Abstract
The microtubule-associated protein tau, which becomes hyperphosphorylated and pathologically aggregates in a number of these diseases, is extremely sensitive to manipulations of chaperone signaling. For example, Hsp90 inhibitors can reduce the levels of tau in transgenic mouse models of tauopathy. Because of this, we hypothesized that a number of Hsp90 accessory proteins, termed co-chaperones, could also affect tau stability. Perhaps by identifying these co-chaperones, new therapeutics could be designed to specifically target these proteins and facilitate tau clearance. Here, we report that the co-chaperone Cdc37 can regulate aspects of tau pathogenesis. We found that suppression of Cdc37 destabilized tau, leading to its clearance, whereas Cdc37 overexpression preserved tau. Cdc37 was found to co-localize with tau in neuronal cells and to physically interact with tau from human brain. Moreover, Cdc37 levels significantly increased with age. Cdc37 knockdown altered the phosphorylation profile of tau, an effect that was due in part to reduced tau kinase stability, specifically Cdk5 and Akt. Conversely, GSK3β and Mark2 were unaffected by Cdc37 modulation. Cdc37 overexpression prevented whereas Cdc37 suppression potentiated tau clearance following Hsp90 inhibition. Thus, Cdc37 can regulate tau in two ways: by directly stabilizing it via Hsp90 and by regulating the stability of distinct tau kinases. We propose that changes in the neuronal levels or activity of Cdc37 could dramatically alter the kinome, leading to profound changes in the tau phosphorylation signature, altering its proteotoxicity and stability.
Keywords: Alzheimer Disease, Chaperone Chaperonin, Heat Shock Protein, Protein Kinases, Tau
Introduction
The microtubule-associated protein tau is an unusual client of the chaperone network due to its intrinsically disordered nature. Disordered proteins are highly susceptible to aggregation, and tau is no exception. The classical function of chaperones is to properly fold proteins into their functional state; however, it is becoming increasingly clear that chaperones also are essential mediators of aggregation kinetics of disordered or misfolded proteins, particularly in neurons (1–3). The recent structural characterization of full-length tau showed that the tau protein can undergo many transitional conformations (4), and each of these conformations may represent a potentially toxic entity. Accumulation of these misfolded tau intermediates in the human brain causes tauopathies, the most common being Alzheimer disease. Although some conformations are certainly necessary for tau function, it is likely that others are pathogenic in the brain. Chaperones facilitate these conformational changes by directly affecting the secondary or tertiary structure of tau; however, post-translational modifications to tau like phosphorylation and proteolysis can also be influenced by chaperones. Such biochemical changes to tau may tip the scales of its folding landscape, favoring the formation of pathogenic tau conformations over normal functional conformations. Thus, chaperones may be able to influence tau stability both directly and indirectly, for better or for worse.
One of the primary ATPases of the chaperone family that is linked to tau stability is (Hsp90 (heat shock protein of 90 kDa). In addition to regulating tau directly, Hsp90 also regulates a number of kinases that can directly phosphorylate tau (5, 6). This coordination of kinase triage decisions by Hsp90 requires the co-chaperone Cdc37 (cell division cycle protein 37). The Hsp90/Cdc37 machine is essential for the maturation of a number of kinases, including Akt and cyclin-dependent kinases, which have been linked to tau phosphorylation (5, 7–10). In fact, Hsp90 inhibition was shown not only to clear tau but also to regulate activation of the tau kinase Cdk5 (cyclin-dependent kinase 5) (11). In Alzheimer disease, aberrant hyperactivation of Cdk5 is thought to contribute to tau pathogenesis (12). On the basis of this evidence, we hypothesized that Cdc37 would be involved in tau stability and perhaps pathobiology by multiple mechanisms. Indeed, we found that neuronal Cdc37 is able to regulate phosphorylation of tau and ultimately its stability.
MATERIALS AND METHODS
Antibodies, siRNAs, Plasmids, and Chemicals
PHF-1 antibodies (anti-phospho-Ser-396/Ser-404 tau) were provided by Peter Davies (Albert Einstein College of Medicine, Yeshiva University, New York, NY). Anti-Cdc37 and anti-α-synuclein antibodies were obtained from Cell Signaling (Danvers, MA). Anti-p23 antibody JJ3 was provided by Dr. David O. Toft (Mayo Clinic). Antibody 12E8 (anti-phospho-Ser-262/Ser-356 tau) was provided by P. Seubert (Elan Pharmaceuticals, San Francisco, CA). Anti-total tau and mouse anti-Cdc37 antibodies were obtained from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). Anti-GAPDH antibody was obtained from BIODESIGN International (Saco, ME). Anti-actin antibody was obtained from Sigma. Anti-phospho-Thr-231 tau antibody was from Abcam (Cambridge, MA). Anti-phospho-Ser-199/Ser-202 tau antibody was from AnaSpec (San Jose, CA). Secondary antibodies were obtained from Southern Biotech (Birmingham, AL). All antibodies were used at a 1:1000 dilution with the exception of antibody PHF-1, which was used at a 1:200 dilution. Secondary antibodies conjugated to fluorophore (Molecular Probes Alexa Fluor) were purchased from Invitrogen. The CDC37 gene was PCR-amplified from a human cDNA library (Invitrogen) and cloned into the pCMV6 plasmid. All other clones used were in the pcDNA3.1 plasmid. All siRNAs were obtained from Qiagen, and their sequences are listed in Table 1. siRNA efficiency for protein knockdown was validated by Western blotting (see Fig. 1A). The Hsp90 inhibitor 17-(allylamino)-17-demethoxygeldanamycin (17-AAG)2 was purchased from Sigma.
TABLE 1.
siRNA sequences used in this study
| Gene name | Common name | Target sequence |
|---|---|---|
| PTGES3 | p23 | CAGCTTAGGGAAAGAGAATAA |
| CDC37 | CDC37 | CACCAGACAATCGTCATGCAA |
| Non-silencing control | Proprietary (Qiagen) |
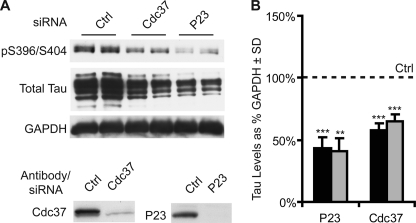
FIGURE 1.
Cdc37 is a potential tau modifier. A, HeLa cells stably overexpressing V5-tagged wild-type human tau were transiently transfected with the indicated siRNAs or scrambled negative control siRNA (Ctrl) for 72 h and analyzed by Western blotting. p23 siRNA was used as a positive reducing control. Knockdown for each gene was confirmed by Western blotting. B, Western blot quantitation by densitometry. The levels of phospho-tau (black bars) were calculated from phospho-Ser-396/Ser-404 tau in A after GAPDH normalization and are presented as a percentage of the control (dashed line). Total tau levels (gray bars) were calculated from total tau in A after GAPDH normalization. Statistical analyses using multiple experiments for p23 (n = 5) and Cdc37 (n = 12) siRNAs across cell models demonstrated that p23 and Cdc37 siRNAs significantly reduced both phospho-tau and total tau levels. **, p < 0.01; ***, p < 0.001.
Cell Culture and Western Blot Analysis
HeLa cells stably transfected with V5-tagged 4R0N tau were maintained as described previously (13). BE(2)-M17 cells were maintained in Opti-MEM plus 10% FBS (Invitrogen). Transfections in cells were done as described previously (14). Briefly, plasmid transfections were done utilizing Lipofectamine 2000 reagent (Invitrogen). siRNAs were transfected with siLentFect (Bio-Rad). Cells were harvested in M-PER buffer (Pierce) containing 1× protease inhibitor mixture (Calbiochem), 1 mm phenylmethylsulfonyl fluoride, and 1× phosphatase inhibitor I and II mixtures (Sigma). Measurements of tau levels in cell culture were performed by Western blot analysis.
Human Brain Tissues
Alzheimer disease patient and normal (control) medial temporal gyruses were homogenized and processed for co-immunoprecipitation with anti-Cdc37 antibody as described (14). Samples were analyzed by Western blotting.
Primary Neurons
Forebrain neurons were obtained from the cortex and hippocampus of P1 wild-type mice as described (15). Primary neurons were grown for 1 week on poly-l-lysine-coated 8-chamber slides in Neurobasal medium with vitamin B27 supplement. They were fixed and stained as described below (see “Immunohistochemistry”).
Immunocytochemistry
M17 human neuroblastoma cells were plated onto a chamber slide (Fisher) and maintained in the medium described above. Cells were processed for immunocytochemistry as described (13). Rabbit anti-tau and mouse anti-Cdc37 primary antibodies (1:100) were used. Alexa Fluor 488-conjugated (anti-rabbit; 1:1000) and Alexa Fluor 594-conjugated (anti-mouse; 1:1000) secondary antibodies were added for detection. Imaging and co-localization scatter analysis were performed with a Zeiss AxioVision imager.
Phos-tag Analysis
Recombinant 4R0N tau (14) or cell lysates treated with or without alkaline phosphatase (400 units for 30 min at 30 °C; New England Biolabs) were resolved on 9% SDS-polyacrylamide gels prepared with 75 μm acrylamide-pendant Phos-tag ligand and 150 μm MgCl2 according to the instructions provided by the Phos-tag Consortium. Gels were electrophoresed at 5 mA/gel for 10–12 h. Prior to transfer, gels were first equilibrated in methanol-free transfer buffer containing 1 mm EDTA for 10 min and then in transfer buffer without EDTA for 10 min. Transfer was performed overnight at 30 V at 4 °C.
Quantification and Statistical Analyses
Replicate densitometric values were obtained from repeated experiments, Western blots, and densitometric variability. S.D. values were calculated from replicate blots/experiments. For analyses across M17 and HeLaC3 cell models (see Fig. 1), S.E. was calculated where n ≥ 3. Statistical significance was determined with a heteroscedastic two-tailed Student's t test.
RESULTS
Cdc37 siRNA Reduces Tau Levels
Cdc37 was initially identified as a potential tau modifier using siRNA in a cell culture system that constitutively overexpresses V5-tagged human 4R0N tau (HeLaC3). We chose to investigate Cdc37 based on its well documented role in regulating kinase stability. Cells were transfected with a scrambled siRNA (control) or siRNA targeting p23 or Cdc37. As reported previously (16), p23 siRNA reduced tau levels. Cdc37 knockdown also reduced tau levels (Fig. 1). p23 knockdown reduced both total tau and phospho-tau. Cdc37 knockdown caused slightly greater reductions in phospho-tau compared with total tau. This was consistent with our hypothesis that Cdc37 might be able to regulate tau through multiple mechanisms, both as an Hsp90 co-chaperone and as a regulator of tau kinases.
Cdc37 Knockdown and Overexpression Reciprocally Regulate Mutant Tau Species, but Not α-Synuclein
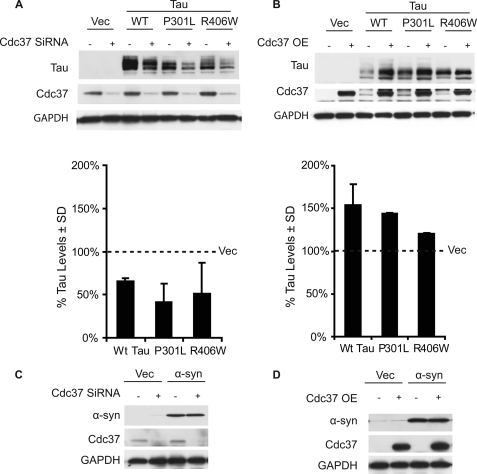
To determine whether Cdc37 knockdown could affect tau mutants associated with human tauopathies (i.e. frontotemporal dementia and parkinsonism linked to chromosome 17 and progressive supranuclear palsy) in the same way that it was affecting wild-type tau, HeLa cells were transfected with Cdc37 siRNA for 24 h and then cDNAs coding for the indicated tau variants (WT, P301L, or R406W). Lysates were analyzed by Western blotting after an additional 48-h incubation. Cdc37 knockdown reduced the levels of all tau species (Fig. 2A). Conversely, overexpression of Cdc37 in a similar experiment preserved both wild-type tau and P301L tau but was not as effective at stabilizing R406W tau (Fig. 2B). Similar studies with a protein that is known to aggregate in Parkinson disease, α-synuclein, showed that Cdc37 modulation did not affect its levels (Fig. 2, C and D), consistent with our previous findings that α-synuclein is not sensitive to chaperone modulation (17). Moreover, this confirmed that Cdc37 was not nonspecifically regulating transcription of tau because both tau and α-synuclein cDNAs were overexpressed using the same vector driven by a CMV promoter.
FIGURE 2.
Cdc37 regulates tauopathy-associated tau mutants, but not synuclein. A, HeLa cells were transfected with Cdc37 siRNA for 24 h; these same cells were then transfected with WT, P301L, or R406W tau or vector (Vec) plasmid DNA. Lysates were analyzed by Western blotting. The quantitation plot of the Western blot after GAPDH normalization shows that Cdc37 knockdown reduced various tau species. B, HeLa cells were cotransfected with Cdc37 and the indicated tau plasmids for 48 h. The quantitation plot of the Western blot after GAPDH normalization shows that overexpression (OE) of Cdc37 increased tau levels. C, HeLa cells were transfected with Cdc37 siRNA for 24 h and then transfected with α-synuclein. D, HeLa cells were cotransfected with Cdc37, α-synuclein, and vector. The levels of α-synuclein protein were unaffected by knockdown (C) or overexpression (D) of Cdc37. Error bars indicate S.D., and values are shown as a percent of cells transfected with vector alone following normalization to the loading control.
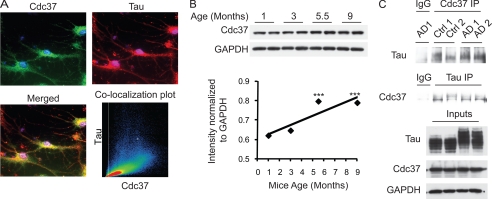
Cdc37 Increases with Age, Co-localizes with Endogenous Tau in Neurons, and Physically Interacts with Tau from Human Brain
To test whether reducing Cdc37 levels could also affect endogenous tau, M17 neuroblastoma cells were transfected with Cdc37 siRNA for 72 h, and lysates were analyzed by Western blotting. Knockdown of Cdc37 reduced the levels of phosphorylated tau and, to a lesser extent, total tau (Fig. 3A). Conversely, overexpression of Cdc37 preserved tau in these same cells (Fig. 3B). Immunofluorescence staining was then used to show that endogenous Cdc37 co-localized with endogenous tau in M17 cells (Fig. 3C) and murine primary neurons derived from wild-type mice (Fig. 4A). Next, changes in Cdc37 protein levels due to age were assessed by Western blotting. Significant increases in Cdc37 levels were apparent in adult mice (5.5 and 9 months old) compared with juvenile mice (1 and 3 months old) (Fig. 4B). Co-immunoprecipitation studies using both non-diseased (control) and Alzheimer disease (AD) human brain lysates showed association of tau with Cdc37 (Fig. 4C). Cdc37 proteins levels were similar in control and AD human brains. These studies confirmed that Cdc37 is very likely able to affect both normal and abnormal tau processing in vivo. Although the levels of Cdc37 were not affected by disease, this does not exclude the possibility that its activity can be changed, as indicated by its increased expression with age. Thus, we began to investigate the mechanisms used by Cdc37 to regulate tau stability.
FIGURE 3.
Cdc37 regulates endogenous tau and co-localizes with tau. A, M17 human neuroblastoma cells were transfected with Cdc37 siRNA for 72 h. The quantification plot of the Western blot from three separate experiments illustrates that knockdown of Cdc37 reduced endogenous tau. Ctrl, control; pTau, phospho-tau; tTau, total tau. B, M17 cells were transfected with the indicated amounts of Cdc37, and lysates were analyzed by Western blotting. Tau levels increased with increasing amounts of Cdc37. C, immunofluorescence staining of Cdc37 (green) and total tau (red) in M17 cells showed very strong co-localization of these two proteins. Scatter plot analysis using the Zeiss AxioVision imager showed Mander's coefficients of >0.96.
FIGURE 4.
Cdc37 increases with age and physically interacts with tau from normal and AD brains. A, immunofluorescence staining of Cdc37 (green) and total tau (red) in primary neurons isolated from mice showed very strong co-localization of these two proteins. Scatter plot analysis using the Zeiss AxioVision imager showed Mander's coefficients of >0.93. B, brain lysates from juvenile (1 and 3 months old) and adult (5.5 and 9 months old) mice were analyzed for Cdc37 levels by Western blotting. A significant increase in Cdc37 levels was seen in adult mice compared with juvenile mice. ***, p < 0.0005. C, AD (AD1 and AD2) and age-matched normal (Ctrl 1 and Ctrl 2) human brain tissue lysates were immunoprecipitated (IP) with mouse anti-Cdc37, nonspecific mouse IgG, or rabbit anti-tau antibodies. Samples were analyzed by Western blotting.
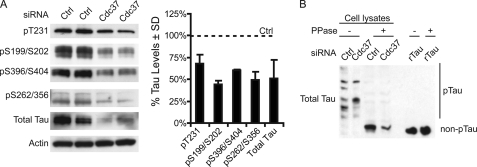
Cdc37 Alters the Phosphorylation Dynamics of Tau
In Fig. 1, knockdown of Cdc37 appeared to marginally reduce phosphorylated tau species more than total tau. Western blot analysis confirmed that a number of additional phospho-tau species were reduced by Cdc37 knockdown (Fig. 5A). These data supported the hypothesis that Cdc37 could selectively regulate phospho-tau. To test this, a dinuclear metal complex of 1,3-bis[bis(pyridin-2-ylmethyl)amine]propan-2-olate that has been reported to act as a phosphate-binding tag (Phos-tag) in aqueous solution was used in combination with SDS-PAGE analysis (18). This phosphate affinity electrophoresis protocol (termed Phos-tag for short) allows for separation of phosphorylated proteins from their non-phosphorylated counterparts (19–21). To establish whether Cdc37 was affecting tau phosphorylation and/or stability, fresh lysates were prepared and incubated with λ-phosphatase (λ-PPase) or buffer for 30 min. Recombinant tau also treated with or without λ-phosphatase was used as a control. Surprisingly, all tau from the cell lysates appeared to be phosphorylated compared with samples treated with λ-PPase (Fig. 5B). Cdc37 siRNA did reduce total tau levels as shown by the reduction in dephosphorylated tau; however, Cdc37 knockdown also altered the phosphorylation pattern of tau such that distinct phospho-tau species were preserved and even enriched by Cdc37 siRNA, whereas others were depleted. The absence of proteolytic cleavage of recombinant tau treated with λ-PPase confirmed enzyme purity. Cdc37 knockdown and high d-glucose treatment caused synergistic reductions in phospho-tau levels (supplemental Fig. 1), consistent with previous findings regarding the effects of d-glucose on tau phosphorylation (22).
FIGURE 5.
Cdc37 suppression alters the phosphorylation dynamics of tau. A, tau-overexpressing stable HeLa cells (HeLaC3) were transfected with Cdc37 siRNA for 72 h. Lysates were analyzed by Western blotting. Quantification of the Western blot showed that Cdc37 knockdown reduced phospho-Thr-231, phospho-Ser-199/Ser-202, phospho-Ser-396/Ser-404, and phospho-Ser-262/Ser-356 tau. B, recombinant tau (rTau) and lysates from HeLaC3 cells transfected with either Cdc37 or control (Ctrl) siRNA were treated with 400 units of λ-PPase or buffer for 30 min at 30 °C. Samples were analyzed using Manganese (II)-Phos-tag SDS-PAGE. Western blotting using an antibody against total tau showed changes in the tau phosphorylation profile and reductions in total tau levels. Recombinant tau was unaffected by Phos-tag or λ-PPase treatment. pTau, phospho-tau.
Cdc37 Prevents Tau Degradation by Hsp90 Inhibition
Previous work showed that Hsp90 inhibition with 17-AAG reduced phospho-tau levels in vivo (16, 23). We speculated that Cdc37 might modulate Hsp90 inhibitor efficacy for phospho-tau. M17 cells were transfected with Cdc37 siRNA and then treated with 1 μm 17-AAG for 24 h. Indeed, reducing Cdc37 synergized with Hsp90 inhibition to reduce tau levels more potently than either condition alone (Fig. 6A). A similar result was obtained in HeLaC3 cells (Fig. 6B). We then tested whether Cdc37 overexpression could prevent tau degradation by 17-AAG. HeLaC3 cells were transfected with a Cdc37 overexpression clone and treated with 17-AAG for 24 h. Indeed, Cdc37 overexpression blocked tau reductions by 17-AAG (Fig. 6C).
FIGURE 6.
Cdc37 modulates Hsp90 inhibitor efficacy for tau. A, M17 cells were transfected with Cdc37 siRNA for 48 h and then treated with the Hsp90 inhibitor 17-AAG for 24 h. Lysates were analyzed by Western blotting. The quantification plot suggests that Cdc37 knockdown dramatically increased the efficacy of the 17-AAG drug for endogenous tau reduction. pTau, phospho-tau. B, HeLa cells stably expressing V5-tagged tau (HeLaC3) were transfected with Cdc37 siRNA and treated with 17-AAG as indicated. The quantification plot shows that Cdc37 knockdown (gray bars) increased the efficacy of the 17-AAG drug in a dose-dependent manner compared with cells transfected with control siRNA (black bars). C, HeLaC3 cells were transfected with the Cdc37 plasmid for 24 h and then treated with 17-AAG for 24 h. The quantification plot shows that Cdc37 overexpression (OE) prevented tau from 17-AAG-mediated degradation. Tau immunoreactivity was assessed with antibody PHF-1. Error bars indicate S.D., and percent values were calculated as a percent of the loading control (actin or GAPDH) or as a percent of cells transfected under control (Ctrl) conditions following normalization to the loading control.
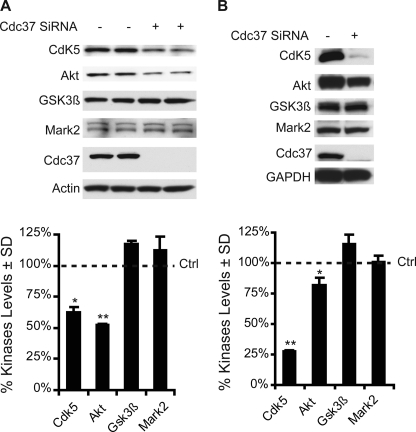
Cdc37 Reduces Cdk5 and Akt Levels, but Not GSK3β and Mark2 Levels
Fig. 5 shows that knockdown of Cdc37 selectively altered the phosphorylation pattern of tau. Given the role of Cdc37 in stabilizing kinases, the levels of tau kinases were assessed following Cdc37 knockdown. Western blot analysis for a panel of tau kinases showed that only the levels of endogenous Cdk5 and Akt were significantly reduced by Cdc37 siRNA; the levels of GSK3β and Mark2 (microtubule affinity-regulating kinase 2) were largely unchanged (Fig. 7A). When these kinases were overexpressed and Cdc37 was suppressed, Cdk5 levels and, to a lesser extent, Akt levels were again reduced, whereas GSK3β and Mark2 levels remained largely unchanged (Fig. 7B). Thus, Cdc37 favors stabilization of Cdk5 and Akt, which may compete with GSK3β for tau phosphorylation at proline-directed Ser/Thr sites.
FIGURE 7.
Cdc37 regulates the stability of specific tau kinases. A, tau-overexpressing stable HeLa cells (HeLaC3) were transfected with Cdc37 siRNA for 72 h. Lysates were analyzed by Western blotting for endogenous kinases. Quantification showed that Cdc37 knockdown significantly reduced the endogenous levels of Cdk5 and Akt kinases. Endogenous GSK3β and Mark2 kinases were largely unaffected by Cdc37 knockdown. B, HeLa cells were transfected with Cdc37 siRNA for 24 h and then transfected with the indicated kinases for 48 h. Lysates were analyzed by Western blotting. Quantification showed that Cdc37 knockdown significantly reduced overexpressed Cdk5 and, to a lesser extent, Akt levels. Overexpressed GSK3β and Mark2 kinases were largely unchanged by Cdc37 knockdown. Error bars indicate S.D., and values are shown as a percent of cells transfected under control (Ctrl) conditions following normalization to the loading control. *, p < 0.05; **, p < 0.01 (n = 3).
DISCUSSION
In AD, cognitive dysfunction and neuronal loss are critically linked to the intracellular accumulation of tau into filamentous tangles (24–28). Tau pathology is also found in ∼15 other neurodegenerative diseases, some caused by mutations in the tau gene (29). More recently, genome-wide association studies have shown that tau expression is increased in sporadic Parkinson disease, further emphasizing the potential value of developing strategies to reduce tau protein levels (30). Abnormal processing and accumulation of tau are necessary for tangle formation. Chaperones such as Hsp90 are essential for the processing of tau and participate in its accumulation (11, 14, 23, 31–38). Modulating Hsp90 directly may be clinically relevant for protein folding and aggregation disorders (16, 39–41); however, Hsp90 acts in concert with other proteins termed co-chaperones to determine whether tau is degraded or recycled, and these co-chaperones could be more specific drug targets. We hypothesized that the diverse enzymatic functions of the co-chaperones could be manipulated to regulate tau (14, 16, 42). We sought to identify those co-chaperones that worked with Hsp90 to specifically stabilize rather than degrade tau.
Cdc37 is an Hsp90 co-chaperone that would be a central mediator of tau biology based on it known function of regulating kinases; our siRNA analyses confirmed this (Figs. 1–3). Indeed, the suppression of Cdc37 levels was shown here for the first time to alter the phosphorylation dynamics of tau, leading to its clearance. We also showed that Cdc37 increased in the brain with age, co-localized with tau in neurons, and physically interacted with tau from AD brain tissue. Cdc37 overexpression stabilized tau and was even able to prevent tau degradation by Hsp90 inhibition. Interestingly, Cdc37 siRNA reduced the levels of only Cdk5 and Akt, whereas the levels of GSK3β and Mark2 were largely unaffected and actually appeared to be slightly increased. GSK3β and Mark2 are more closely tied to cell polarity and differentiation (43, 44), whereas Akt and Cdk5 are linked to cell division and apoptosis (45, 46). Thus, Cdc37 may be critically linked to regulating cell division rather than cell differentiation.
The implications of Cdc37 serving as a neuronal differentiation versus division switch for tau could be profound; Cdc37 may shift the balance of kinases in the brain. Changes in the levels of Cdc37 or altering its interaction with Hsp90 could change the phosphorylation profile of tau by modulating Cdk5 and Akt levels. Because Cdk5 and GSK3β phosphorylate tau at the same consensus sites (proline-directed Ser/Thr sites), Cdc37 may change the phosphorylation signature on tau, driving either its toxicity or it clearance. Further investigation into the impact that modulation of Cdc37 levels has on tau aggregation and toxicity in vivo will elucidate these mechanisms. Moreover, such studies could explain how neurons determine whether tau is phosphorylated by GSK3β or Cdk5 despite overlapping consensus motifs. Cdc37 may be at the heart of this process.
Because Cdc37 is known to facilitate a pro-folding Hsp90 complex, we hypothesized that Cdc37, working with known tau kinases and Hsp90, could stabilize phosphorylated tau. We previously demonstrated that phospho-tau is a preferred client for Hsp90 (16). Indeed, silencing Cdc37 enhanced whereas Cdc37 overexpression prevented Hsp90 inhibitor-mediated phospho-tau clearance (Fig. 6). Cdc37 is known to arrest ATP catalysis by Hsp90 during client loading (47). Thus, the client is protected despite the lack of ATP hydrolysis. 17-AAG competes with ATP binding on Hsp90. Because Cdc37 prevents ATP catalysis, 17-AAG is unable to inhibit Hsp90 in the presence of excess Cdc37, explaining why tau was protected from degradation by Hsp90 inhibition in the presence of Cdc37. These findings confirm that Cdc37 is capable of preserving phospho-tau at the Hsp90 scaffold, where it also is able to preserve Cdk5 and Akt, which have both been linked to tau pathogenesis, creating a perfect storm for accumulation of aberrantly phosphorylated tau.
In conclusion, strategies aimed at inhibiting the interaction of Cdc37 with Hsp90 may be highly effective for reducing tau levels and altering its hyperphosphorylation profile in the brains of patients suffering from AD and other tauopathies. The role of chaperones in tau processing is becoming increasingly important as processes such as ubiquitination (35), chaperone-mediated autophagy (38, 48), and caspase cleavage (49, 50) continue to be linked to tau pathogenesis. The identification of specific chaperones like Cdc37 that affect tau in distinct ways will not only elucidate mechanisms involved in tau processing but will also lead to new therapeutic targets that could be used to effectively treat these devastating diseases.
Supplementary Material
Acknowledgments
We thank Dr. Benjamin Wolozin for providing the GSK3β expression plasmid, Dr. Tom Beach (Sun Health) for human tissue, and Dr. Li-Huei Tsai for the Cdk5 plasmid.
This work was supported, in whole or in part, by National Institutes of Health Grant AG031291 from NIA. This work was also supported by the Rosalinde and Arthur Gilbert Foundation/American Federation for Aging Research, CurePSP, and Alzheimer's Association Grants IIRG-09-130689 and NIRG-10-174517.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- 17-AAG
- 17-(allylamino)-17-demethoxygeldanamycin
- AD
- Alzheimer disease
- λ-PPase
- λ-phosphatase.
REFERENCES
- 1. Hartl F. U., Hayer-Hartl M. (2009) Nat. Struct. Mol. Biol. 16, 574–581 [DOI] [PubMed] [Google Scholar]
- 2. Balch W. E., Morimoto R. I., Dillin A., Kelly J. W. (2008) Science 319, 916–919 [DOI] [PubMed] [Google Scholar]
- 3. Ben-Zvi A., Miller E. A., Morimoto R. I. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 14914–14919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mukrasch M. D., Bibow S., Korukottu J., Jeganathan S., Biernat J., Griesinger C., Mandelkow E., Zweckstetter M. (2009) PLoS Biol. 7, e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Basso A. D., Solit D. B., Chiosis G., Giri B., Tsichlis P., Rosen N. (2002) J. Biol. Chem. 277, 39858–39866 [DOI] [PubMed] [Google Scholar]
- 6. Miyata Y., Nishida E. (2004) Mol. Cell. Biol. 24, 4065–4074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Smith J. R., Clarke P. A., de Billy E., Workman P. (2009) Oncogene 28, 157–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cicero S., Herrup K. (2005) J. Neurosci. 25, 9658–9668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gould C. M., Kannan N., Taylor S. S., Newton A. C. (2009) J. Biol. Chem. 284, 4921–4935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pei J. J., Khatoon S., An W. L., Nordlinder M., Tanaka T., Braak H., Tsujio I., Takeda M., Alafuzoff I., Winblad B., Cowburn R. F., Grundke-Iqbal I., Iqbal K. (2003) Acta Neuropathol. 105, 381–392 [DOI] [PubMed] [Google Scholar]
- 11. Luo W., Dou F., Rodina A., Chip S., Kim J., Zhao Q., Moulick K., Aguirre J., Wu N., Greengard P., Chiosis G. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 9511–9516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cruz J. C., Tseng H. C., Goldman J. A., Shih H., Tsai L. H. (2003) Neuron 40, 471–483 [DOI] [PubMed] [Google Scholar]
- 13. Jinwal U. K., O'Leary J. C., 3rd, Borysov S. I., Jones J. R., Li Q., Koren J., 3rd, Abisambra J. F., Vestal G. D., Lawson L. Y., Johnson A. G., Blair L. J., Jin Y., Miyata Y., Gestwicki J. E., Dickey C. A. (2010) J. Biol. Chem. 285, 16798–16805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jinwal U. K., Koren J., 3rd, Borysov S. I., Schmid A. B., Abisambra J. F., Blair L. J., Johnson A. G., Jones J. R., Shults C. L., O'Leary J. C., 3rd, Jin Y., Buchner J., Cox M. B., Dickey C. A. (2010) J. Neurosci. 30, 591–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Abisambra J. F., Fiorelli T., Padmanabhan J., Neame P., Wefes I., Potter H. (2010) PLoS ONE 5, e8556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dickey C. A., Kamal A., Lundgren K., Klosak N., Bailey R. M., Dunmore J., Ash P., Shoraka S., Zlatkovic J., Eckman C. B., Patterson C., Dickson D. W., Nahman N. S., Jr., Hutton M., Burrows F., Petrucelli L. (2007) J. Clin. Invest. 117, 648–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jinwal U. K., Miyata Y., Koren J., 3rd, Jones J. R., Trotter J. H., Chang L., O'Leary J., Morgan D., Lee D. C., Shults C. L., Rousaki A., Weeber E. J., Zuiderweg E. R., Gestwicki J. E., Dickey C. A. (2009) J. Neurosci. 29, 12079–12088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kinoshita E., Takahashi M., Takeda H., Shiro M., Koike T. (2004) Dalton Trans. 1189–1193 [DOI] [PubMed] [Google Scholar]
- 19. Kinoshita E., Kinoshita-Kikuta E., Takiyama K., Koike T. (2006) Mol. Cell. Proteomics 5, 749–757 [DOI] [PubMed] [Google Scholar]
- 20. Kinoshita-Kikuta E., Aoki Y., Kinoshita E., Koike T. (2007) Mol. Cell. Proteomics 6, 356–366 [DOI] [PubMed] [Google Scholar]
- 21. Kinoshita E., Kinoshita-Kikuta E., Matsubara M., Yamada S., Nakamura H., Shiro Y., Aoki Y., Okita K., Koike T. (2008) Proteomics 8, 2994–3003 [DOI] [PubMed] [Google Scholar]
- 22. Hatakeyama S., Matsumoto M., Kamura T., Murayama M., Chui D. H., Planel E., Takahashi R., Nakayama K. I., Takashima A. (2004) J. Neurochem. 91, 299–307 [DOI] [PubMed] [Google Scholar]
- 23. Dickey C. A., Dunmore J., Lu B., Wang J. W., Lee W. C., Kamal A., Burrows F., Eckman C., Hutton M., Petrucelli L. (2006) FASEB J. 20, 753–755 [DOI] [PubMed] [Google Scholar]
- 24. Oddo S., Caccamo A., Shepherd J. D., Murphy M. P., Golde T. E., Kayed R., Metherate R., Mattson M. P., Akbari Y., LaFerla F. M. (2003) Neuron 39, 409–421 [DOI] [PubMed] [Google Scholar]
- 25. Frautschy S. A., Baird A., Cole G. M. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 8362–8366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Roberson E. D., Scearce-Levie K., Palop J. J., Yan F., Cheng I. H., Wu T., Gerstein H., Yu G. Q., Mucke L. (2007) Science 316, 750–754 [DOI] [PubMed] [Google Scholar]
- 27. Braak H., Braak E. (1991) Acta Neuropathol. 82, 239–259 [DOI] [PubMed] [Google Scholar]
- 28. Mukaetova-Ladinska E. B., Garcia-Siera F., Hurt J., Gertz H. J., Xuereb J. H., Hills R., Brayne C., Huppert F. A., Paykel E. S., McGee M., Jakes R., Honer W. G., Harrington C. R., Wischik C. M. (2000) Am. J. Pathol. 157, 623–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hardy J., Orr H. (2006) J. Neurochem. 97, 1690–1699 [DOI] [PubMed] [Google Scholar]
- 30. Simón-Sánchez J., Schulte C., Bras J. M., Sharma M., Gibbs J. R., Berg D., Paisan-Ruiz C., Lichtner P., Scholz S. W., Hernandez D. G., Krüger R., Federoff M., Klein C., Goate A., Perlmutter J., Bonin M., Nalls M. A., Illig T., Gieger C., Houlden H., Steffens M., Okun M. S., Racette B. A., Cookson M. R., Foote K. D., Fernandez H. H., Traynor B. J., Schreiber S., Arepalli S., Zonozi R., Gwinn K., van der Brug M., Lopez G., Chanock S. J., Schatzkin A., Park Y., Hollenbeck A., Gao J., Huang X., Wood N. W., Lorenz D., Deuschl G., Chen H., Riess O., Hardy J. A., Singleton A. B., Gasser T. (2009) Nat. Genet. 41, 1308–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shimura H., Miura-Shimura Y., Kosik K. S. (2004) J. Biol. Chem. 279, 17957–17962 [DOI] [PubMed] [Google Scholar]
- 32. Shimura H., Schwartz D., Gygi S. P., Kosik K. S. (2004) J. Biol. Chem. 279, 4869–4876 [DOI] [PubMed] [Google Scholar]
- 33. Carrettiero D. C., Hernandez I., Neveu P., Papagiannakopoulos T., Kosik K. S. (2009) J. Neurosci. 29, 2151–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Petrucelli L., Dickson D., Kehoe K., Taylor J., Snyder H., Grover A., De Lucia M., McGowan E., Lewis J., Prihar G., Kim J., Dillmann W. H., Browne S. E., Hall A., Voellmy R., Tsuboi Y., Dawson T. M., Wolozin B., Hardy J., Hutton M. (2004) Hum. Mol. Genet. 13, 703–714 [DOI] [PubMed] [Google Scholar]
- 35. Dickey C. A., Yue M., Lin W. L., Dickson D. W., Dunmore J. H., Lee W. C., Zehr C., West G., Cao S., Clark A. M., Caldwell G. A., Caldwell K. A., Eckman C., Patterson C., Hutton M., Petrucelli L. (2006) J. Neurosci. 26, 6985–6996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dickey C. A., Koren J., Zhang Y. J., Xu Y. F., Jinwal U. K., Birnbaum M. J., Monks B., Sun M., Cheng J. Q., Patterson C., Bailey R. M., Dunmore J., Soresh S., Leon C., Morgan D., Petrucelli L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3622–3627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dou F., Netzer W. J., Tanemura K., Li F., Hartl F. U., Takashima A., Gouras G. K., Greengard P., Xu H. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 721–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang Y., Martinez-Vicente M., Krüger U., Kaushik S., Wong E., Mandelkow E. M., Cuervo A. M., Mandelkow E. (2009) Hum. Mol. Genet. 18, 4153–4170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Waza M., Adachi H., Katsuno M., Minamiyama M., Sang C., Tanaka F., Inukai A., Doyu M., Sobue G. (2005) Nat. Med. 11, 1088–1095 [DOI] [PubMed] [Google Scholar]
- 40. Sittler A., Lurz R., Lueder G., Priller J., Lehrach H., Hayer-Hartl M. K., Hartl F. U., Wanker E. E. (2001) Hum. Mol. Genet. 10, 1307–1315 [DOI] [PubMed] [Google Scholar]
- 41. Auluck P. K., Chan H. Y., Trojanowski J. Q., Lee V. M., Bonini N. M. (2002) Science 295, 865–868 [DOI] [PubMed] [Google Scholar]
- 42. Wang X., Venable J., LaPointe P., Hutt D. M., Koulov A. V., Coppinger J., Gurkan C., Kellner W., Matteson J., Plutner H., Riordan J. R., Kelly J. W., Yates J. R., 3rd, Balch W. E. (2006) Cell 127, 803–815 [DOI] [PubMed] [Google Scholar]
- 43. Hur E. M., Zhou F. Q. (2010) Nat. Rev. Neurosci. 11, 539–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Marx A., Nugoor C., Panneerselvam S., Mandelkow E. (2010) FASEB J. 24, 1637–1648 [DOI] [PubMed] [Google Scholar]
- 45. Lalioti V., Pulido D., Sandoval I. V. (2010) Cell Cycle 9, 284–311 [DOI] [PubMed] [Google Scholar]
- 46. Kim D., Dan H. C., Park S., Yang L., Liu Q., Kaneko S., Ning J., He L., Yang H., Sun M., Nicosia S. V., Cheng J. Q. (2005) Front. Biosci. 10, 975–987 [DOI] [PubMed] [Google Scholar]
- 47. Roe S. M., Ali M. M., Meyer P., Vaughan C. K., Panaretou B., Piper P. W., Prodromou C., Pearl L. H. (2004) Cell 116, 87–98 [DOI] [PubMed] [Google Scholar]
- 48. Dolan P. J., Johnson G. V. (2010) J. Biol. Chem. 285, 21978–21987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rissman R. A., Poon W. W., Blurton-Jones M., Oddo S., Torp R., Vitek M. P., LaFerla F. M., Rohn T. T., Cotman C. W. (2004) J. Clin. Invest. 114, 121–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Guillozet-Bongaarts A. L., Garcia-Sierra F., Reynolds M. R., Horowitz P. M., Fu Y., Wang T., Cahill M. E., Bigio E. H., Berry R. W., Binder L. I. (2005) Neurobiol. Aging 26, 1015–1022 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.