Abstract
Our laboratory has reported that deoxyribonucleoside triphosphate (dNTP) pools in rat tissue mitochondria are highly asymmetric, with dGTP predominating, and that the imbalance probably contributes toward the high spontaneous mutation rate of the mitochondrial genome. Ferraro et al. (Ferraro, P., Nicolosi, L., Bernardi, P., Reichard, P., and Bianchi, V. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 18586–18591) have challenged these findings, based upon their studies of mouse liver mitochondria. Moreover, they have identified a potential artifact in the DNA polymerase-based assay for dNTPs, based upon overestimation of dGTP when GTP levels in extracts are much higher than dGTP levels. We measured ribonucleoside triphosphate (rNTP) pools in rat mitochondrial extracts and found that GTP pools exceed dGTP pools by 50-fold or less, not enough to interfere with the dGTP assay. Analysis of dNTP pools in state 3 mitochondria, after incubation with ADP and oxidizable substrates, gave similar results. We confirmed our earlier finding that rat mitochondrial dNTP pools are highly asymmetric. dNTP pools in cytosolic extracts are uniformly low, suggesting that the dNTP pool asymmetry arises within the mitochondrion. Moreover, we found rat tissue rNTP pools to be even more highly asymmetric, with ATP, for example, at least 2 orders of magnitude more abundant than CTP in liver extracts. This finding raises the possibility that transcription of the mitochondrial genome is more error-prone than transcription in the nucleus.
Keywords: ADP, ATP, Cell Metabolism, DNA Synthesis, HPLC, Mitochondrial Metabolism, Nucleoside Nucleotide Metabolism
Introduction
In 2005 our laboratory reported that pool sizes of the four canonical deoxyribonucleoside triphosphates (dNTPs)2 are highly asymmetric within mitochondria from rat tissues (1), with dGTP representing nearly 90% of total dNTP in mitochondria from some tissues. This finding contrasted with dNTP pool measurements in whole cell extracts, where dGTP is usually found to represent just 5–10% of total dNTP (2, 3).
The mitochondrial asymmetry that we described was of interest for two reasons. First, it is sufficiently large to affect fidelity of DNA replication, as determined by in vitro analysis of replication errors committed by DNA polymerase γ (1). Second, the large size of the dGTP pool, plus oxidizing conditions within the mitochondrion, suggested that dGTP could be a target for oxidation to 8-oxo-dGTP, a nucleotide thought to stimulate mutagenesis following its incorporation into DNA (4). Indeed, we detected substantial 8-oxo-dGTP pools in mitochondrial extracts from some rat tissues, levels high enough to be mutagenic, again, as determined through in vitro fidelity assays (5). Hence, the unexpectedly large intramitochondrial dGTP pools could be contributing in two ways toward the relatively high spontaneous mutation rate for mitochondrial genes: first, insertion and proofreading errors caused by unbalanced dNTP concentrations (6); and second, oxidative mutagenesis caused by incorporation of 8-oxo-dGTP.
Our reports of asymmetric mitochondrial dNTP pools were challenged by Ferraro et al. (7, 8) on two grounds. First was the possibility of ATP depletion during mitochondrial isolation and extraction, with secondary effects upon other nucleoside triphosphates (7). The second criticism pertained to technical aspects of the DNA polymerase-based assay procedure for dNTPs, used both in our laboratory and that of Ferraro et al. (8). In view of the significance of our findings for understanding mutagenesis of mitochondrial genes, it seemed important for us to evaluate these issues insofar as they affected our previously reported results. As described in this paper, we have done so, and we reiterate our findings of extensive mitochondrial dNTP asymmetry. In addition, we have determined mitochondrial ribonucleoside triphosphate (rNTP) pools and find them also to be strikingly asymmetric.
EXPERIMENTAL PROCEDURES
Materials
[3H]dNTPs for the dNTP assay were purchased from Moravek Biochemicals. DNA polymerase Klenow fragment was purchased from U. S. Biochemicals, and Taq polymerase was purchased from Fermentas Life Sciences.
Isolation of Mitochondria
All animal procedures were approved by the Institutional Animal Care and Use Committee of Oregon State University. Young adult male Wistar rats, one animal per experiment, were anesthetized with isoflurane and decapitated. Organs were removed and chilled immediately in ice-cold 0.9% NaCl. Liver was removed most rapidly, with the organ being placed in ice-cold saline within about 30 s after decapitation. After mincing with scissors, each organ mince was suspended in isolation buffer (1). Homogenization and mitochondrial isolation by differential centrifugation were carried out as described previously (1). During isolation, care was taken to remove lipid from the sides of centrifuge tubes, and pipetting mitochondrial suspensions was carried out only with large mouth pipettes, to minimize mechanical damage to mitochondria. All extractions and experimental procedures with mitochondrial preparations were carried out within 4 h after sacrifice of an animal.
Nucleotide Extraction
A mitochondrial pellet representing 1 g of tissue was suspended in 1 ml of ice-cold 60% aqueous methanol. The suspension was heated for 3 min in a water bath at 90°, then chilled and sonicated for 20 s. The suspension was stored at −20 °C with occasional agitation, for at least 2 h. The suspension was then centrifuged for 30 min at 15,000 rpm in a microcentrifuge at 4 °C. The clear supernatant was taken to dryness under vacuum, and the residue was dissolved in 200 μl of water. The extract was clarified by additional centrifugation if necessary. For occasional experiments requiring more material the procedure was scaled up appropriately.
dNTP Assay
We assayed for dNTPs by the DNA polymerase-based method as described by Sherman and Fyfe (9) and modified in our laboratory (10). Radioactivity in DNA polymerase reaction mixtures was determined in duplicate, and duplicate readings agreed within 10%.
rNTP Assay
We assayed for ribonucleoside triphosphates (rNTPs) by ion-exchange HPLC, using a Whatman Partisil 10 SAX column, 4.6 × 250 mm, fitted to a Hitachi Model D-7000 HPLC system. The sample size was 50–100 μl. The gradient was from 0.3 m to 0.7 m ammonium phosphate, pH 3.6, over 45 min at a flow rate of 1.0 ml/min, followed by return to 0.3 m over 5 min. Absorbance was monitored at 260 nm.
Adenine Nucleotide Assay
Adenine nucleotides were monitored by reversed-phase HPLC with ion pairing, using an AlltimaTM C18 5-μm column, 4.6 × 250 mm. Buffer A was 8 mm tetrabutylammonium hydroxide, 0.01 m potassium phosphate, pH 7.0, and 0.25% methanol. Buffer B was 2 mm tetrabutylammonium hydroxide, 0.10 m potassium phosphate, pH 7.0, and 30% methanol. Elution was carried out with a gradient from 70% A/30% B to 40% A/60% B over 45 min, followed by 5 min at 40% A/60% B, followed by return to 70% A/30% B over 5 min, all at a flow rate of 0.7 ml/min. Absorbance was monitored at 260 nm.
Nucleotide pool sizes are reported as pmol/mg protein (dNTPs) or nmol/mg protein (rNTPs), with protein determined by the Bradford method as previously described (1). Pool sizes were converted when desired to approximate millimolar concentrations by dividing by 0.82 μl/mg protein, a value representing an average from several published determinations of milligrams of protein per microliters of mitochondrial fluid volume (1).
RESULTS AND DISCUSSION
Adenine Nucleotides in Respiring Mitochondria
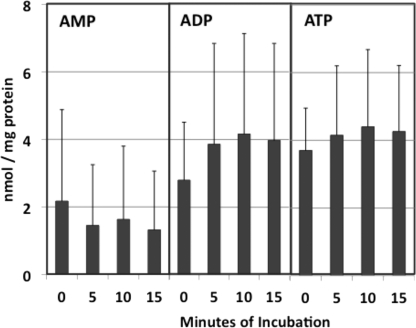
As Ferraro et al. (7) pointed out, if significant ATP depletion occurs during dissection and homogenization of tissue, then pool sizes of other nucleoside triphosphates may be affected as well. If so, then allowing isolated mitochondria to respire in the presence of ADP and oxidizable substrates might restore conditions approximating those in vivo. Reports that isolated rat liver mitochondria robustly incorporate exogenous nucleotides into DNA and RNA for some time in vitro (11, 12) are consistent with this premise. Accordingly, we carried out adenine nucleotide analyses in rat liver mitochondria after incubation in an isotonic buffer containing glutamate, malate, and ADP, in other words, in state 3 mitochondria. Combined results from three experiments are shown in Fig. 1. ATP and ADP levels increased by 20–30% during 10 min of incubation, while AMP declined. We noted significant variability among individual animals. However, in six experiments involving ATP measurements, we observed the ATP pool to rise from 4.75 ± 1.15 to 6.10 ± 1.73 nmol/mg protein over the 10-min incubation. This latter value is close to previously reported values for ATP pools in isolated rat liver mitochondria. Hamman and Haynes (13) reported a value of 6 nmol/mg protein, whereas Schwenke et al. (14) reported the intramitochondrial ATP concentration as 7.49 mm. Given an average value from several studies of 0.82 μl/mg protein for the mitochondrial fluid volume (1), this latter value corresponds to an ATP pool size of about 6.1 nmol/mg protein. In both of these studies (13, 14) the ADP pool was reported to be equal to or slightly higher than the ATP pool, just as we have seen (Fig. 1).
FIGURE 1.
Adenine nucleotide pools in rat liver mitochondria incubated under state 3 conditions. Four rat liver mitochondrial pellets, each representing 0.75 g of liver, were suspended in 1.0 ml each of a buffer containing 70 mm sucrose, 220 mm mannitol, 5 mm MOPS, 5 mm KH2PO4, 5 mm MgCl2, 1 mm EDTA, 10 mm potassium glutamate, 2.5 mm sodium malate, 1 mg/ml BSA, and 1.0 mm ADP, adjusted to pH 7.4. Incubation was carried out with shaking at 30 °C. At each indicated time one of the incubation mixtures was removed and rapidly chilled. After the last incubation mixture had been removed, the mitochondria in each sample were pelleted by a 30-min centrifugation at 15,000 rpm, then extracted and assayed for adenine nucleotides as described under “Experimental Procedures.” Data represent average values ± S.D. (error bars) from three experiments.
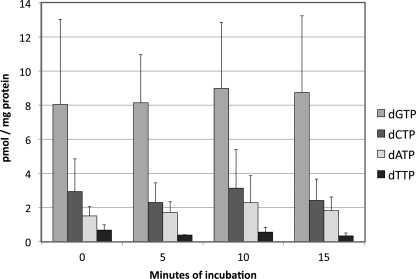
If these experiments suggest that ATP is partially depleted during the mitochondrial isolation procedure, how do these conditions affect dNTP pools? As shown in Fig. 2, we find changes in these pools to be slight over a 15-min incubation under the state 3 conditions mentioned above. These values are consistent with those we have reported previously (1, 5). dGTP is the most abundant dNTP in these mitochondria, followed by dCTP, then dATP, then dTTP.
FIGURE 2.
dNTP pools in rat liver mitochondria incubated under state 3 conditions. Rat liver mitochondria were incubated under state 3 conditions as described in the legend to Fig. 1, and extracts were analyzed for dNTPs. Each data point represents the average value ± S.D. (error bars) from three experiments.
Enzymatic Assay for dNTPs
In the Sherman-Fyfe procedure for dNTP analysis (9) oligonucleotides of defined base sequence are used as template and primer for DNA polymerase reactions, normally using Klenow fragment. A dNTP in an extract is incorporated into DNA in the presence of a radiolabeled counternucleotide, present in molar excess. For example, as used in our laboratory (10), the counternucleotide for the dGTP assay is [3H]dTTP. The sequence of the template-primer is such that the pmol of radiolabel incorporated from dTTP during a polymerase reaction is proportional to the amount of unlabeled dGTP in an extract being analyzed. Ferraro et al. (8) observed that Klenow fragment has relaxed specificity for guanine and cytosine dNTPs; at high rNTP concentrations, GTP can be incorporated into reaction products in addition to dGTP, and this leads to overestimation of dGTP. Ferraro et al. suggested that this potential artifact could explain our observations of very high dGTP levels in mitochondria. This artifact could be circumvented by using Taq polymerase instead of Klenow, for its much higher selectivity toward deoxyribonucleotides.
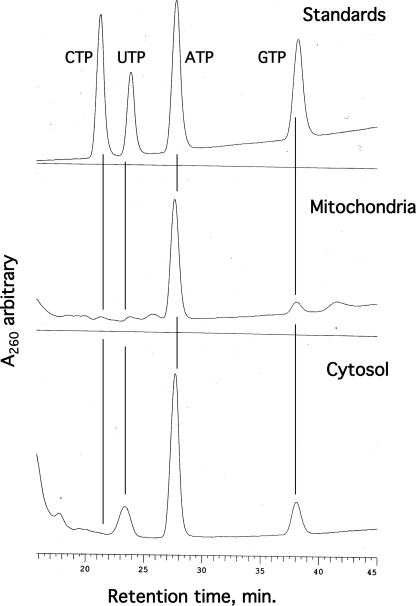
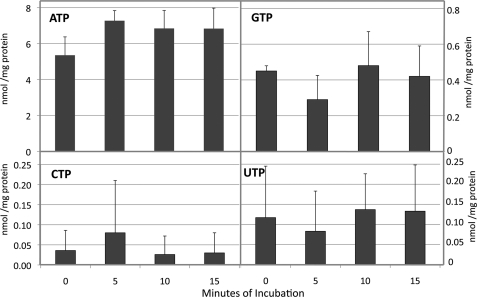
In the whole cell extracts analyzed by Ferraro et al. (8) the GTP pool exceeded that of dGTP by as much as 2000-fold, making this a significant source of error. To evaluate this problem for mitochondrial analysis, it was essential to quantitate mitochondrial GTP pools and ask whether they exceed the dGTP pools enough to make this a significant source of error. Fig. 3 shows HPLC resolution of rNTPs in rat liver and cytosolic extracts. Surprisingly, we found mitochondrial rNTPs to be even more highly asymmetric than values we have reported for dNTPs; CTP and UTP were almost too low for detection. More to the point, we are able to quantitate GTP from data of this kind. First, however, we needed to correct for the possibility that rNTPs other than ATP are affected by transient anaerobiosis during mitochondrial isolation (7), by measuring rNTP pools in state 3 mitochondria. Fig. 4 shows data comparable with those of Fig. 2. Again, we incubated isolated rat liver mitochondria in isotonic buffer containing glutamate, malate, and ADP. As seen in Fig. 1, ATP accumulated during the 15 min of incubation, in this case by about 30%. Levels of the other three rNTPs changed in minor ways if at all. The average GTP value from the four extracts analyzed was 0.42 nmol/mg protein. In Fig. 2 the corresponding average pool size for dGTP was 8.5 pmol/mg protein. Thus, in these experiments the GTP pool exceeded the dGTP pool by about 49-fold. Is this sufficient to bias the results of our dGTP assays?
FIGURE 3.
HPLC resolution of ribonucleoside triphosphates. Extracts of rat liver mitochondria and cytosol were subjected to HPLC as described under “Experimental Procedures,” and peaks were compared with standard rNTPs. The cytosolic extract was prepared by adding 100% methanol to an aliquot of postmitochondrial supernatant to 60% and then working up as described for extraction of mitochondria.
FIGURE 4.
rNTP pools in rat liver mitochondria incubated under state 3 conditions. Conditions were as described in the legend to Fig. 2, except that extracts were analyzed for rNTPs. Average values from four experiments ± S.D. (error bars) are shown.
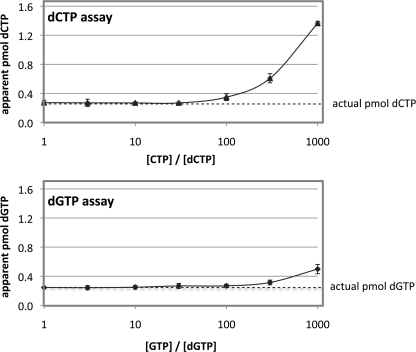
To answer that question we carried out the dGTP assay on 0.25 pmol of a dGTP standard, in the presence of excess GTP in varying amounts. The same experiment was conducted with dCTP and CTP because that is subject to the same error. As shown in Fig. 5, the dCTP assay is more sensitive to rNTP excess than the dGTP assay, as reported also by Ferraro et al. (8). More to the point, with the dGTP assay using the Klenow fragment, GTP excess biased the dGTP value only at GTP/dGTP ratios of 100 or higher. Hence, at the GTP levels we observe in mitochondrial extracts, GTP does not interfere significantly with the dGTP assay.
FIGURE 5.
Effect of CTP and GTP excess on the analysis of dCTP and dGTP, respectively. Standard reaction mixtures for the dNTP assay were run with 0.25 pmol of either dCTP or dGTP as shown, with CTP added to the dCTP assay mixtures and GTP to the dGTP assay mixtures in the amounts shown.
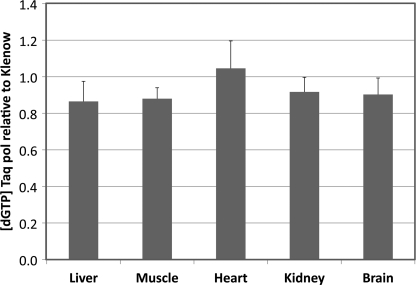
The same conclusion was reached in an experiment in which mitochondrial extracts from five different rat tissues were subjected to the dGTP assay using Taq polymerase, which is more selective for dNTPs vis-à-vis rNTPs than is the Klenow fragment (8). As shown in Fig. 6, the two extracts gave similar results when assayed for dGTP with either polymerase. In most of the extracts the value with Klenow was slightly higher than that with Taq, but the results were within experimental variation. We conclude from these experiments that, at least in mitochondrial extracts, the excess of GTP over dGTP does not lead to significant overestimation of dGTP pools.
FIGURE 6.
dGTP analysis of rat tissue mitochondrial extracts with either Klenow fragment or Taq polymerase. Extracts were analyzed by the standard dGTP assay using either Klenow fragment or Taq polymerase. Data represent six independent determinations for each assay ± S.D. (error bars).
Mitochondrial Nucleoside Triphosphate Pool Asymmetries
If the incubation of freshly isolated mitochondria with oxidizable substrates and ADP restores them to near physiological conditions, then the data of Figs. 2 and 4 suggest that the slight ATP depletion occurring during tissue dissection and fractionation does not significantly affect pool sizes of the other seven canonical nucleoside triphosphates. Hence, the values determined in freshly isolated mitochondria represent reasonably well the intramitochondrial nucleotide concentrations in living cells. ATP would be underestimated by 20–30% in these extracts. Tables 1 and 2 present such data for extracts from freshly isolated mitochondria. From these data we can estimate the intramitochondrial GTP/dGTP ratio in each tissue as follows: heart, 2.3; kidney, 11.3; brain, 12.3; liver, 26.2; skeletal muscle, 3.0. Thus, for none of the five tissues investigated does the GTP pool appear high enough to interfere with the enzymatic assay for dGTP.
TABLE 1.
Mitochondrial dNTP pools in rat tissues
| Tissue | na | dNTP pool pmol/mg protein ± S.D. |
|||
|---|---|---|---|---|---|
| dATP | dTTP | dCTP | dGTP | ||
| Heart | 5 | 3.30 ± 0.95 | 1.14 ± 0.31 | 4.30 ± 0.97 | 454 ± 184 |
| Kidney | 5 | 2.60 ± 1.83 | 2.16 ± 1.59 | 4.70 ± 2.89 | 97 ± 62 |
| Brain | 5 | 2.52 ± 1.62 | 3.78 ± 2.64 | 4.72 ± 3.40 | 77 ± 62 |
| Liver | 6 | 2.40 ± 1.88 | 1.49 ± 1.85 | 6.35 ± 5.43 | 18.7 ± 14.9 |
| Skeletal muscle | 7 | 1.63 ± 1.17 | 0.45 ± 0.48 | 1.22 ± 1.14 | 46 ± 57 |
a n = number of animals analyzed.
TABLE 2.
Mitochondrial rNTP pools in rat tissues
| Tissue | na | rNTP pool nmol/mg protein ± S.D. |
|||
|---|---|---|---|---|---|
| ATP | UTP | CTP | GTP | ||
| Heart | 5 | 4.46 ± 1.40 | 0.01 ± 0.01 | <0.01 | 1.05 ± 0.28 |
| Kidney | 5 | 2.57 ± 1.87 | 0.09 ± 0.06 | 0.02 ± 0.02 | 1.10 ± 0.95 |
| Brain | 5 | 1.87 ± 0.91 | 0.05 ± 0.04 | 0.05 ± 0.06 | 0.95 ± 0.27 |
| Liver | 6 | 4.78 ± 2.93 | 0.11 ± 0.06 | 0.05 ± 0.05 | 0.49 ± 0.36 |
| Skeletal muscle | 4 | 3.57 ± 2.69 | 0.01 ± 0.03 | 0.01 ± 0.02 | 0.14 ± 0.25 |
a n = number of animals analyzed.
The degree of dNTP asymmetry reported in Table 1 is similar to what we have previously reported (1, 5). Heart has the largest dGTP pool and liver the lowest, whereas kidney, skeletal muscle, and brain have intermediate values. Some of the values listed in Table 1, particularly the heart dGTP value, are considerably higher than what we have reported previously. This is partly because of the greater care we are using now in homogenizing tissue and isolating mitochondria and partly because all of our current experiments were carried out with freshly isolated mitochondria. Measured dNTP pools, particularly dGTP, decline significantly over just a few days of storage at −20 °C of either mitochondrial extracts or pelleted mitochondria.
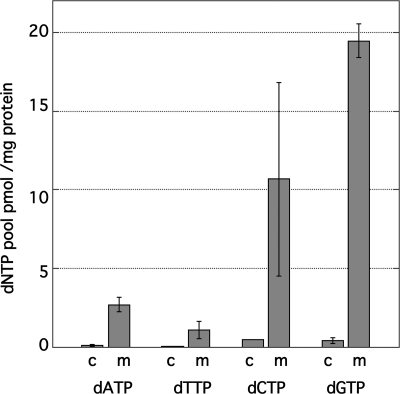
What might be the biochemical basis for the large dGTP excess we observe in mitochondria from all five rat tissues we have investigated? Our attempts to date to compare relative rates of dNTP synthesis and degradation have been inconclusive. However, analysis of dNTPs in cytosol suggests that the relevant metabolic reactions take place within the mitochondria. As seen in Fig. 7, we find all four dNTP levels to be comparable and low in the postmitochondrial supernatant, representing the cytosol, during rat liver mitochondrial isolation. Similar results were seen with the other four tissues we have analyzed (data not shown).
FIGURE 7.
Distribution of dNTPs in rat liver mitochondria and cytosol. Cytosolic extracts were prepared as described in the legend to Fig. 3. m, mitochondrial; c, cytosolic. Averages from three experiments ± S.D. (error bars) are shown.
With respect to the ribonucleoside triphosphates, it is no surprise to find ATP as the most abundant rNTP and no surprise to see substantial GTP pools because GTP would be needed for mitochondrial protein synthesis. However, the low CTP and UTP pools were unexpected, given that all four rNTPs are needed for mitochondrial transcription. These values differ markedly from rNTP pools in whole cell extracts. Traut (15) summarized data from several hundred studies of nucleotide levels in whole cell extracts and reported average intracellular rNTP levels (with considerable variation) as follows: ATP, 3.15 mm; GTP, 0.47 mm; UTP, 0.57 mm; CTP, 0.28 mm, for an ATP/CTP ratio of only 1 order of magnitude.
Our finding that intramitochondrial ATP concentrations in rats exceed those of CTP by 2 orders of magnitude in most cases suggests that mitochondrial transcription may be more error-prone than transcription of nuclear genes because of mismatches forced by the high concentration of ATP. How this might affect mitochondrial function is unknown at this stage. In this context, it would be interesting to know concentrations of rNTPs within mammalian cell nuclei, because Traut's data are for whole cell extracts, and cytosolic rNTP levels must be considerable. However, because of the porous nature of the nuclear envelope, isolating nuclei with small-molecule pools intact presents technical challenges.
Finally, because mitochondrial transcription occurs within the nucleoid, one might speculate that pools are compartmentalized, such that local concentrations of rNTPs are more nearly equal at transcription sites. At this point we do not know enough about the structure of the nucleoid (16) to predict that it could provide a barrier for physical compartmentation of small molecules.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM073744. This work was also supported by Army Research Office Grant 55953-LS.
- dNTP
- deoxyribonucleoside 5′-triphosphate
- rNTP
- ribonucleoside 5′-triphosphate.
REFERENCES
- 1. Song S., Pursell Z. F., Copeland W. C., Longley M. J., Kunkel T. A., Mathews C. K. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 4990–4995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mathews C. K., Ji J. (1992) BioEssays 14, 295–301 [DOI] [PubMed] [Google Scholar]
- 3. Martomo S. A., Mathews C. K. (2002) Mutat. Res. 499, 197–211 [DOI] [PubMed] [Google Scholar]
- 4. Kamiya H. (2003) Nucleic Acids Res. 31, 517–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pursell Z. F., McDonald J. T., Mathews C. K., Kunkel T. A. (2008) Nucleic Acids Res. 36, 2174–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mathews C. K. (2006) FASEB J. 20, 1300–1314 [DOI] [PubMed] [Google Scholar]
- 7. Ferraro P., Nicolosi L., Bernardi P., Reichard P., Bianchi V. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 18586–18591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ferraro P., Franzolin E., Pontarin G., Reichard P., Bianchi V. (2010) Nucleic Acids Res. 38, e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sherman P. A., Fyfe J. A. (1989) Anal. Biochem. 180, 222–226 [DOI] [PubMed] [Google Scholar]
- 10. Mathews C. K., Wheeler L. J. (2009) in Mitochondrial DNA: Methods and Protocols, 2nd Ed. (Stuart J. A. ed) pp. 371–381, Humana Press, Totowa, NJ [Google Scholar]
- 11. Enríquez J. A., Ramos J., Pérez-Martos A., López-Pérez M. J., Montoya J. (1994) Nucleic Acids Res. 22, 1861–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Enríquez J. A., Fernández-Silva P., Pérez-Martos A., López-Pérez M. J., Montoya J. (1996) Eur. J. Biochem. 237, 601–610 [DOI] [PubMed] [Google Scholar]
- 13. Hamman H. C., Haynes R. C., Jr. (1983) Arch. Biochem. Biophys. 223, 85–94 [DOI] [PubMed] [Google Scholar]
- 14. Schwenke W. D., Soboll S., Seitz H. J., Sies H. (1981) Biochem. J. 200, 405–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Traut T. W. (1994) Mol. Cell. Biochem. 140, 1–22 [DOI] [PubMed] [Google Scholar]
- 16. Bogenhagen D. F., Rousseau D., Burke S. (2008) J. Biol. Chem. 283, 3665–3675 [DOI] [PubMed] [Google Scholar]