure in mammals. The specific mitochondrial uncoupling that is characteristic of brown adipocytes creates a specialized cell type adapted to promoting energy expenditure in response to cold or overfeeding. In contrast, white adipocytes are specialized in the accumulation of metabolic energy in the form of lipids. Brown adipocytes respond to noradrenergic stimulation through β-adrenoreceptors in the cell surface, which bind norepinephrine and signal through adenylate cyclase to increase cAMP and activate protein kinase A. Ultimately, this signaling cascade lead to activation of hormone-sensitive lipase, induction of the expression of the gene for uncoupling protein-1 (Ucp1)2 and other genes involved in thermogenesis, and activation of the Ucp1-dependent uncoupling of mitochondria (1). Acquisition of the cellular machinery typical of brown adipocyte thermogenic function is a highly plastic process. In fact, pre-adipocytes or even white adipocytes may acquire brown adipocyte properties in response to developmental or environment regulators. Several molecular agents have been reported to promote brown adipocyte differentiation, central among them is Pgc-1α.
Pgc-1α is a transcriptional coactivator that plays a major role in the acquisition of the specific brown adipocyte phenotype. Pgc-1α coactivates nuclear receptors and transcription factors and thereby activates genes involved in thermogenesis (e.g. Ucp1), lipid oxidation, and mitochondrial oxidation, which are associated with the specific thermogenic function of brown adipose tissue (2). For instance, when white adipocytes are forced to express high levels of Pgc-1α, they acquire the features of brown adipocytes, including expression of the brown fat-specific Ucp1 gene (3). Cells lacking Pgc-1α acquire a partial brown adipocyte phenotype but remain insensitive to noradrenergic activation of thermogenesis (4). Thus, the control of Pgc-1α gene expression can ultimately determine the acquisition of a brown adipocyte phenotype in an adipose cell. The identification of factors that control Pgc-1α expression and activity is an important prerequisite for developing tools to modulate brown adipocyte activity in the organisms.
Sirtuins are protein deacetylases that act on histones, transcription factors, and transcriptional co-regulators, modulating their activities by regulating their degree of acetylation (5). Among them, Sirt3 is present in mitochondria and possibly in nuclei as well (6, 7). In mitochondria, Sirt3 controls fatty acid oxidation by deacetylating enzymes involved in fatty acid oxidation, such as long-chain acyl-CoA dehydrogenase (8) and acetyl-CoA synthase (9, 10). Other roles reported for Sirt3 include protection against cardiac hypertrophy (11, 12) and tumor suppression (13). Several reports have indicated that SIRT3 gene polymorphisms are associated with human aging (14, 15).
Sirt3 is highly expressed in brown adipose tissue, in contrast with its low expression in white fat. Sirt3 gene expression is impaired in brown fat of rodents under conditions of diminished thermogenic activity such as obesity, and it has been proposed that Sirt3 is involved in the control of cAMP-mediated gene expression in brown fat (16).
In the present study, we report that PGC-1α is a major controller of the transcription of the Sirt3 gene. By this means, it contributes to the acquisition of the thermogenic capacity of the brown adipose cell.
EXPERIMENTAL PROCEDURES
Cell Culture
Primary brown adipocytes were differentiated in culture as described previously (17). Preadipocytes were isolated from interscapular, cervical, and axillary brown adipose tissue depots from 21-day-old Swiss mice, or from litter-matched wild-type and Sirt3-null mice. Sirt3-null mice (strain name: B6; 129S5-SIRT3Gt(neo)218Lex) were obtained from MMRRC (Mutant Mouse Regional Resource Center) and had been backcrossed into the C57BL/6JOla Hsd line (Harlan) for six generations. Where indicated, differentiated brown adipocytes were treated for 6 h with 0.5 μm norepinephrine or for 24 h with 1 mm dibutyryl-cAMP. The HIB-1B brown adipocyte cell line was cultured as reported elsewhere (18). SGBS human adipocyte cells were cultured and differentiated as already reported (19), and experiments were performed on day 12 after induction of differentiation, a time at which more than 70% of cells were differentiated.
Mouse embryonic fibroblasts (MEFs) were obtained from 13-day-old embryos and were cultured and differentiated as described elsewhere (20) with some modifications. MEFs from wild-type and Sirt3-null mice were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS) at 37 °C in a humidified 5% CO2 atmosphere. Two days after reaching confluence, MEFs were differentiated into adipocytes in a DMEM containing 10% FBS, 0.5 mm 3-isobutyl-1-methylxanthine, 1.7 μm insulin, 1 μm dexamethasone, and 10 μm rosiglitazone. Beginning on day 10, MEFs were cultured in DMEM containing 1.7 μm insulin and 1 μm dexamethasone until lipid accumulation was observed. Experiments were performed when 80–90% of the cells were differentiated, determined based on the acquisition of adipocyte morphology. Where indicated, cells were exposed to the Errα antagonist/inverse agonist XCT-790 (Sigma) at a final concentration 5 μm for 48 h.
Western Blot Analysis
Cell extracts were prepared by homogenization in a buffer containing 20 mm NaHepes, pH 8.5, 25 mm MgCl2, 1% Igepal CA-630 (Sigma), 1 mm EDTA, a mixture of protease inhibitors (Complete-Mini, Roche Diagnostics) and 0.1% phenylmethylsulfonyl fluoride. Proteins (30 μg/lane) were separated by 12 or 8% SDS-PAGE and transferred to Immobilon-P membranes (Millipore). Immunological detection was performed with specific antibodies against mouse Sirt3 (ab56214, Abcam, UK) and Pgc-1α (H300, Santa Cruz Biotechnology, Inc., Santa Cruz, CA). For detection, an enhanced chemiluminescence system (ECL, Amersham Biosciences) was employed. The intensity of the signals was quantified by densitometry (Phoretics 1D Software, Phoretic International, UK). Coomassie Blue staining of the gels was performed to assess equal total protein loading.
Cloning of the Sirt3 Promoter and Sirt3 Promoter-Reporter Constructs
The Sirt3 promoter construct was created by amplifying a 978-base pair (bp) fragment of the mouse Sirt3 gene corresponding to the −956 to +22 bp upstream region by PCR using primers 5′-CTC AAG GGC AGG GCC AGA AAC C-3′ (forward) and 5′-CTG GAA TTC CAA TGC CAC AAC C-3′ (reverse). The fragment obtained was cloned into the PGEM-T Easy Vector (Promega, Madison, WI) and subsequently cloned into the PGL3-Basic Vector (Promega) using SmaI and MluI restriction enzymes (-956SIRT3-Luc). A shorter version (110 bp) of this construct was prepared by cutting with the KspI restriction enzyme to yield -85SIRT3-Luc. The -956SIRT3-Luc variants -956SIRT3-ERR1Mut, -956SIRT3-ERR2Mut, and -9565SIRT3-ERR1+2Mut containing point mutations were generated using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). Complementary oligonucleotides containing the desired mutation flanked by unmodified nucleotide sequence (5′-CGG GTT GCG GTC GTC AAC TTA ACC GCG TTC TTG ACT TCC GC-3′ for the ERR1 mutation, and 5′-GGG CAT GCT GGG AGC GTC AGC CTA GCA GCA CGG GTT GCG GTC G-3′ for the ERR2 mutation) were used. The fidelity of cloning and mutagenesis procedures were made by direct DNA sequencing.
Cell Transfection
For reporter assays, HIB-1B cells in 24-well plates were transfected with 0.3 μg of SIRT3-Luc reporter plasmid, 0.5 ng of the expression vector pRL-CMV (Promega), and where indicated, 0.06 μg of the expression vectors for PGC-1α, nuclear respiratory factor 2-α and -β (NRF2α and NRF2β), peroxisome proliferator-activated receptor-α and -γ, thyroid receptor-α and -β, alone or in combination with Pgc-1α. Cells were transfected using FuGENE6 (Roche Diagnostics) and incubated for 48 h prior to assaying for luciferase activity. Firefly and Renilla luciferase activities were measured in a Turner Designs Luminometer using the Dual Luciferase Reporter assay system (Promega). Firefly luciferase activity was expressed relative to Renilla luciferase activity to normalize for transfection efficiency. Each point was assayed in triplicate.
Adenoviral-mediated Gene Transfer
Adenoviral vectors expressing green fluorescent protein (GFP), Errα, Pgc-1α, and interfering small hairpin RNA (shRNA) for mouse Errα have been described (21, 22). For adenoviral-mediated gene transfer, differentiated SGBS adipocytes, MEF-derived adipocytes, or HIB-1 brown adipocytes were infected with adenoviral vectors driving Pgc-1α (AdCMV-PGC-1α, provided by Dr. B. Spiegelman), Errα, shRNA-ERRα, or GFP (AdCMV-GFP, control) at a multiplicity of infection of 100 for 4 h. Experiments were performed after further incubation in differentiation medium for 48 h. Transduction efficiency based on GFP fluorescence was ∼80%.
Quantification of Transcript Levels
Total RNA was extracted using NucleoSpin (Macherey Nagel, Düren, Germany). Reverse transcription was performed in a total volume of 20 μl using random hexamer primers (Applied Biosystems, Foster City, CA) and 0.5 μg of total RNA. Real time quantitative PCR was conducted in 20-μl reaction mixtures containing 1 μl of cDNA, 10 μl of TaqMan Universal PCR Master Mix (Applied Biosystems), and probes from Assays-on-Demand Gene Expression Assay Mix (Applied Biosystems). TaqMan Gene Expression Assays primers for the transcripts of murine genes Sirt3 (Mm00452129), Ucp1 (Mm004940969), Dio2 (Mm00515664), Fabp4 (Mm00445880), Pgc-1α (Mm00447183), and Errα (Mm00433143) and the human genes SIRT3 (Hs00202030), UCP1 (Hs00222453), DIO2 (Hs00255341), and FABP4 (Hs00609791) were used. Each assay was performed in duplicate, and the mean value was used to calculate mRNA expression for the gene of interest and the housekeeping reference gene (18S rRNA; Hs99999901). The amount of the gene of interest in each sample was normalized to that of the reference control using the comparative (2−ΔCT) method following the manufacturer's instructions.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation experiments (ChIP) were performed as described elsewhere (22). Differentiated brown adipocytes in primary culture and HIB-1B cells were used. When indicated, HIB-1B cells were transfected with luciferase reporter plasmids, as described above. Immunoprecipitation was carried out using anti-Pgc-1α (H300) antibody (Santa Cruz Biotechnology, Inc.), anti-HA antibody (ab9110, Abcam, Cambridge, UK), or an equal amount of an unrelated immunoglobulin (Sigma). Purified DNA was amplified using primers 5′-CAC GGA AGT GCT CGC TCA-3′ (forward) and 5′-GGG GAA GTT TAG CGG AAG TC-3′ (reverse) encompassing the proximal promoter region (from −107 to +18) of the Sirt3 gene, to generate a 125-bp fragment (proximal promoter region) or using primers 5′-AAG TGG CAG GCT CTT TGT GT-3′ (forward) and 5′-CAA AAG GCT CCA CCT GAA AG-3′ (reverse) to generate a 122-bp fragment encompassing an unrelated region, distant from the promoter (+17033/+17155) in the Sirt3 gene, used as control. Input DNA and immunoprecipitated DNA were analyzed by quantitative PCR using SYBR Green fluorescent dye. The protein-bound DNA was calculated as a ratio to input DNA. When indicated, bound fragments were amplified by PCR (30 cycles of 30 s at 95 °C, 30 s at 60 °C, and 30 s at 72 °C) and visualized electrophoresing on a 1.5% agarose gel with ethidium bromide staining.
RESULTS
Sirt3 Is Expressed in Association with Brown Adipocyte Differentiation and Is Required for the Induction of Thermogenesis-related Gene Expression
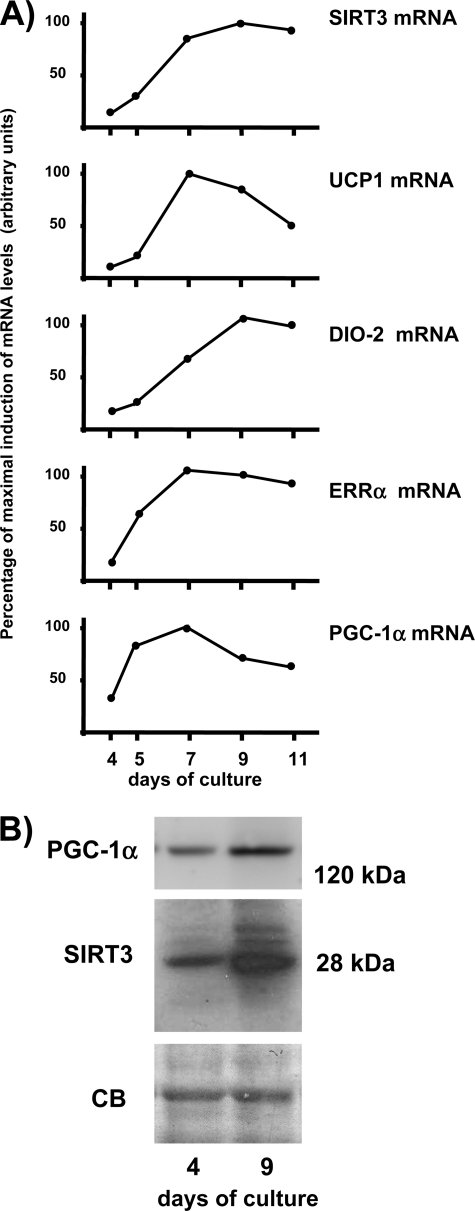
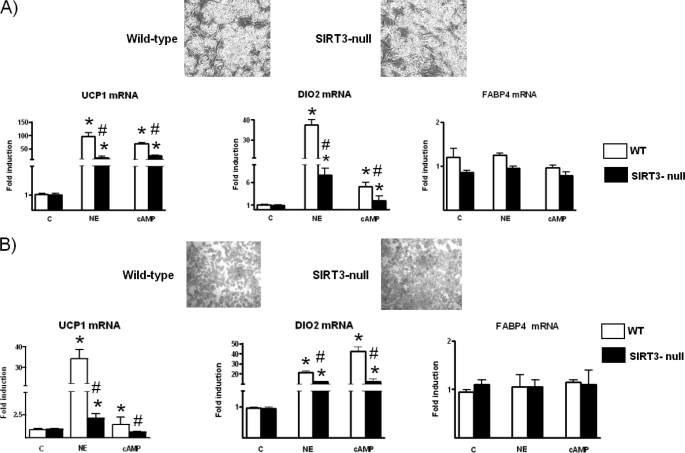
Fig. 1 shows the Sirt3 gene expression is dramatically induced during the brown adipocyte differentiation process, in parallel with up-regulation of the thermogenesis marker genes such as Ucp1. The expression profile showed that induction of Sirt3 mRNA was delayed with respect to the induction of Pgc-1α mRNA and Errα mRNA. In fact, on day 5 of differentiation Pgc-1α and Errα mRNA levels had reached more than one-half their maximum levels in differentiated brown adipocytes (day 9), whereas SIRT3 mRNA levels were still much lower. Induction of Sirt3 mRNA in association with differentiation was also observed for the Sirt3 protein, which was much more abundant in differentiated brown adipocytes than in preadipocytes (Fig. 1B), as well as for Pgc-1α protein. Thus, Sirt3 gene expression appeared as a component of the pattern of gene expression associated with brown adipocyte differentiation. However, in contrast with Ucp1 and other genes directly related to thermogenesis, Sirt3 gene expression was not induced by norepinephrine (data not shown). To ascertain the role of Sirt3, brown adipocytes were differentiated in primary culture from preadipocytes obtained from Sirt3-null mice and compared with parallel cultures from wild-type littermates. Acquisition of brown adipocyte morphology and accumulation of lipid were essentially unaltered when preadipocytes were isolated from mice lacking Sirt3 (Fig. 2A). Accordingly, there were no differences between wild-type and Sirt3-null differentiated adipocytes in the basal expression of genes related to thermogenesis (Ucp1 and Dio2) and overall adipogenesis (fatty acid binding protein-4, Fabp4) (Fig. 2A), genes related to mitochondriogenesis (cytochrome c oxidase subunit IV) and the β3-adrenoreceptor gene (data not shown). In contrast, the responsiveness of the Ucp1 and Dio2 genes to norepinephrine exposure was significantly reduced in differentiated brown adipocytes from Sirt3-null mice compared with that in wild-type brown adipocytes (Fig. 2A). The transcript for Fabp4, a gene associated with white or brown adipocyte differentiation and not specifically related to thermogenesis, was not up-regulated by norepinephrine and was unaffected by the absence of Sirt3. Parallel experiments in which brown adipocytes were exposed to dibutyryl-cAMP led to similar observations. As a second model of study, we performed similar experiments in adipocytes differentiated from MEFs obtained from Sirt3-null embryos. Adipocytes derived from MEFs exhibit an intermediate brown to white phenotype and a high propensity toward the acquisition of brown adipocyte features and expression of thermogenic genes (23).3 As for primary brown adipocytes, the lack of Sirt3 did not alter the acquisition of adipocyte morphology or basal levels of expression of the genes studied (Fig. 2B). However, again, the responsiveness of thermogenic genes (Ucp1 and Dio2) to induction by norepinephrine or cAMP, was significantly decreased in Sirt3-null adipocytes, and there was no effect on the non-brown fat-specific gene Fabp4. Collectively, these results indicate that Sirt3 is required for the full thermogenic competence of brown adipocytes.
FIGURE 1.
A, expression of Sirt3 and thermogenic gene transcripts in brown adipocytes differentiating in culture. Points are the means of two to three independent experiments at each time of culture and are expressed as percentages relative to the mean value at the time of maximum levels of expression (defined as 100%). B, representative immunoblot (of three independent cell culture experiments) of Sirt3 and Pgc-1α protein levels in brown preadipocytes (day 4) and differentiated brown adipocytes (day 9). CB, Coomassie Blue staining of the gel showing equal loading.
FIGURE 2.
Expression of Ucp1, Dio2, and Fabp4 in wild-type and Sirt3-null brown adipocytes and Sirt3-null MEF-derived adipocytes. The tops of panels A and B show micrographs representative of differentiated primary cultured brown adipocytes and MEF-derived adipocytes, respectively, from wild-type and Sirt3-null mice. Bars are mean ± S.E. of four to five independent cultures (*, p < 0.05, NE or cAMP versus controls; #, p < 0.05, wild-type versus Sirt3-null cells). NE, 0.5 μm norepinephrine for 6 h; cAMP, 1 mm dibutyryl-cAMP for 24 h. c, control.
Pgc-1α Induces Sirt3 Gene Expression in Adipocytes
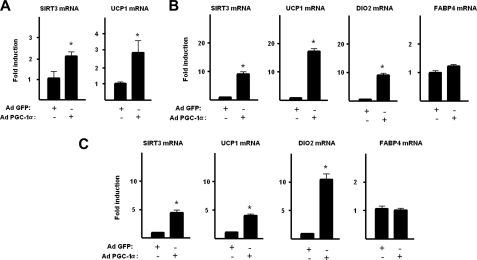
Given that Sirt3 appears as an essential component of the acquisition of full thermogenic activity in brown adipocytes, we analyzed the action of Pgc-1α, a master regulator of acquisition of thermogenic brown adipocyte phenotype and of its sensitivity to noradrenergic activation (4). For this purpose we transduced HIB-1B brown preadipocytes with an adenoviral vector driving Pgc-1α. This procedure led to a 2.7 ± 0.3-fold increase in the levels of the Pgc-1α protein in these cells. Pgc-1α overexpression caused a significant induction of Sirt3 mRNA expression in parallel with the induction of Ucp1 mRNA, the marker gene of the thermogenically differentiated status of HIB-1B brown adipocytes (Fig. 3A). Considering that Pgc-1α is known to be capable of driving white adipocytes into a brown adipocyte phenotype (3), we analyzed the effects of Pgc-1α overexpression on human SGBS white adipocytes. In this cell system, an induction of 3.5 ± 0.5-fold in the levels of the Pgc-1α protein was achieved. Pgc-1α effectively caused a coordinated induction of marker genes of brown versus white adipocyte phenotype (UCP1 and DIO2) accompanied by a marked induction of SIRT3 gene expression (Fig. 3B). No changes were observed in the expression of the FABP4 gene, which is known to be commonly expressed in brown and white adipocytes. MEF-derived adipocytes, another cellular model, was used. Adenoviral driven overexpression of Pgc-1α in these cells (3.1 ± 0.4-fold increase in Pgc-1α protein) again dramatically induced the expression of Sirt3 in parallel with the induction of the Ucp1 and Dio2 genes, without affecting expression of the Fabp4 gene (Fig. 3C). These findings indicate that the Sirt3 gene belongs to the cluster of genes under control of the Pgc-1α that determines the thermogenesis-related brown versus white differential phenotype.
FIGURE 3.
Effects of Pgc-1α overexpression on the expression of Sirt3 and thermogenic genes. HIB-1B brown preadipocytes (A), SGBS adipocytes (B), and MEF-derived adipocytes (C) were transduced with an adenoviral vector driving the expression of Pgc-1α or GFP (control) (see “Experimental Procedures”). Results are presented as mean ± S.E. of four to five independent experiments (*, p < 0.05).
Sirt3 Expression Is Required for Induction of the Brown Fat Phenotype in Response to Pgc-1α in Adipocytes
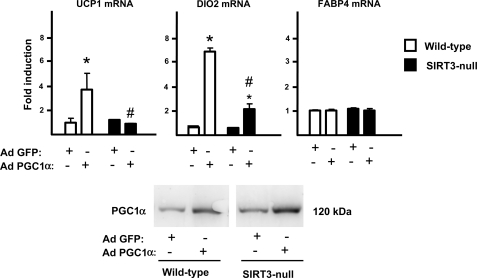
To establish the role of Sirt3 in the Pgc-1α-mediated induction of the brown fat pattern of gene expression, we compared the effects of Pgc-1α overexpression in Sirt3-null MEF-derived adipocytes to those in wild-type MEF-derived adipocytes. In this experimental setting, similar levels of increased Pgc-1α expression were attained in both types of cells using adenoviral mediated Pgc-1α overexpression (2.9 ± 0.3-fold induction in wild-type MEFs and 3.2 ± 0.4-fold induction in Sirt3-null MEFs; see also Fig. 3, bottom). The induction of Ucp1 and Dio2 mRNA expression by Pgc-1α was dramatically impaired in the Sirt3-null cells with respect to wild-type cells, whereas Fabp4 gene expression was insensitive to the absence of Sirt3 (Fig. 4). These results indicate that the induction of Sirt3 by Pgc-1α is required for a full acquisition of the gene expression pattern characteristic of thermogenically competent brown adipocytes.
FIGURE 4.
Effects of PGC-1α on the expression of thermogenic genes in wild-type or Sirt3-null adipocytes. Wild-type and Sirt3-null MEF-derived adipocytes were transduced with an adenoviral vector driving the expression of Pgc-1α or GFP (control) (see “Experimental Procedures”). Results are expressed as mean ± S.E. of four to five independent experiments (*, p < 0.05, differences due to Pgc-1α for each type of cell; #, p < 0.05, wild-type versus Sirt3-null cells) (top). A representative example of a similar overexpression of PGC-1α protein achieved by adenoviral mediated transfer of wild-type and Sirt3-null cells (bottom).
Transcriptional Activation of the Sirt3 Gene by Pgc-1α
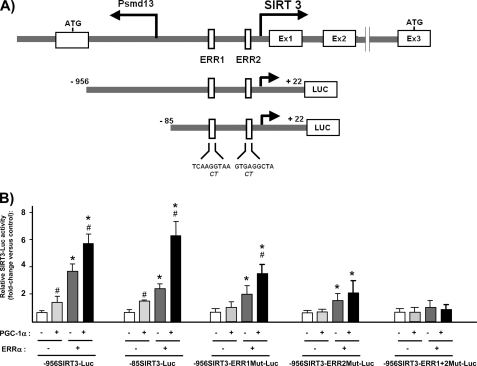
To establish whether Pgc-1α acts as a direct transcriptional coactivator of the Sirt3 gene, we first created a Sirt3 promoter-reporter construct by amplifying the 5′ promoter region of the Sirt3 gene from mouse genomic DNA by PCR, and then cloned it into a promoterless luciferase expression vector (see “Experimental Procedures”). The putative structure of the 5′ transcriptional regulatory region of the Sirt3 gene is depicted in Fig. 5A. The Sirt3 gene is closely apposed and in inverse orientation to the gene for Psmd13, a poorly characterized component of the proteasome system (24). The upstream translation initiation site of the Sirt3 gene (as defined in NM_001177804.1 and upstream to the most 5′ defined exonic region) is only 85 bp distant from the putative transcriptional initiation site of the Psmd13 gene. This structure is similar to that of the human SIRT3 gene (24), with the exception that the distance between the transcription initiation sites of the two genes is shorter in mice. A region encompassing 978 bp of the upstream region of the mouse Sirt3 gene (−956/22), including part of the first non-translated exon of the Psmd13 gene but excluding the Psmd13 gene translation initiation site) was cloned into the luciferase reporter plasmid pGL3. This construct exhibited strong promoter activity when transfected into the brown adipocyte cell line HIB-1B, within the range of the activity of gene promoters typically expressed in brown adipocytes (2.6-fold compared with the 4.5-kb UCP1-Luc promoter activity, 3.0-fold compared with the 2-kb PGC-1α-Luc promoter activity, data not shown). The 956SIRT3-Luc construct was co-transfected with a Pgc-1α expression vector alone or in combination with an expression vector for Nrf1, Nrf2α, Nrf2β, peroxisome proliferator-activated receptor-α, peroxisome proliferator-activated receptor-γ, Errα, thyroid receptor-α or -β, transcription factors, and nuclear receptors known to be potentially coactivated by Pgc-1α. Pgc-1α transfection alone caused a mild but significant induction of Sirt3 promoter activity (Fig. 5B). The co-transfection of the indicated transcription factors and nuclear receptors failed to modify the Sirt3 promoter activity in response to Pgc-1α (data not shown), with the exception of Errα. Co-transfection with the expression vector for Errα markedly increased the transcriptional activity of the Sirt3 promoter, as well as it enhanced the activation by Pgc-1α (Fig. 5B). A Sirt3 promoter-reporter construct containing only the −85/+22 bp proximal reporter region (-85SIRT3-Luc) was activated to essentially the same extent by Pgc-1α and Errα as the construct containing the longer promoter region. Computer-assisted analysis of the proximal promoter region revealed the presence of two sites, one at −45/−36 (ERR1) and one at −22/−14 (ERR2), with sequences potentially capable of binding members of the nuclear hormone receptor superfamily, including Errα. Individually mutating ERR1 or ERR2 sites to blunt receptor binding partially reduced Pgc-1α/Errα-induced Sirt3 gene promoter activity, whereas mutating both sites completely abolished the action of co-transfected Pgc-1α/Errα (Fig. 5B). Collectively, these results suggest that Pgc-1α acts in concert with Errα to promote Sirt3 gene transcription and identify two cis-acting sites in the Sirt3 promoter that mediate Pgc-1α action.
FIGURE 5.
Effects of Pgc-1α and Errα on Sirt3 promoter activity. A, schematic representation of the mouse Sirt3 promoter region (top) and luciferase promoter constructs (bottom). Mutations in the ERR1 and ERR2 sites are shown in italics under the wild-type sequence. B, effects of co-transfection of expression vectors for Pgc-1α and/or Errα on the relative luciferase activity of the indicated Sirt3 promoter constructs. Bars are mean ± S.E. of five to seven independent experiments. Statistically significant differences (p < 0.05) due to Errα co-transfection are shown as *, and those due to Pgc-1α are shown by #.
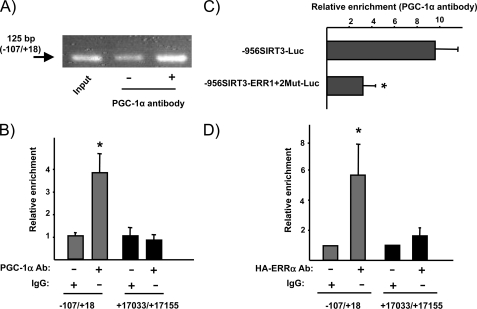
To determine whether Pgc-1α binds specifically to the Sirt3 promoter, we performed ChIP assays in primary cultured brown adipocytes. Using a specific antibody to immunoprecipitate Pgc-1α and primers that spanned the proximal region of the Sirt3 promoter, we found a significant enrichment in the expected PCR product (Fig. 6), whereas no such enrichment was found following amplification of an Sirt3 gene region far from the promoter (negative control). This indicated that Pgc-1α does indeed bind the endogenous Sirt3 promoter. Moreover, ChIP was also performed in HIB-1B cells transfected with the 956SIRT3-Luc and 956SIRT3-ERR1+2Mut-Luc constructs, after co-transfection with the Pgc-1α expression vector. Enrichment was evident when cells were transfected with the wild-type version of the Sirt3 promoter construct but was strongly impaired when cells had been transfected with the construct in which the Errα binding sites had been mutated (Fig. 6B). In addition, we analyzed the binding of Errα to the Sirt3 proximal promoter region. For this purpose, we transfected HIB-1B cells with an expression vector driving HA-Errα. ChIP assays indicated a substantial enrichment in Errα binding to the proximal promoter region of the endogenous Sirt3 gene (Fig. 6C).
FIGURE 6.
Chromatin immunoprecipitation of Pgc-1α and Errα binding to the proximal Sirt3 gene promoter region. A, representative PCR after ChIP of Pgc-1α binding to the proximal promoter region of the endogenous Sirt3 gene in brown adipocytes. The arrow indicates the −125-bp PCR product from the mouse Sirt3 gene promoter. Input was 1/10 diluted for amplification. B, quantitative analysis of ChIP amplification of the Pgc-1α binding to the proximal promoter region of the endogenous Sirt3 gene (−107/+18) in comparison with a control, non-promoter, region of the Sirt3 gene (+17033/+17155) in brown adipocytes. The enrichment due to protein-bound DNA was calculated as a ratio to input DNA. C, quantitative analysis of ChIP amplification of the Pgc-1α binding to the proximal promoter region of the transfected wild-type -956SIRT3 promoter region and the mutated version in the ERR1 and ERR2 sites in HIB-1B cells co-transfected with the Pgc-1α expression vector. D, quantitative analysis of ChIP amplification of the binding of Errα to the Sirt3 gene promoter region in comparison with the non-promoter region (see above). Expression vector for the HA-tagged version of Errα was transfected to HIB-1B cells before the assay. Bars are mean ± S.E. of three to four independent assays (*, p < 0.05, differences due to specific enrichment due to binding of Pgc-1α or Errα in B and D, and for the comparison of Pgc-1α binding between wild-type and ERR1+2-mutated Sirt3 promoter construct, in C).
ERRα Is Required for Sirt3 Gene Expression in Response to Pgc-1α
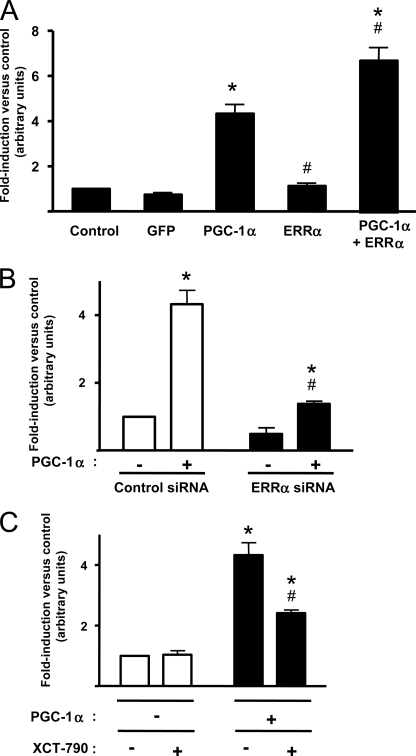
To ascertain whether the endogenous Sirt3 gene, like the transfected Sirt3 promoter, is also induced by Pgc-1α/Errα, we transduced MEFs with adenoviral vectors for Errα, alone or in combination with that for Pgc-1α. Errα caused a mild but significant induction of Sirt3 gene expression, whereas the combination of ERRα plus PGC-1α yielded maximal induction (Fig. 7A). To confirm the role of Errα in the induction of the Sirt3 gene by Pgc-1α, we transduced MEFs with a vector driving the expression of Errα siRNA. This procedure reduced the Errα expression to ∼30% of original levels, and significantly reduced the capacity of Pgc-1α to induce Sirt3 gene expression (Fig. 7B). As a complementary approach, we used XCT-790, an inhibitor of Errα transcriptional activity (25). Treatment of cells with XCT-790 caused a significant reduction in the capacity of Pgc-1α to induce Sirt3 gene expression, thus confirming the involvement of Errα in Pgc-1α-mediated regulation of the Sirt3 gene (Fig. 7C).
FIGURE 7.
Involvement of Errα in the responsiveness of Sirt3 gene expression to Pgc-1α. A, effects of adenoviral mediated overexpression of Pgc-1α and/or Errα on Sirt3 gene expression in MEF-derived adipocytes. B, effects of impairment of endogenous Errα expression via adenoviral mediated expression of Errα siRNA on the capacity of Pgc-1α to induce Sirt3 gene expression. C, effects of the Errα inhibitor 5 μm XCT-790 on the action of Pgc-1α on Sirt3 gene expression. Bars are mean ± S.E. of five to seven independent experiments. Statistically significant differences (p < 0.05) due to Pgc-1α are shown as *, and those due to Errα overexpression (A), Errα siRNA (B), or XCT-790 (C) are shown by #.
DISCUSSION
Complex regulation of gene expression is required to program cells to acquire the phenotype of differentiated, thermogenically competent brown adipocytes. This differentiation process allows brown adipocytes to perform the specialized thermogenic function of the cell in response to noradrenergic activation. Pgc-1α is a transcriptional coactivator known to play a master role in eliciting the coordinate induction of gene expression that leads to brown adipocyte differentiation. Increasing Pgc-1α levels can drive white adipocytes to acquire a brown fat phenotype (3). Moreover, Pgc-1α is required for brown adipocytes to acquire the sensitivity to noradrenergic-stimulated thermogenic activation (4). Our present findings, which constitute the first analysis of Sirt3 gene regulation in brown adipocytes, indicate that Pgc-1α induces Sirt3 gene expression and this effect is essential for brown adipocyte full differentiation. The observations indicating that Pgc-1α is unable to activate the full program of thermogenic gene expression in cells that lack Sirt3 indicates that the induction of Sirt3 may be a mechanism that contributes to the ability of Pgc-1α to promote the widespread cellular effects that lead to the differentiation of fully thermogenic competent brown adipocytes. The specific requirement for Sirt3 in the acquisition of sensitivity to thermogenic activation by brown adipocytes is in complete agreement with the observation of impaired responses to noradrenergic activation in cells with impaired Pgc-1α expression (4).
Sirt3 is a protein deacetylase of the sirtuin family, and is considered to control intracellular biological processes by de-acetylating target proteins involved in metabolism and gene expression. It is localized to the mitochondria and, it is controversial whether under specific circumstances, to the nucleus as well (6). The capacity of Sirt3 to influence nuclear gene expression has been reported in several cell models, including brown adipocytes (16, 26). It is unknown whether this is due to signaling from mitochondrial to nuclei (the so-called mitochondrial reverse signaling) or it should be considered a direct consequence of the potential presence of Sirt3 in nuclei, as proposed by some authors (7, 12). Shi et al. (16) reported that Sirt3 acts by increasing cAMP-response element-binding protein phosphorylation, but the specific targets of SIRT3 deacetylation among the actors involved in the cascade of noradrenergic induction of gene expression have not been identified to date. In any case, our present findings indicate that Sirt3 expression is an essential component of the specific brown adipocyte phenotype, and is functionally required for brown adipocytes to respond to Pgc-1α-mediated thermogenic activation.
Although few studies have been conducted on the thermoregulation of Sirt3-null mice in vivo, Lombard et al. (27) reported that the response to short-term cold exposure was unaltered in these mice. However, we observed that the thermal stress-induced postnatal expression of the Ucp1 gene is significantly reduced in brown fat from Sirt3-KO mice (28). Further studies will be needed to determine whether inactivation of the Sirt3 gene in vivo influences the overall capacity of adaptive thermogenesis in mice under distinct physiological conditions. Moreover, the possibility that other sirtuins (e.g. Sirt1) might compensate for the lack of Sirt3 cannot be excluded. Although it is not known whether Sirt1 affects BAT differentiation or cAMP responsiveness, chronic activation of Sirt1 in rodents has been reported to promote brown adipose tissue thermogenic activity (29).
Although Pgc-1α can potentially coactivate multiple nuclear receptors and transcription factors, its action at the Sirt3 promoter appears to involve mainly the orphan nuclear receptor Errα. The identification of Errα as the transcription factor being coactivated by Pgc-1α, and involved in driving the induction of the Sirt3 gene is consistent with the known role of Errα in brown adipocyte function as well as on the transcriptional control of protein preferentially expressed in the mitochondria. Errα appears to be essential for adaptation to cold environment, as evidenced by the failure of mice lacking Errα to maintain body temperature when exposed to cold (30). Errα is needed for the high levels of oxidative capacity characteristic of brown adipose tissue, and thus for providing the energy necessary for thermogenesis. Errα fulfills this role by acting directly at genes important for brown fat thermogenesis: for instance, it is one of the transcription factors that interact with Pgc-1α to mediate Pgc-1α action on the Ucp1 gene (31). The present findings demonstrate that Errα is essential for the promotion of Sirt3 gene expression by Pgc-1α. During the preparation of this article, Kong et al. (32) reported that Pgc-1α controls Sirt3 gene expression in the context of muscle cells. This confirms that the Pgc-1α-Sirt3 regulatory axis may be important not only in brown adipocytes but also in other cell types in which mitochondrial functions are regulated for physiological purposes other than thermogenesis. In summary, we conclude that Pgc-1α controls the expression of Sirt3 in brown adipocytes, and this process may be a relevant intracellular mechanisms by which Pgc-1α conducts its master role in the induction of brown adipocyte phenotype.
Acknowledgments
We thank Dr. B. Spiegelman for HIB-1B cells and PGC-1α expression vectors, Dr. Wabitsch for SGBS white adipocytes, and Drs. R. Evans, H. H. Samuels, S. Green, and S. McKnight for expression vectors.
The work was supported by Grant SAF2008-01896 from the Ministerio de Ciencia e Innovación, Spain, and Grant 2009SGR-284 from the Generalitat de Catalunya.
A. Giralt and J. Diaz-Delfin, unpublished observations.
- Ucp1
- uncoupling protein 1
- Pgc-1α
- peroxisome proliferator-activated receptor-γ coactivator-1α
- Sirt3
- silent mating type information regulation 2, homolog 3
- Nrf
- nuclear respiratory factor
- ERR
- estrogen-related receptor
- Dio2
- type II 5′-deiodinase
- Fabp4
- fatty acid binding protein-4
- MEF
- mouse embryonic fibroblast
- FABP
- fatty acid-binding protein.
REFERENCES
- 1. Cannon B., Nedergaard J. (2004) Physiol. Rev. 84, 277–359 [DOI] [PubMed] [Google Scholar]
- 2. Handschin C., Spiegelman B. M. (2006) Endocr. Rev. 27, 728–735 [DOI] [PubMed] [Google Scholar]
- 3. Tiraby C., Tavernier G., Lefort C., Larrouy D., Bouillaud F., Ricquier D., Langin D. (2003) J. Biol. Chem. 278, 33370–33376 [DOI] [PubMed] [Google Scholar]
- 4. Uldry M., Yang W., St-Pierre J., Lin J., Seale P., Spiegelman B. M. (2006) Cell Metab. 3, 333–341 [DOI] [PubMed] [Google Scholar]
- 5. Bao J., Sack M. N. (2010) Cell Mol. Life Sci. 67, 3073–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huang J. Y., Hirschey M. D., Shimazu T., Ho L., Verdin E. (2010) Biochim. Biophys. Acta 1804, 1645–1651 [DOI] [PubMed] [Google Scholar]
- 7. Scher M. B., Vaquero A., Reinberg D. (2007) Genes Dev. 21, 920–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hirschey M. D., Shimazu T., Goetzman E., Jing E., Schwer B., Lombard D. B., Grueter C. A., Harris C., Biddinger S., Ilkayeva O. R., Stevens R. D., Li Y., Saha A. K., Ruderman N. B., Bain J. R., Newgard C. B., Farese R. V., Jr., Alt F. W., Kahn C. R., Verdin E. (2010) Nature 464, 121–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schwer B., Bunkenborg J., Verdin R. O., Andersen J. S., Verdin E. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 10224–10229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hallows W. C., Lee S., Denu J. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 10230–10235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pillai V. B., Sundaresan N. R., Kim G., Gupta M., Rajamohan S. B., Pillai J. B., Samant S., Ravindra P. V., Isbatan A., Gupta M. P. (2010) J. Biol. Chem. 285, 3133–3144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sundaresan N. R., Gupta M., Kim G., Rajamohan S. B., Isbatan A., Gupta M. P. (2009) J. Clin. Invest. 119, 2758–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim H. S., Patel K., Muldoon-Jacobs K., Bisht K. S., Aykin-Burns N., Pennington J. D., van der Meer R., Nguyen P., Savage J., Owens K. M., Vassilopoulos A., Ozden O., Park S. H., Singh K. K., Abdulkadir S. A., Spitz D. R., Deng C. X., Gius D. (2010) Cancer Cell 17, 41–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rose G., Dato S., Altomare K., Bellizzi D., Garasto S., Greco V., Passarino G., Feraco E., Mari V., Barbi C., BonaFe M., Franceschi C., Tan Q., Boiko S., Yashin A. I., De Benedictis G. (2003) Exp. Gerontol. 38, 1065–1070 [DOI] [PubMed] [Google Scholar]
- 15. Bellizzi D., Rose G., Cavalcante P., Covello G., Dato S., De Rango F., Greco V., Maggiolini M., Feraco E., Mari V., Franceschi C., Passarino G., De Benedictis G. (2005) Genomics 85, 258–263 [DOI] [PubMed] [Google Scholar]
- 16. Shi T., Wang F., Stieren E., Tong Q. (2005) J. Biol. Chem. 280, 13560–13567 [DOI] [PubMed] [Google Scholar]
- 17. Carmona M. C., Hondares E., Rodríguez de la Concepción M. L., Rodríguez-Sureda V., Peinado-Onsurbe J., Poli V., Iglesias R., Villarroya F., Giralt M. (2005) Biochem. J. 389, 47–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Klaus S., Choy L., Champigny O., Cassard-Doulcier A. M., Ross S., Spiegelman B., Ricquier D. (1994) J. Cell Sci. 107, 313–319 [DOI] [PubMed] [Google Scholar]
- 19. Wabitsch M., Brenner R. E., Melzner I., Braun M., Möller P., Heinze E., Debatin K. M., Hauner H. (2001) Int. J. Obes. Relat. Metab. Disord. 25, 8–15 [DOI] [PubMed] [Google Scholar]
- 20. Hansen J. B., Jørgensen C., Petersen R. K., Hallenborg P., De Matteis R., Bøye H. A., Petrovic N., Enerbäck S., Nedergaard J., Cinti S., te Riele H., Kristiansen K. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 4112–4117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Herzog B., Cardenas J., Hall R. K., Villena J. A., Budge P. J., Giguère V., Granner D. K., Kralli A. (2006) J. Biol. Chem. 281, 99–106 [DOI] [PubMed] [Google Scholar]
- 22. Hondares E., Mora O., Yubero P., Rodriguez de la Concepción M., Iglesias R., Giralt M., Villarroya F. (2006) Endocrinology 147, 2829–2838 [DOI] [PubMed] [Google Scholar]
- 23. Mercader J., Palou A., Bonet M. L. (2010) Obesity 18, 655–662 [DOI] [PubMed] [Google Scholar]
- 24. Bellizzi D., Dato S., Cavalcante P., Covello G., Di Cianni F., Passarino G., Rose G., De Benedictis G. (2007) Genomics 89, 143–150 [DOI] [PubMed] [Google Scholar]
- 25. Busch B. B., Stevens W. C., Jr., Martin R., Ordentlich P., Zhou S., Sapp D. W., Horlick R. A., Mohan R. (2004) J. Med. Chem. 47, 5593–5596 [DOI] [PubMed] [Google Scholar]
- 26. Jacobs K. M., Pennington J. D., Bisht K. S., Aykin-Burns N., Kim H. S., Mishra M., Sun L., Nguyen P., Ahn B. H., Leclerc J., Deng C. X., Spitz D. R., Gius D. (2008) Int. J. Biol. Sci. 4, 291–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lombard D. B., Alt F. W., Cheng H. L., Bunkenborg J., Streeper R. S., Mostoslavsky R., Kim J., Yancopoulos G., Valenzuela D., Murphy A., Yang Y., Chen Y., Hirschey M. D., Bronson R. T., Haigis M., Guarente L. P., Farese R. V., Jr., Weissman S., Verdin E., Schwer B. (2007) Mol. Cell. Biol. 27, 8807–8814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Villarroya F., Giralt A., Planavila A., Hondares E., Iglesias R., Giralt M. (2009) J. Physiol. Sci. 59, Suppl. 1, 279 [Google Scholar]
- 29. Feige J. N., Lagouge M., Canto C., Strehle A., Houten S. M., Milne J. C., Lambert P. D., Mataki C., Elliott P. J., Auwerx J. (2008) Cell Metab. 8, 347–358 [DOI] [PubMed] [Google Scholar]
- 30. Villena J. A., Hock M. B., Chang W. Y., Barcas J. E., Giguère V., Kralli A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 1418–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Debevec D., Christian M., Morganstein D., Seth A., Herzog B., Parker M., White R. (2007) Mol. Endocrinol. 21, 1581–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kong X., Wang R., Xue Y., Liu X., Zhang H., Chen Y., Fang F., Chang Y. (2010) PLoS. One 5, e11707. [DOI] [PMC free article] [PubMed] [Google Scholar]