Abstract
The NAD-dependent histone deacetylase Sirt1 is a negative regulator of T cell activation. Here we report that Sirt1 inhibits T cell activation by suppressing the transcription of Bcl2-associated factor 1 (Bclaf1), a protein required for T cell activation. Sirt1-null T cells have increased acetylation of the histone 3 lysine 56 residue (H3K56) at the bclaf1 promoter, as well as increasing Bclaf1 transcription. Sirt1 binds to bclaf1 promoter upon T cell receptor (TCR)/CD28 stimulation by forming a complex with histone acetyltransferase p300 and NF-κB transcription factor Rel-A. The recruitment of Sirt1, but not p300, requires Rel-A because blocking Rel-A nuclear translocation in T cells and siRNA-mediated knockdown of Rel-A can inhibit Sirt1 binding to bclaf1 promoter. Although knockdown of either p300 or GCN5 partially suppressed global H3K56 acetylation, only p300 knockdown specifically attenuated H3K56 acetylation at the bclaf1 promoter. Lastly, knockdown of Bclaf1 suppresses the hyperactivation observed in Sirt1−/− T cells, indicated by less IL-2 production in CD4+ T cells and reduced proliferation. Therefore, Sirt1 negatively regulates T cell activation via H3K56 deacetylation at the promoter region to inhibit transcription of Bclaf1.
Keywords: Histone Deacetylase, Immunology, Immunosuppressor, Sirtuins, Transcription Regulation, T Cell Activation
Introduction
The silent information regulator 2 (sir2) gene was first discovered in yeast as a transcriptional repressor (1). Mammals have seven orthologs of Sir2, which are named Sirt1 to Sirt7, respectively (2, 3). The sirtuins can function as NAD-dependent histone deacetylases, removing acetyl groups on histones to inhibit access of transcription factors to DNA. Histone deacetylases work along with histone acetyl transferases in an antagonistic manner to regulate gene transcription (3). Some sirtuins, such as Sirt1, also have non-histone substrates, and the deacetylation of these substrates is thought to regulate protein activity (12). Sirt1 in particular is reported to be involved in various biological processes, including aging, metabolism, and development (4, 5). Recently, studies indicated that Sirt1 plays an important role in immune regulation (6–11). Sirt1-null mice develop autoantibodies against nuclear antigens, resulting in the accumulation of immune complexes in the kidney and liver (9, 11). Sirt1-null mice also have hyper-responsive T cells and are more susceptible to experimental autoimmune encephalomyelitis (11). Myeloid-specific deletion of Sirt1 induces inflammatory signaling in response to environmental stress (7). In addition, Sirt1 is a target of HIV Tat protein, which can lead to hyperactivation of T lymphocytes during HIV infections (8). These studies suggest that Sirt1 plays an important role in both adaptive and innate immune response and is a physiological target of viruses, possible resulting in the attenuation of the adaptive immune response.
In addition to Sirt1 acting as an inhibitor of NF-κB (12), our group recently showed that Sirt1 can negatively regulate T cell activation by antagonizing the activation of transcription factor AP-1, which binds to the IL-2 promoter and mediates IL-2 production (11). Nonetheless, the mechanism by which Sirt1 regulates T cell activation is not entirely understood. During the study of Sirt1 in regulating T cell activation, we observed a 3–5-fold increase of Bcl2-associated factor 1 (Bclaf1) expression in Sirt1-deficient CD4+ T cells when compared with wild-type T cells.
Bclaf1 was originally identified in a screen of proteins that interact with the adenoviral Bcl2 homolog E1B19K (13). Initial studies reported that Bclaf1 functions as an inducer of apoptosis (14, 15), but subsequent studies showed that Bclaf1 plays critical roles in a wide range of processes that are not normally associated with Bcl2 family members, including RNA processing and stabilization, lung development, T cell activation, and control of the lytic infection program in Kaposi sarcoma-associated herpesvirus (16–18). Adoptive transfer of Bclaf1-null fetal liver cells into Rag2−/− mice revealed that Bclaf1 is indispensable for T cell development. Interestingly, peripheral T cells reconstituted from Bclaf1-null cells failed to respond to TCR/CD282 stimuli even in the presence of IL-2, indicating a requirement of Bclaf1 for the clonal expansion of T cells upon antigen stimulation (19). Here we report that Sirt1 forms a complex with Rel-A and p300 at the bclaf1 promoter region to regulate Bclaf1 expression by suppressing H3K56 acetylation and that the suppression of Bclaf1 attenuates T cell activation.
MATERIALS AND METHODS
Cells, Reagents, and Mice
Human embryonic kidney (HEK) 293 cells were cultured in DMEM containing 10% FBS. Sirt1+/+ and Sirt1−/− mouse embryonic fibroblasts were isolated at embryonic days 10–12. Antibodies against Bclaf1, Rel-A, Sirt1, Myc, GCN5, p300, and HA were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies specific to acetyl-H3K56 and histone H3 were purchased from Cell Signaling Technology (Danvers, MA). Anti-CD3 and anti-CD28 were from eBioscience (San Diego, CA). The anti-Actin and anti-FLAG antibodies were from Sigma. Sirt1-null mice were used as reported (20). All mice used in this study were maintained and used at the Northwestern University mouse facility under pathogen-free conditions according to institutional guidelines and animal study proposals approved by the Institutional Animal Care and Use Committee.
Plasmids
bclaf1 promoter region was amplified by PCR using primers as shown in supplemental Table 1. DNA fragment was subcloned into pGL3-luc vector (Invitrogen). Mouse GCN5 cDNA was amplified by PCR and cloned into pCMV-Myc vector. Sirt1, Rel-A, and p300 expression plasmids were used as described (21). E2F1 expression plasmids were purchased from Addgene (Cambridge, MA) and used as reported (22).
Isolating Mouse Naive CD4+ T Cells, Cell Proliferation Assay, and Intercellular Cytokine Staining
CD4+ T cells were isolated from the lymph nodes and spleens of 8–10-week-old wild-type and Sirt1−/− mice. These cells were maintained in RPMI 1640 medium supplemented with 10% FBS, 100 units/ml penicillin, 200 μg/ml streptomycin, and 0.25 μg/ml amphotericin and stimulated with anti-CD3 plus anti-CD28 (1 μg/ml each). Upon stimulation with anti-CD3 or anti-CD3 plus anti-CD28 antibodies, the proliferation of stimulated cells was determined by [3H]thymidine incorporation assay. IL-2 production in the stimulated CD4 T cells was analyzed by intracellular cytokine staining upon an additional 4 h of stimulation with phorbol myristate acetate (10 ng/ml) plus ionomycin (1 μm) in the presence of 10 μg/ml brefeldin A. Cells were fixed and permeabilized, and intracellular staining was performed with anti-IL-2-APC (eBioscience) as described (23).
Gene Transfection, Immunoprecipitation, and Western Blotting
HEK293 cells were transfected with different combinations of plasmid DNA as indicated in the corresponding figure legends with Lipofectamine 2000 (Invitrogen) as described (24). Transiently transfected HEK 293 cells were washed with ice-cold PBS, resuspended in radioimmunoprecipitation assay buffer that contains 20 mm Tris-HCl (pH 7.5), 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1% Nonidet P-40, 1% sodium deoxycholate, 2.5 mm sodium pyrophosphate, 1 mm β-glycerophosphate, 1 mm Na3VO4 in the presence of a protease inhibitor mixture (Roche Applied Science), and incubated on ice for 15 min. Insoluble fractions were removed by centrifugation (15,000 × g, 15 min). Supernatants were precleaned with protein G-Sepharose at 4 °C for 15 min and then incubated with each indicated antibody (1 μg/ml) for 1 h followed by incubation with protein G-Sepharose beads for additional 2 h. The protein G-Sepharose beads were washed four times with lysis buffer dissolved with 4× Laemmli buffer and boiled for 5 min. Supernatants were subjected to SDS-PAGE and transferred to nitrocellulose membrane. After blocking with 5% (w/v) skim milk in Tris-buffered saline containing 0.1% Tween 20, the membrane was incubated overnight at 4 °C with the indicated primary antibodies followed by HRP-conjugated secondary antibody. Membranes were then washed and visualized with enhanced chemiluminescence (ECL). When necessary, membranes were stripped using stripping buffer (Bio-Rad) and reprobed with various antibodies.
Dual-Luciferase Assay
HEK293 or MEF cells in 12-well plates were transfected with Bclaf1-luciferase and control pRL-TK (Promega, Madison, WI) plasmids along with various expression plasmids as indicated (24). The pRL-TK plasmid contains the Renilla reniformis (sea pansy) luciferase gene under the transcriptional control of the herpesvirus thymidine kinase promoter and constitutively expresses low levels of renillar luciferase. Transfected cells were lysed, and the luciferase activity in cell lysates was analyzed using a Dual-Luciferase reporter assay kit (Promega). Luciferase activity was measured as relative light units using a luminometer (Turner BioSystems, Inc., Sunnyvale, CA).
Lentivirus-based Bclaf1 Knockdown in Mouse Primary CD4 T Cells
Bclaf1 shRNA plasmids bearing the 21-mer fragment were subcloned into the lentiviral expression vectors as described (25) and co-transfected with packaging plasmids (Invitrogen) into HEK293 cells. Supernatants of the transfected cells were collected and used to infect CD4+ T cells isolated from Sirt1−/− and Sirt1+/+ mice as reported. 48 h after infection, cells were analyzed by intracellular staining for IL-2 production, and GFP+ cells were sorted for proliferation assay.
Chromatin Immunoprecipitation (ChIP) Assay
Primary T cells from Sirt1−/− and Sirt1+/+ mice were stimulated with anti-CD3 plus anti-CD28 for 16 h, cross-linked with 1% formaldehyde, and lysed with SDS lysis buffer. Cell lysates were sonicated, and 5% of cell lysate was removed and used to determine the total amount of target DNA in input. Remaining cell lysates were diluted in ChIP dilution buffer. Immunoprecipitation was performed with each of the indicated antibodies (4 μg) at 4 °C overnight. Immune complexes were then mixed with salmon sperm DNA/protein agarose 50% slurry at 4 °C for 1 h. After immunoprecipitates were washed sequentially with low salt buffer, high salt buffer, LiCl wash buffer, and Tris EDTA, DNA-protein complexes were eluted with elution buffer, and cross-linking was reversed. Genomic DNA was extracted using phenol/chloroform, and ethanol-precipitated DNA was resuspended in Tris EDTA. PCR was performed with specific primers as listed in supplemental Table 1.
RESULTS
Sirt1 Suppresses Bclaf1 Expression in T Cells
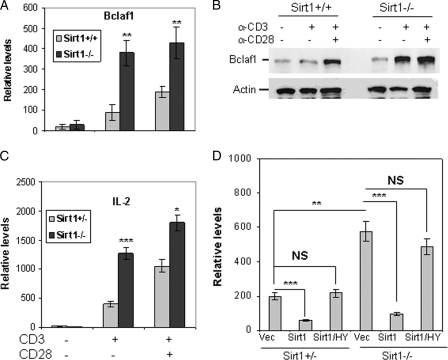
We have recently found that Sirt1 functions as a negative regulator of T cell activation (11). While studying the role of Sirt1 in T cell gene regulation, we found that Sirt1 suppressed the transcription of Bclaf1, a Bcl2-associated factor. As shown in Fig. 1A, the mRNA level of Bclaf1 in Sirt1-null T cells is about 3–4-fold higher than that in wild-type T cells upon TCR/CD28 stimuli. As controls, Bclaf1 mRNA levels are indistinguishable between naive Sirt1−/− and wild-type T cells. Similar to our previous report (11), IL-2 mRNA level is higher in Sirt1−/− T cells than that in wild-type control T cells upon TCR/CD28 stimulation (Fig. 1B), confirming that Sirt1 is a negative regulator for T cell activation. Notably, the full scales of both Bclaf1 and IL-2 mRNA transcription depend on CD28-mediated co-stimulatory signaling in wild-type T cells in contrast to the independence to CD28 stimulation for their transcription in Sirt1-null T cells. These results suggest that Sirt1 is a negative regulator for Bclaf1 gene transcription and that Sirt1 suppression of Bclaf1 transcription is regulated by TCR/CD28 signaling. To support this, we further demonstrated that Bclaf1 protein expression levels are increased in Sirt1-null T cells when compared with that in control wild-type T cells upon TCR/CD28 stimulation (Fig. 1C). A significant increase of Bclaf1 protein level was also detected in Sirt1−/− MEFs. Reconstitution with Sirt1, but not with its deacetylase activity-inactive mutant Sirt1(HY), into Sirt1-null MEF inhibited Bclaf1 expression (Fig. 1D). Therefore, the deacetylase catalytic activity of Sirt1 is required for its suppression of Bclaf1 expression.
FIGURE 1.
Sirt1 inhibits Bclaf1 expression in T cells. A and B, naive CD4+ T cells from Sirt1+/+ and Sirt1−/− mice were stimulated with anti-CD3 or CD3 plus anti-CD28 antibodies for 16 h. Total RNA was isolated, and the levels of Bclaf1 (A) and IL-2 (B) were determined by real-time PCR. C, primary T cells were stimulated with anti-CD3 or with anti-CD3 plus anti-CD28 for 24 h. Bclaf1 protein expression in the stimulated cells was analyzed by Western blotting using anti-Bclaf1 (top panel) antibody. The same membrane was reblotted with anti-actin as a loading control (bottom panel). D, Sirt1+/+ and Sirt1−/− MEF cells were transfected with Sirt1 or Sirt1/HY mutant. The mRNA expression levels of endogenous Bclaf1 in the transfected cells were analyzed by real-time PCR using β-actin mRNA as an internal control. vec, vector. Student's t test was used for statistic analysis, NS, no significant difference. Error bars indicate mean ± S.D.; *, p < 0.05; **, p < 0.01; ***, p < 0.005.
Hyperacetylation of Histone H3K56 in bclaf1 Promoter Region in Sirt1-null T Cells
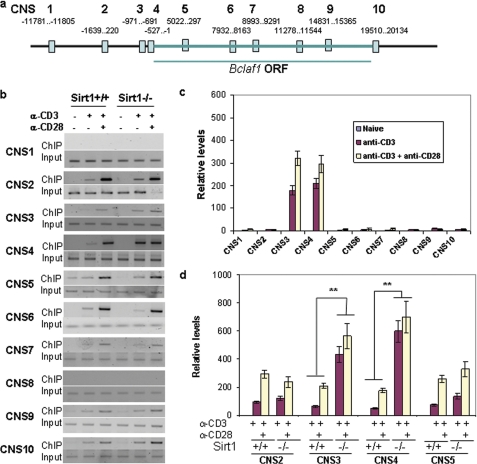
As a histone deacetylase, Sirt1 may regulate Bclaf1 gene transcription at the epigenetic level in T cells. Because histone acetylation is often localized within conserved non-coding sequences (CNS) (26), we used an online-based mVISTA alignment program to compare DNA sequences of the bclaf1 locus between mouse and human. Regions containing fragments longer than 200 bp, with at least 80% homology between paired sequences, were identified as CNS (27). We found 10 CNS in the bclaf1 locus (Fig. 2A), which may represent potential regulatory elements. Sirt1 has been recently found to deacetylate histone H3 at the Lys-56 residue (28, 29); we therefore asked whether Sirt1 inhibits Bclaf1 transcription by deacetylating histone H3K56. ChIP assay using anti-acetyl-H3K56 antibody revealed a background level of acetylated histone H3 binding to all CNS regions in naive T cells (Fig. 2B). Except for CNS1 and CNS8, TCR stimulation induced histone H3K56 acetylation at the chromatin of the other eight CNS regions, which is further enhanced by CD28-mediated co-stimulatory signaling (Fig. 2B and supplemental Fig. 1). Interestingly, Sirt1 is specifically recruited to the CNS3 and CNS4 regions of the bclaf1 promoter (Fig. 2C), suggesting that Sirt1 may regulate Bclaf1 transcription by deacetylating histones at these regions. In fact, the hyperacetylation of H3K56 in CNS3 and CNS4 of the bclaf1 locus was detected in Sirt1-deficient T cells upon TCR/CD28 stimuli. Unlike wild-type T cells, in which a full-scale H3K56 acetylation on bclaf1 promoter requires anti-CD28 stimulation, Sirt1-null T cells achieved a full-scale H3K56 acetylation at the bclaf1 promoter even in the absence of CD28 signaling (Fig. 2, B and D), indicating that Sirt1 suppresses Bclaf1 expression by deacetylating histone H3 at the Lys-56 residue in T cells. In addition, our data suggest that Sirt1 has a minor effect on the global acetylation of H3K56 in T cells as analyzed by Western blotting because acetylated H3K56 levels, but not total histone H3 protein levels, were slightly increased in Sirt1-null T cells when compared with those in wild-type T cells upon TCR and CD28 stimulation (data not shown). The partial increase suggests that other histone deacetylases are also involved in suppressing H3K56 acetylation.
FIGURE 2.
Hyperacetylation of histone H3K56 at bclaf1 promoter in Sirt1-null T cells. A, a schematic of CNS in bclaf1 locus. Numbers indicate the exact regions of each CNS. B and C, naive CD4+ T cells from Sirt1+/+ mice were stimulated with anti-CD3 or with anti-CD3 plus anti-CD28 for 16 h. ChIP assay was performed using anti-acetyl-H3K56 antibody (B) and anti-Sirt1 antibody (C), respectively. The bound DNA fragments were used as templates for analyzing each CNS region by PCR or real-time PCR. D, naive CD4+ T cells from Sirt1+/+ mice were stimulated with anti-CD3 or with anti-CD3 plus anti-CD28 for 16 h. The H3K56 acetylation at CNS2–5 regions was analyzed ChIP with anti-acetyl-H3K56 and real-time PCR. Student's t test was used for statistic analysis. Error bars indicate mean ± S.D.; **, p < 0.01.
The Acetyltransferase p300 Catalyzes Histone H3K56 Acetylation in T Cells
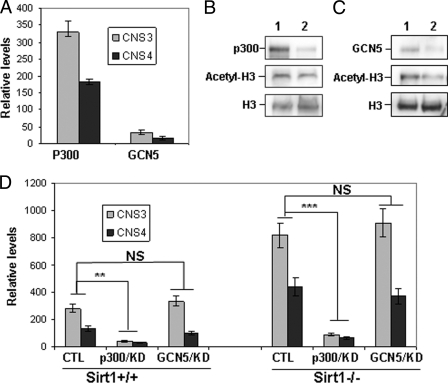
In mammals, both p300 and GCN5 are involved in catalyzing H3K56 acetylation (28–31). We therefore asked which acetyltransferase is involved in H3K56 acetylation of the bclaf1 promoter in T cells. ChIP analysis with anti-p300 antibody revealed a direct binding of p300 acetyltransferase to bclaf1 promoter in T cells upon TCR/CD28 stimulation, indicating that the binding of p300 to bclaf1 promoter is regulated by TCR and CD28-mediated signaling (Fig. 3A). However, a direct binding of GCN5 with bclaf1 promoter was not detected in T cells by ChIP analysis with anti-GCN5 antibody (Fig. 3A).
FIGURE 3.
The acetyltransferase p300 regulates H3K56 acetylation at bclaf1 promoter. A, mouse primary T cells were stimulated with anti-CD3 or with anti-CD3 plus anti-CD28 for 16 h. The binding of GCN5 and p300 to bclaf1 promoter in T cells was analyzed by ChIP using specific antibodies against each protein followed by real-time PCR. B and C, MEF cells were transfected with siRNA specific to p300 (B) or GCN5 (C). Three days after transfection, cells were collected and lysed. The expression levels p300 and GCN5 proteins were determined by Western blotting (top panels). The levels of acetyl-H3K56 (middle panels) and total H3 proteins (bottom panels) in p300 (B) and GCN5 (C) knockdown cells were analyzed by Western blotting. D, the levels of acetyl-H3K56 at the chromatin of bclaf1 promoter in p300 and GCN5 knockdown (KD) cells were analyzed by ChIP assay. CTL, control. Student's t test was used for statistic analysis, NS, no significant difference. Error bars indicate mean ± S.D.; **, p < 0.01; ***, p < 0.005.
To determine whether p300 is responsible in mediating the de novo acetylation of histone H3K56, we used siRNA to knock down p300 in mouse MEF cells. P300 siRNA inhibited more than 90% of p300 expression, and p300 knockdown inhibited the global H3K56 acetylation by about 50% (Fig. 3B), suggesting that other acetyltransferases are also involved in catalyzing global histone H3K56 acetylation. In fact, suppression of GCN5 expression also partially inhibited total histone H3K56 acetylation in MEFs (Fig. 3C). Interestingly, p300 knockdown significantly inhibited H3K56 acetylation in bclaf1 promoter in both wild-type and Sirt1-null MEF cells, but GCN5 knockdown did not affect H3K56 acetylation on the bclaf1 promoter (Fig. 3D). Therefore, both GCN5 and p300 are involved in the global H3K56 acetylation, but only p300 is involved in H3K56 acetylation at the bclaf1 promoter region.
Bclaf1 Expression Is Regulated by TCR/CD28-mediated NF-κB Transcriptional Activation
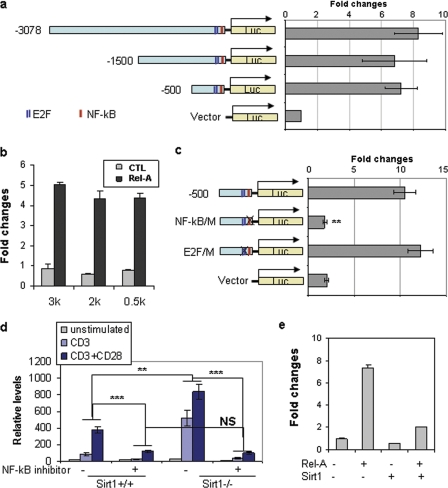
Both CNS3 and CNS4 locate in the optimal promoter region of bclaf1 locus (−1500 bp). Analyzing this region revealed one NF-κB-binding site and two E2F1-binding sites, all of which locate within the −500-bp region (Fig. 4A), suggesting a possibility of the involvement of NF-κB and E2F1 in Bclaf1 gene transcription. To test this, we subcloned the 3-kbp region (−3078 to + 76) into pGL3 luciferase vector. In contrast to a basal level pGL3-luciferase activity, this 3-kbp fragment mediated significant luciferase activity. A serial deletion of this 3-kbp region identified that the −500 to +76 region carries transcriptional activity (Fig. 4A). Co-transfection of Rel-A, an NF-κB family transcription factor, but not E2F1, dramatically enhanced Bclaf1-luciferase activity (Fig. 4B and supplemental Fig. 2). Mutation of the NF-κB-binding site completely abolished Rel-A-mediated Bclaf1-luciferase activity, whereas mutation of the E2F1-binding sites had little effect (Fig. 4C).
FIGURE 4.
The transcription factor NF-κB is involved in Bclaf1 transcription in T cells. A and B, luciferase (Luc) plasmids that carry bclaf1 promoter regions were co-transfected with control TK-luciferase plasmids. The luciferase activity in transfected cells was analyzed. -Fold changes when compared with control TK-luciferase activities are shown (A). Bclaf1-luciferase plasmids in A were co-transfected with or without Rel-A expression plasmid. The -fold changes of luciferase activity using different plasmids were normalized to TK-luciferase activities (B). C, the binding sites of NF-κB and E2F1 in bclaf1 promoter were mutated, and their luciferase activities were analyzed in transiently transfected HEK293 cells. D, primary CD4 T cells from Sirt1+/+ and Sirt1−/− mice were stimulated with anti-CD3 or with anti-CD3 plus anti-CD28 for 16 h in the presence or absence of 20 μm NF-κB inhibitor JSH-23. Bclaf1 mRNA levels were analyzed by real-time PCR. E, Sirt1 expression plasmids were co-transfected with Bclaf1-luc and Rel-A expression plasmids. The luciferase activities in transfected cells were analyzed. Error bars represent data (mean ± S.D.) from three independent experiments. Student's t test was used for statistic analysis, NS, no significant difference. **, p < 0.01; ***, p < 0.005.
We then used ChIP assay and analyzed the binding of Rel-A to bclaf1 promoter to investigate whether NF-κB is involved in Bclaf1 transcription in T cells. Only a basal level of Rel-A binding to bclaf1 promoter was detected in naive CD4+ T cells. TCR stimulation dramatically induced Rel-A binding to bclaf1 promoter, which is further enhanced by CD28 signaling, indicating that NF-κB is involved in TCR/CD28-induced Bclaf1 transcription in T cells (Fig. 4D). Sirt1 has been found to inhibit both NF-κB and E2F1 transcriptional activities by directly interacting with Rel-A and E2F1 (12, 32). Therefore, we asked whether Sirt1 inhibits Bclaf1 transcription by suppressing NF-κB. In fact, the promoter binding of Rel-A in Sirt1-null T cells was increased over that of wild-type T cells upon TCR/CD28 stimuli (Fig. 4D), suggesting that Sirt1 inhibits Rel-A promoter DNA binding activity. Treatment of T cells with an NF-κB inhibitor, which blocks Rel-A nuclear translocation, inhibited Bclaf1 transcription both in wild-type and in Sirt1-null T cells (Fig. 4D). Furthermore, co-expression of Sirt1 inhibited Rel-A-driven Bclaf1-luciferase activity (Fig. 4E). Collectively, our data suggest that Rel-A is a transcription factor for Bclaf1 gene expression and that Sirt1 inhibits Bclaf1 transcription by suppressing Rel-A.
Sirt1 Binds to bclaf1 Promoter through NF-κB Transcription Factor Rel-A
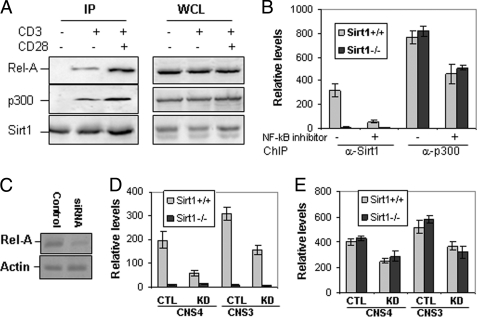
Both Sirt1 and p300 have been found to regulate NF-κB transcriptional activity by physically interacting with Rel-A (12). We therefore asked whether p300, Sirt1, and Rel-A form a complex in primary T cells. Both Rel-A and p300 proteins were found to co-immunoprecipitate with Sirt1 in mouse primary T cells upon stimulation with anti-CD3 or anti-CD3 plus anti-CD28, but not in naive T cells (Fig. 5A), indicating that TCR/CD28-mediated signaling is required to form the p300·Sirt1·Rel-A complex for Bclaf1 expression in T cells.
FIGURE 5.
Rel-A, Sirt1, and p300 form a complex in T cells. A, primary T cells from wild-type mice were stimulated with anti-CD3 or anti-CD3 plus anti-CD28 for 2 h and lysed. The cell lysate was immunoprecipitated (IP) with anti-Sirt1 antibody. Sirt1-bound Rel-A (left top panel) and p300 (left middle panel) were detected with anti-Rel-A and anti-p300 antibodies, respectively. The same membrane was reprobed with anti-Sirt1 as a control (left bottom panel). The expression levels of Rel-A, Sirt1, and p300 in the whole cell lysates (WCL) were determined by Western blotting as controls (right three panels). B, mouse CD4 T cells were stimulated with anti-CD3 or anti-CD3 plus anti-CD28 for 16 h in the presence or absence of 10 μm NF-κB inhibitor. The binding of Sirt1 and p300 to bclaf1 promoter CNS4 was analyzed by ChIP assay followed by real-time PCR. C–E, MEF cells were transfected with control (lane 1) or Rel-A-specific siRNA (lane 2). The efficiency of siRNA-mediated knockdown was determined by Western blotting (C). The effects of Rel-A knockdown (KD) on binding of Sirt1 (D) or p300 (E) to bclaf1 promoter CNS4 and CNS3 regions in Sirt1+/+ and Sirt1−/− MEF cells were determined by ChIP assay followed by real-time PCR. CTL, control. Error bars represent data of triplicate wells (mean ± S.D.), and representative data from three independent experiments are shown.
Because Sirt1 lacks DNA-binding domains, we asked whether Sirt1 is recruited to bclaf1 promoter by binding to Rel-A. To test this hypothesis, we treated T cells with an NF-κB inhibitor, which blocks Rel-A nuclear translocation. As shown in Fig. 5B, inhibition of Rel-A nuclear translocation dramatically blocked Sirt1 recruitment to bclaf1 promoter. As a control, Sirt1 binding to bclaf1 promoter was not detected in Sirt1-null T cells. In contrast, p300 binding to bclaf1 promoter was only reduced by about 40% with the treatment of NF-κB inhibitor. Loss of Sirt1 function has little effect on the promoter binding activity of p300 in T cells. Therefore, bclaf1 promoter binding by Sirt1 appears to solely depend on Rel-A, whereas additional mechanisms by which p300 binds to bclaf1 promoter exist.
To further determine the role of Rel-A in Sirt1 and p300 binding at the bclaf1 promoter, we used an siRNA approach to knock down Rel-A in MEF cells. As reported by Kawahara et al. (33), siRNA transfection sufficiently inhibited Rel-A expression with more than 90% knockdown as measured by Western blotting (Fig. 5C). Suppression of Rel-A inhibited Sirt1 binding to bclaf1 promoter by more than 80% (Fig. 5D), whereas Rel-A knockdown only inhibited p300 binding to bclaf1 promoter by about 20–30% (Fig. 5E). Similar to that in T cells, Sirt1 deficiency has no effect of p300 binding to bclaf1 promoter, and knockdown of Rel-A in Sirt1-null MEF inhibited p300 binding to bclaf1 promoter by about 50% (Fig. 5E), suggesting that the bclaf1 promoter binding activity of p300 is partially regulated by Rel-A but is independent of its interaction with Sirt1. Rel-A has been identified as a substrate of p300 acetyltransferase and Sirt1 deacetylase; it is therefore possible that Sirt1 suppresses Bclaf1 transcription by directly inhibiting Rel-A acetylation. We then compared Rel-A acetylation levels between Sirt1-deficient and wild-type T cells. As shown in supplemental Fig. 3, a slight increase of Rel-A acetylation, but not its total protein expression levels, was detected, suggesting that Sirt1 is partially involved in regulation of Rel-A acetylation, which in turn could regulate Bclaf1 transcription. Therefore, Sirt1 appears to inhibit Bclaf1 transcription through two mechanisms.
Knockdown of Bclaf1 Expression Inhibits the Activation of Sirt1-null T Cells
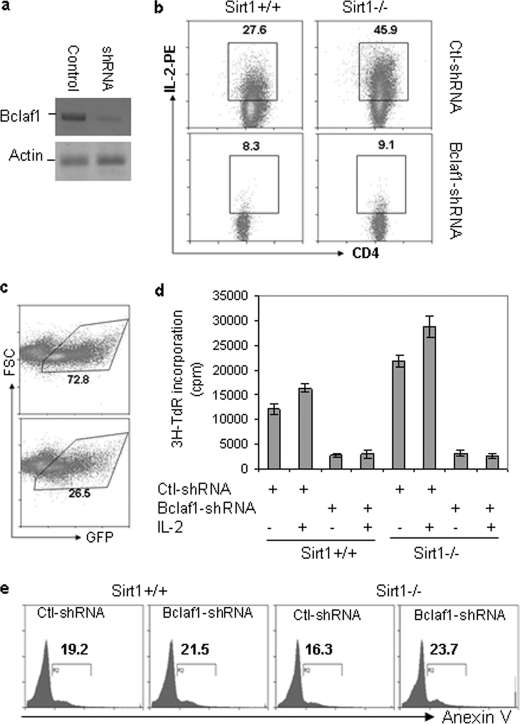
Our data so far suggest that TCR/CD28-mediated Rel-A·Sirt1·p300 complex formation at bclaf1 promoter region regulates Bclaf1 transcription in T cells. To test whether increased Bclaf1 expression is responsible for enhanced activation of Sirt1-null T cells, we used an shRNA approach to knock down Bclaf1 expression in T cells. As shown in Fig. 6A, lentivirus-based shRNA delivery inhibited more than 80% of Bclaf1 expression in primary T cells. Inhibition of Bclaf1 expression by shRNA in both wild-type and Sirt1−/− CD4 T cells dramatically inhibited IL-2 production, providing a direct link between Sirt1-mediated suppression of Bclaf1 expression and Sirt1-mediated suppression of T cell activation (Fig. 6B).
FIGURE 6.
Bclaf1 knockdown inhibits Sirt1-null T cell activation. A, mouse primary T cells were isolated from Sirt1+/+ and Sirt1−/− mice and infected with lentivirus that carries shRNA specific to Bclaf1 or a control shRNA. GFP-positive cells were sorted 2 days after infection. Bclaf1 protein expression in sorted cells was determined by Western blotting with anti-Bclaf1 antibody (top panel). Protein expression of actin was analyzed as a loading control (bottom panel). B and C, mouse primary CD4+ T cells from Sirt1+/+ and Sirt1−/− mice were infected with lentivirus that carries shRNA specific to Bclaf1 or a control (Ctl) shRNA. The production of IL-2 and cell surface CD4 expression in gated GFP+ cells was analyzed by intracellular staining followed by flow cytometry (B), and the percentages of GFP+ cells at 2 days after infection are shown (C). D, GFP+ cells were sorted and restimulated with anti-CD3 (1 μg/ml) or anti-CD3 plus IL-2 (1 ng/ml). Proliferation was analyzed by [3H]thymidine incorporation (3H-TdR). Error bars indicate mean ± S.D. E, GFP+ cells were gated, and apoptotic cells were analyzed by annexin V staining. FSC, forward scatter; 3H-TdR, [3H]thymidine.
We also noticed a significantly reduced percentage of GFP+ CD4 T cells when infected with lentivirus carrying Bclaf1-specific shRNA when compared with those infected with lentivirus carrying scrambled shRNA (Fig. 6C), suggesting that Bclaf1 knockdown inhibits CD4 T cell proliferation or promotes T cell death. Indeed, sorted GFP+ cells with Bclaf1 knockdown showed greatly impaired proliferation upon restimulation with anti-CD3 plus anti-CD28 antibodies (Fig. 6C). In contrast, the percentages of annexin V-positive cells were indistinguishable between Bclaf1 knockdown and control cells (Fig. 6D). Therefore, Bclaf1 is required for T cell proliferation and Sirt1 suppresses T cell activation by inhibiting Bclaf1 transcription.
DISCUSSION
Increasing evidence indicates that Sirt1 can suppress both innate and adaptive immune responses in mice. Despite the identification of NF-κB and AP-1 transcription factors as substrates of Sirt1 deacetylase activity in T cell and macrophages (6–11), the molecular mechanisms underlying how Sirt1 suppresses immune functions remain unclear. In this study, we demonstrate that Sirt1 suppresses NF-κB-dependent transcription of Bclaf1, which is required for T cell activation. Given the fact that Sirt1, p300, and Rel-A are ubiquitously expressed in mammals, we suggest that Sirt1-mediated regulation of Bclaf1 gene expression via H3K56 deacetylation likely serves as a common mechanism in a variety of physiological contexts.
Rel-A translocates to the nucleus of T cells upon TCR/CD28 signaling, and Rel-A translocation is required for the recruitment of Sirt1 to bclaf1 promoter. Based on these results, we suggest a model in which Sirt1 is recruited to bclaf1 promoter via physical interaction with Rel-A and deacetylates histone H3K56 to regulate the transcription of Bclaf1 and possibly other NF-κB target genes. Sirt6 has been found to bind chromatin via a similar mechanism (33). Thus, both Sirt1 and Sirt6 can contribute to the negative feedback of NF-κB-driven gene expression. However, unlike Sirt6, which binds to Rel-A without affecting Rel-A acetylation, Sirt1 can regulate NF-κB transcriptional activity using two mechanisms. Our data here indicate that Sirt1 is recruited by Rel-A and deacetylates H3K56 at NF-κB-binding loci of genes to inhibit NF-κB-mediated transcription. Rel-A knockdown inhibited Sirt1 recruitment to both CNS3 and CNS4 regions. However, NF-κB-binding sites were not identified in the CNS3 region. One explanation is that Rel-A may bind to this region through interaction with other transcription factors. In addition, it is reported that Sirt1 suppresses p300-mediated Rel-A acetylation to inhibit Rel-A binding to promoter DNA (12). In fact, a slightly increased Rel-A acetylation, but not its protein expression, was detected in Sirt1-null T cells. Therefore, Sirt1 appears to inhibit Bclaf1 transcription via both mechanisms. Because NF-κB regulates the expression of a variety of genes involved in cell survival, apoptosis, immunity, and inflammation (34, 35), further studies are needed to understand how Sirt1 and Sirt6 differentially regulate Rel-A activity in response to extracellular stimuli. One possibility is the expression levels of Sirt1 and Sirt6 in different cell types. For example, Sirt1 is highly expressed in T cells in contrast to its low protein levels in B lymphocytes, whereas the opposite is true for Sirt6 (data not shown).
Histone H3K56 acetylation has recently been shown to have a critical role in packaging DNA into chromatin following DNA replication and repair in budding yeast and mammals (28, 31, 36–39). Little is known about the role of H3K56 acetylation in gene transcription. However, a recent study using ChIP-on-chip revealed that H3K56 acetylation is most likely involved in transcription of genes in several pathways in tumorigenesis such as cell cycle, DNA damage response, DNA repair, and apoptosis (31). Our findings here suggest that H3K56 acetylation at the Bcalf1 promoter correlates with increased Bclaf1 transcription. In addition, our previous study demonstrated that activation of sirt1-null T cells does not require co-stimulatory signaling (11). In accordance with our previous findings, TCR stimulation alone is sufficient to up-regulate Bclaf1 expression and H3K56 acetylation specifically at the promoter region of bclaf1 in Sirt1-null T cells. In contrast, the acetylation of CNS regions outside of the promoter region of bclaf1 increased with TCR/CD28 signaling but did not differ between WT and Sirt1-null T cells. This is possibly because TCR signaling alone is sufficient to recruit Sirt1 protein to bclaf1 promoter, and Sirt1 attenuates T cell activation by suppressing TCR-mediated Bclaf1 transcription in T cells.
Despite limited knowledge on the physiological functions of Bclaf1, a recent study using gene-targeted mutation in mice (19) clearly indicates an essential role of Bclaf1 in T cell activation. Although it was initially identified as a Bcl2-interacting protein and its functions are associated with apoptosis, Bclaf1 deficiency has no effects on activation-induced T cell death when stimulated in vitro (19). In this study, we demonstrate that knocking down Bclaf1 using RNAi leads to reduced IL-2-producing T cells and reduced proliferation in response to TCR stimulation, especially in hyperactive, Sirt1-null T cells. The unresponsiveness to TCR/CD28 stimuli of Bclaf1-null T cells is phenotypically different from T cell clonal anergy because further addition of exogenous IL-2 cannot rescue the proliferation of T cells.
In conclusion, our study here demonstrates that Sirt1 inhibits Bclaf1 transcription by deacetylating H3K56 residues at CNS3 and CNS4 within the promoter region. Loss of Sirt1, which causes spontaneous autoimmunity in mice, leads to increased Bclaf1 gene transcription that is responsible for T cell activation. It is still not known how Bclaf1 regulates T cell activation, but the function of Bclaf1 appears to be cell type-specific because loss of Bclaf1 function has no effect on B cell activation (19). Further studies will help us gain a better understanding of the underlying molecular mechanism of Bclaf1 in promoting T cell activation.
Supplementary Material
Acknowledgment
We thank Dr. Michael McBurney for Sirt1-null mice.
This work was supported, in whole or in part, by National Institutes of Health Grants R01AI079056 and R21AG028493 and a “The type I Diabetes Pathfinder Award” (DK083050) (to D. F.). This work was also supported by research funds of Chonbuk National University (to S.-M. L.) and by a grant from the Northwestern University Interdepartmental ImmunoBiology Flow Cytometry Core Facility.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1–3.
- TCR
- T cell receptor
- MEF
- mouse embryonic fibroblast
- CNS
- conserved non-coding sequence(s).
REFERENCES
- 1. Shore D., Squire M., Nasmyth K. A. (1984) EMBO J. 3, 2817–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brachmann C. B., Sherman J. M., Devine S. E., Cameron E. E., Pillus L., Boeke J. D. (1995) Genes Dev. 9, 2888–2902 [DOI] [PubMed] [Google Scholar]
- 3. Michishita E., Park J. Y., Burneskis J. M., Barrett J. C., Horikawa I. (2005) Mol. Biol. Cell 16, 4623–4635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Haigis M. C., Sinclair D. A. (2010) Annu. Rev. Pathol. 5, 253–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gillum M. P., Erion D. M., Shulman G. I. (2010) Trends Mol. Med., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gratchev A., Kzhyshkowska J., Kannookadan S., Ochsenreiter M., Popova A., Yu X., Mamidi S., Stonehouse-Usselmann E., Muller-Molinet I., Gooi L., Goerdt S. (2008) J. Immunol. 180, 6553–6565 [DOI] [PubMed] [Google Scholar]
- 7. Schug T. T., Xu Q., Gao H., Peres-da-Silva A., Draper D. W., Fessler M. B., Purushotham A., Li X. (2010) Mol. Cell Biol. 30, 4712–4721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kwon H. S., Brent M. M., Getachew R., Jayakumar P., Chen L. F., Schnolzer M., McBurney M. W., Marmorstein R., Greene W. C., Ott M. (2008) Cell Host Microbe 3, 158–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sequeira J., Boily G., Bazinet S., Saliba S., He X., Jardine K., Kennedy C., Staines W., Rousseaux C., Mueller R., McBurney M. W. (2008) Exp. Cell Res. 314, 3069–3074 [DOI] [PubMed] [Google Scholar]
- 10. Silvestre R., Silva A. M., Cordeiro-da-Silva A., Ouaissi A. (2009) Immunology 128, 484–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang J., Lee S. M., Shannon S., Gao B., Chen W., Chen A., Divekar R., McBurney M. W., Braley-Mullen H., Zaghouani H., Fang D. (2009) J. Clin. Invest. 119, 3048–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yeung F., Hoberg J. E., Ramsey C. S., Keller M. D., Jones D. R., Frye R. A., Mayo M. W. (2004) EMBO J. 23, 2369–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kasof G. M., Goyal L., White E. (1999) Mol. Cell Biol. 19, 4390–4404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu H., Lu Z. G., Miki Y., Yoshida K. (2007) Mol. Cell Biol. 27, 8480–8491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Haraguchi T., Holaska J. M., Yamane M., Koujin T., Hashiguchi N., Mori C., Wilson K. L., Hiraoka Y. (2004) Eur. J. Biochem. 271, 1035–1045 [DOI] [PubMed] [Google Scholar]
- 16. Ziegelbauer J. M., Sullivan C. S., Ganem D. (2009) Nat. Genet. 41, 130–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee K. M., Hsu IaW., Tarn W. Y. (2010) Nucleic Acids Res. 38, 3340–3350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bracken C. P., Wall S. J., Barré B., Panov K. I., Ajuh P. M., Perkins N. D. (2008) Cancer Res. 68, 7621–7628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McPherson J. P., Sarras H., Lemmers B., Tamblyn L., Migon E., Matysiak-Zablocki E., Hakem A., Azami S. A., Cardoso R., Fish J., Sanchez O., Post M., Hakem R. (2009) Cell Death Differ. 16, 331–339 [DOI] [PubMed] [Google Scholar]
- 20. McBurney M. W., Yang X., Jardine K., Hixon M., Boekelheide K., Webb J. R., Lansdorp P. M., Lemieux M. (2003) Mol. Cell Biol. 23, 38–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Atanassov B. S., Evrard Y. A., Multani A. S., Zhang Z., Tora L., Devys D., Chang S., Dent S. Y. (2009) Mol. Cell 35, 352–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sellers W. R., Novitch B. G., Miyake S., Heith A., Otterson G. A., Kaye F. J., Lassar A. B., Kaelin W. G., Jr. (1998) Genes Dev. 12, 95–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gao B., Lee S. M., Fang D. (2006) J. Biol. Chem. 281, 29711–29718 [DOI] [PubMed] [Google Scholar]
- 24. Chen A., Gao B., Zhang J., McEwen T., Ye S. Q., Zhang D., Fang D. (2009) Mol. Cell Biol. 29, 5348–5356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee S. M., Gao B., Fang D. (2008) Blood 111, 3599–3606 [DOI] [PubMed] [Google Scholar]
- 26. Johnson C. A., O'Neill L. P., Mitchell A., Turner B. M. (1998) Nucleic Acids Res. 26, 994–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cohn R. H., Kedes L. H. (1979) Cell 18, 855–864 [DOI] [PubMed] [Google Scholar]
- 28. Das C., Lucia M. S., Hansen K. C., Tyler J. K. (2009) Nature 459, 113–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yuan J., Pu M., Zhang Z., Lou Z. (2009) Cell Cycle 8, 1747–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hebert C., Roest Crollius H. (2010) Genome Biol. 11, R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vempati R. K., Jayani R. S., Notani D., Sengupta A., Galande S., Haldar D. (2010) J. Biol. Chem. 285, 28553–28564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang C., Chen L., Hou X., Li Z., Kabra N., Ma Y., Nemoto S., Finkel T., Gu W., Cress W. D., Chen J. (2006) Nat. Cell Biol. 8, 1025–1031 [DOI] [PubMed] [Google Scholar]
- 33. Kawahara T. L., Michishita E., Adler A. S., Damian M., Berber E., Lin M., McCord R. A., Ongaigui K. C., Boxer L. D., Chang H. Y., Chua K. F. (2009) Cell 136, 62–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Karin M., Greten F. R. (2005) Nat. Rev. Immunol. 5, 749–759 [DOI] [PubMed] [Google Scholar]
- 35. Hoffmann A., Levchenko A., Scott M. L., Baltimore D. (2002) Science 298, 1241–1245 [DOI] [PubMed] [Google Scholar]
- 36. Maas N. L., Miller K. M., DeFazio L. G., Toczyski D. P. (2006) Mol. Cell 23, 109–119 [DOI] [PubMed] [Google Scholar]
- 37. Schneider J., Bajwa P., Johnson F. C., Bhaumik S. R., Shilatifard A. (2006) J. Biol. Chem. 281, 37270–37274 [DOI] [PubMed] [Google Scholar]
- 38. Tang Y., Holbert M. A., Wurtele H., Meeth K., Rocha W., Gharib M., Jiang E., Thibault P., Verreault A., Cole P. A., Marmorstein R. (2008) Nat. Struct. Mol. Biol. 15, 738–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Thaminy S., Newcomb B., Kim J., Gatbonton T., Foss E., Simon J., Bedalov A. (2007) J. Biol. Chem. 282, 37805–37814 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.