Abstract
Arp2/3 complex is a key actin filament nucleator that assembles branched actin networks in response to cellular signals. The activity of Arp2/3 complex is regulated by both activating and inhibitory proteins. Coronins make up a large class of actin-binding proteins previously shown to inhibit Arp2/3 complex. Although coronins are known to play a role in controlling actin dynamics in diverse processes, including endocytosis and cell motility, the precise mechanism by which they regulate Arp2/3 complex is unclear. We conducted a detailed biochemical analysis of budding yeast coronin, Crn1, and found that it not only inhibits Arp2/3 complex but also activates it. We mapped regions required for activation and found that Crn1 contains a sequence called CA, which is conserved in WASp/Scar proteins, the prototypical activators of Arp2/3 complex. Point mutations in CA abolished activation of Arp2/3 complex by Crn1 in vitro. Confocal microscopy and quantitative actin patch tracking showed that these mutants had defective endocytic actin patch dynamics in Saccharomyces cerevisiae, indicating that activation of Arp2/3 complex by coronin is required for normal actin dynamics in vivo. The switch between the dual modes of regulation by Crn1 is controlled by concentration, and low concentrations of Crn1 enhance filament binding by Arp2/3 complex, whereas high concentrations block binding. Our data support a direct tethering recruitment model for activation of Arp2/3 complex by Crn1 and suggest that Crn1 indirectly inhibits Arp2/3 complex by blocking it from binding actin filaments.
Keywords: Actin, Cell Motility, Cytoskeleton, Endocytosis, Yeast, Arp2/3
Introduction
Regulation of actin polymerization is critical for orchestrating complex cellular processes, including endocytosis, cell division, cellular differentiation, and motility (1). Arp2/3 (actin-related protein 2/3) complex is a key actin regulator that nucleates branched actin filaments and initiates the assembly of highly cross-linked actin networks (2). Arp2/3 complex is tightly regulated, and proteins with both activating and inhibitory influence act on the complex in vivo. The best studied activators are the WASp/SCAR family proteins, which bind directly to Arp2/3 complex and actin monomers, initiating an activating conformational change and recruiting the first actin subunits for the nascent (daughter) filament (3, 4). A second class of activators, referred to as class II nucleation-promoting factors (NPFs),3 lacks actin monomer binding regions and instead binds actin filaments (2). Class II NPFs, best represented by the Src kinase substrate cortactin, are thought to activate by recruiting Arp2/3 complex to the sides of actin filaments, but the precise mechanism is unknown (5, 6). In the absence of NPFs, Arp2/3 complex has weak or no nucleation activity. However, inhibitors of Arp2/3 complex play important roles in controlling the activity of the complex in vivo (7–11), suggesting that multiple layers of regulation of Arp2/3 complex are required for controlling the assembly of cellular actin networks.
In budding yeast, Arp2/3 complex nucleates patches of cortical branched actin networks required for endocytosis (12). Actin patches contain at least six different Arp2/3 regulators (Las17, Pan1, Myo3, Myo5, Abp1, and Crn1) (13). Five of these proteins have been shown to activate the complex in vitro, and in vivo experiments have demonstrated that despite some functional overlap, all five NPFs contribute to actin patch assembly (13–16). Coronin, an Arp2/3 complex inhibitor present in yeast actin patches, plays a role in regulating the dynamics of the patches (13, 17–19), which assemble at the cortex as the endocytic vesicle forms and then disassemble as the vesicle moves into the cytoplasm to fuse with endosomes (20). Actin polymerization is thought to provide force for invagination of endocytic vesicles and may propel the vesicle into the cytoplasm (21). Deletion of coronin increases the duration of both the assembly and the mobile phases, indicating that coronin is involved in fine-tuning actin dynamics during endocytosis (13). Unlike other known cellular Arp2/3 complex inhibitors, coronins have been shown to both inhibit and directly interact with Arp2/3 complex (9, 18). However, the mechanism of inhibition is not known, and it is currently unclear how coronin is involved in tuning actin patch dynamics.
Here we describe the biochemical dissection of the mechanism of regulation of Arp2/3 complex by budding yeast coronin, Crn1. To our surprise, we found that this coronin not only inhibits Arp2/3 complex, but also activates it, and therefore, has dual modes of regulation. The concentration of coronin controls the switch between the regulatory modes; low concentrations activate Arp2/3 complex, and high concentrations inhibit the complex. We show that Crn1 is a type II NPF with a previously undiscovered CA sequence within its central unique region and that mutations of this sequence abolish activation of the complex in vitro and slow patch assembly in vivo. Our data support a model by which at low concentrations, Crn1 recruits the complex to the sides of actin filaments and activates it. Activation is synergistic with Las17, the budding yeast WASp homologue, but does not require Las17. We also propose a model for the switch between regulatory modes of Crn1 in which high concentrations of Crn1 inhibit nucleation by blocking Arp2/3 complex binding sites on actin filaments.
EXPERIMENTAL PROCEDURES
Budding yeast Arp2/3 complex, Crn1, Las17-VCA, and N-WASp-VCA proteins were purified as described in the supplemental material. Actin was purified and labeled with pyrene iodoacetamide, and the increase in fluorescence was measured to monitor polymerization as described previously (22). Sedimentation velocity experiments were carried out in a Beckman XL-I analytical ultracentrifuge as described in the supplemental material. Fluorescence anisotropy competition experiments were carried out as described previously (22) and analyzed as described in the supplemental material. Detailed procedures for all other methods can be found in the supplemental material.
RESULTS
Crn1 Activates Arp2/3 Complex
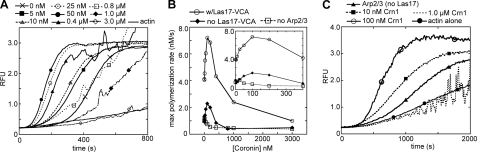
To determine how coronin regulates Arp2/3 complex, we used a pyrene actin polymerization assay to measure the rates of polymerization of actin filaments in the presence of Arp2/3 complex, Las17-VCA, and a range of concentrations of Crn1. Low micromolar concentrations inhibited Arp2/3 complex-mediated nucleation, as reported previously for human and yeast coronins (Fig. 1A) (18, 23). The rate of polymer formation was equal to actin alone when 3.0 μm or more coronin was added to our assays, suggesting that Arp2/3 was completely inhibited under these conditions. At high Crn1 concentrations, we observed fluctuations in the emission values in late stages of the reaction and increased total fluorescence, which we attributed to filament bundling, a previously reported activity of coronin (24).
FIGURE 1.
Low concentrations of Crn1 activate Arp2/3 complex. A, time course of 2 μm 15% pyrene actin polymerization showing the effect of 0–3.0 μm full-length Crn1 on polymerization nucleated by 20 nm Arp2/3 complex and 40 nm GST-Las17-VCA. RFU, relative fluorescence units. B, plot of maximum (max) polymerization rate versus Crn1 concentration for reactions with or without 25 nm GST-Las17-VCA and 10 nm Arp2/3 complex. C, Crn1 activates Arp2/3 complex in the absence of VCA. Conditions are the same as in panel A, except that GST-Las17-VCA was excluded from the reaction. Note that Arp2/3 complex has some nucleating activity even in the absence of Las17-VCA, as observed previously (54, 55).
Surprisingly, we found that low concentrations of coronin increased the rate of polymer formation (Fig. 1, A and B). The maximal rate of actin assembly increased and the lag phase decreased with more coronin added, up to an optimal concentration range of 50–100 nm. In the presence of Las17-VCA, the maximal rate of polymer formation was ∼2-fold higher at 100 nm coronin than with no coronin. Las17-VCA was not required because coronin alone increased the polymerization rate in the presence of Arp2/3 complex, although not as efficiently as Las17-VCA (Fig. 1, B and C). As the rate increase is dependent on the presence of Arp2/3 complex (supplemental Fig. S1) but did not require Las17-VCA, we concluded that Crn1 activates Arp2/3 complex.
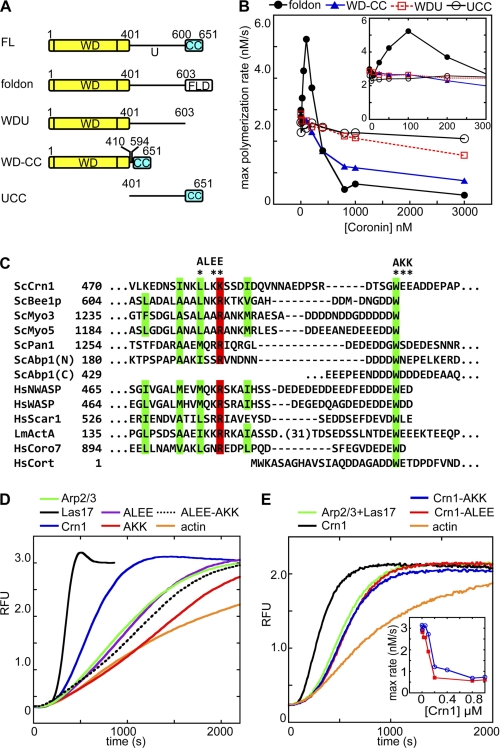
The Unique Region of Crn1 Contains C and A Sequences Required for Activation
To determine how Crn1 activates Arp2/3 complex, we mapped regions of Crn1 required for the increased polymerization rate. Crn1 contains a conserved N-terminal WD repeat domain that binds to actin filaments (25, 26), a C-terminal predicted coiled coil, and a non-conserved central region of 197 amino acids predicted to be unstructured (27) (Fig. 2A). Deletion of either the coiled-coil (Crn1-WDU) or the WD repeat domain (Crn1-UCC) abolished activation (Fig. 2B). A Crn1 construct (Crn1WD-CC) lacking the unique region also failed to activate Arp2/3 complex (Fig. 2B), so all three regions of Crn1 contribute to activation. Humphries et al. (18) demonstrated that Arp2/3 complex and Crn1 interact directly in pulldown experiments and that the C-terminal half of Crn1 (residues 400–651, which include the entire unique region and the coiled-coil domain) co-immunoprecipitates with Arp2/3 complex in Saccharomyces cerevisiae lysate. Therefore, we hypothesized that residues in the C-terminal region might directly interact with Arp2/3 complex and contribute to activation. We examined the Crn1 sequence and identified motifs in the unique region similar to the C (central) and A (acidic) sequences of WASp/Scar family proteins (Fig. 2C), which are known to bind and activate Arp2/3 complex (28, 29). The acidic sequence in Crn1 spans residues 503–508 and is composed of the sequence WEEADD. Mutation of this sequence to AKKADD (Crn1-AKK) abolished activation (Fig. 2, D and E). In WASp/Scar proteins, the C region is thought to form an amphipathic helix that interacts with Arp2/3 complex (30) and promotes an activating conformational change (31, 32). We mutated three residues within the putative C-region of Crn1 (L480A/K482E/K483E) and found that this mutant, Crn1-ALEE, did not activate Arp2/3 complex alone or with Las17-VCA (Fig. 2, D and E). We conclude that Crn1 contains sequences homologous to WASp/Scar C and A regions within its unique domain and that these regions are required for activation.
FIGURE 2.
Mapping regions of Crn1 required for inhibition and activation of Arp2/3 complex. A, Crn1 constructs used in mapping experiments. WD, WD repeat domain; U, unique region; CC, coiled-coil domain; FLD, foldon, 27-amino acid trimerization domain from bacteriophage T4 fibritin. FL, full length. B, plot of maximum (max) polymerization rate versus coronin concentration for four different Crn1 constructs in the presence of Las17-VCA. Conditions are as in Fig. 1B. C, sequence alignment of CA regions from Arp2/3 activators from S. cerevisiae (Sc), human (Hs), and Listeria monocytogenes (Lm). Conserved hydrophobic (green) and basic (red) residues are indicated by colored boxes. Crn1 mutants characterized in this work (ALEE and AKK) are indicated with asterisks. D, neither Crn1-AKK nor Crn1-ALEE activates Arp2/3 complex in the absence of Las17-VCA. Wild-type, AKK, ALEE, or ALEE-AKK coronin (40 nm) or GST-Las17-VCA (40 nm) was added to actin polymerization reactions with 2.0 μm 15% pyrene actin and 20 nm Arp2/3 complex, and the time course of polymerization was monitored. RFU, relative fluorescence units. E, neither Crn1-AKK nor Crn1-ALEE activates Arp2/3 complex with Las17-VCA present. Conditions are: 4 μm 15% pyrene actin, 50 nm Crn1 (WT, AKK, or ALEE), 40 nm GST-Las17-VCA, and 20 nm Arp2/3 complex. Inset: plot of maximum polymerization rate for a broad range of Crn1-AKK (red line) or Crn1-ALEE (blue line) concentrations calculated from polymerization time courses. Reactions contained 2 μm 15% pyrene actin with 20 nm Arp2/3 complex, 40 nm GST-Las17-VCA, and 0–1 μm Crn1-AKK or Crn1-ALEE.
Activation of Arp2/3 Is Required for Normal Actin Patch Dynamics in Budding Yeast
In wild-type S. cerevisiae, actin and actin-binding proteins assemble to their peak concentrations in the patches in about 8 s, after which the patches separate from the cortex and move into the cytoplasm (15, 19, 21). In the crn1Δ strain, the duration of the assembly phase of an average actin patch is increased by 2 s (13, 19). To determine whether activation of Arp2/3 complex by Crn1 influences the dynamics of patch assembly, we deleted CRN1 from a haploid strain and reintegrated mutant or wild-type CRN1 genes under control of their native promoters into the leu2-3,112 locus (see supplemental Table S1 for all strains used in this study). To visualize actin patches in live cells, we added a C-terminal GFP tag to Abp1, an actin filament-binding protein that localizes to actin patches with the same kinetics as actin, Arp2/3 complex, and a number of other actin-binding proteins (15, 33). We used confocal microscopy to image live cells and then used previously developed automated patch tracking software to follow the trajectories of hundreds of individual actin patches (13).
Consistent with previous reports, we found that the average duration of the assembly phase of patches increased from 8.0 to 9.8 s in the crn1Δ strain (Table 1). The ALEE and AKK point mutants, which abolished activation of Arp2/3 complex, caused a small but significant increase in the duration of the assembly phase when compared with wild type (0.8 and 0.9 s, respectively), demonstrating that Arp2/3 complex activation by coronin plays a role in fine-tuning the dynamics of actin patch assembly. When compared with other Arp2/3 complex activators, the change in the duration of the assembly phase is greater than observed when the acidic region of Pan1 is deleted (∼0.5 s) but less than observed when the acidic region of Las17 is deleted (∼4 s) (13). Neither the ALEE nor the AKK Crn1 mutants lengthened the assembly phase as much as the coronin knock-out, so activation of Arp2/3 complex is not the only role coronin plays in actin patch assembly.
TABLE 1.
Actin patch average assembly times and total lifetimes in wild type and mutant strains
The assembly phase length and total patch lifetime were calculated as described in the supplemental material. Means ± S.E. are listed.
| Strain | Assembly phase | Total patch lifetime | No. of observations | p value |
|---|---|---|---|---|
| s | s | |||
| WT | 7.98 ± 0.18 | 12.35 ± 0.19 | 275 | |
| crn1Δ | 9.82 ± 0.20 | 14.62 ± 0.26 | 238 | <0.001 |
| crn1-wd-cc | 9.04 ± 0.18 | 14.36 ± 0.24 | 256 | <0.001 |
| crn1-akk | 8.80 ± 0.21 | 13.39 ± 0.22 | 194 | <0.038 |
| crn1-alee | 8.94 ± 0.16 | 13.35 ± 0.22 | 190 | 0.003 |
Low Concentrations of Crn1 Recruit Arp2/3 Complex to Actin Filaments
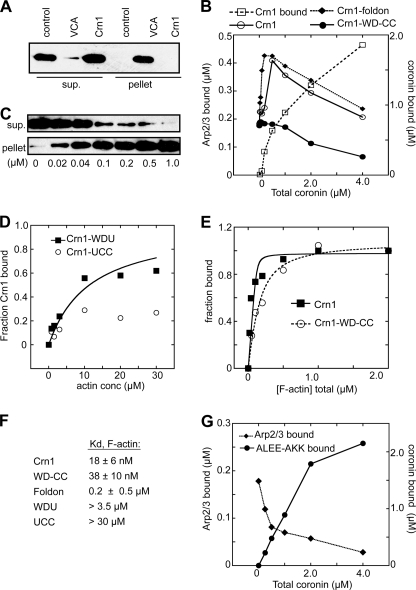
Because Crn1 was previously only known to inhibit Arp2/3 complex, we sought to determine the mechanism of activation to better understand the molecular basis of Arp2/3 complex regulation. We first asked whether actin monomer recruitment is important for activation. Monomer recruitment is mediated by the V sequence of the VCA region in type I NPFs, which binds directly to actin monomers and is required for activation by type I NPFs (29, 34, 35). We did not find a V sequence in Crn1, and actin monomers did not interact with GST-Crn1 in a pulldown assay (Fig. 3A), so it is unlikely that Crn1 activates by recruiting monomers to Arp2/3 complex.
FIGURE 3.
Crn1 recruits Arp2/3 complex to actin filaments. A, GST pulldown assay showing that Crn1 does not bind monomeric actin. Actin (1.0 μm) was incubated with 10 μm GST-Crn1 or GST-N-WASp-VCA bound to glutathione-Sepharose beads or with beads alone (control) before pelleting. B, copelleting assays showing the effect of increasing concentrations of Crn1 on sedimentation of Arp2/3 complex with actin filaments. Arp2/3 complex at 0.5 μm and 0–4 μm Crn1 were incubated with 2.0 μm polymerized actin. Pelleted coronin and Arp2/3 complex were quantified by densitometry. The dashed line with open squares indicates the amount of Crn1 pelleted with F-actin at each concentration. C, copelleting assay showing binding of Crn1 to actin filaments. Phalloidin-stabilized actin filaments were incubated with 40 nm His-Crn1 and pelleted, and supernatant and pellet were blotted with an anti-His antibody. sup., supernatant. D, binding isotherm from actin filament copelleting assays showing the effect of increasing concentrations (conc) of actin filaments on Crn1 sedimentation. Actin filaments were incubated with Crn1-WDU (0.5 μm) or Crn1-UCC (3.0 μm) before pelleting. Data were fit as described under “Experimental Procedures.” E, binding isotherm for Crn1 and Crn1-WD-CC as described in C. F, tabulated binding affinities of Crn1 constructs for actin filaments. G, copelleting assays showing the effect of increasing concentrations of Crn1-ALEE-AKK on sedimentation of Arp2/3 complex with actin filaments assays. Conditions were identical to B. Plots shown in panels B, D, E, and G are representative of at least three separate experiments.
We next explored the possibility that Crn1 activates Arp2/3 complex by recruiting it to the sides of actin filaments. Actin filament binding is necessary for activation of the complex (3, 36, 37), but the complex has low affinity and binds with a slow on-rate to actin filaments (KD ∼1–5 μm (3, 38)), whereas Crn1 binds tightly (24). In addition, Humphries et al. (18) reported that 2.0 μm Crn1 increased the amount of Arp2/3 complex that copellets with actin filaments. We conducted copelleting experiments with a range of concentrations of Crn1 and found that copelleting of Arp2/3 complex increased ∼2-fold as the Crn1 concentration was increased to 500 nm and then decreased as more Crn1 was added (Fig. 3B). Therefore, at low concentrations, Crn1 recruits Arp2/3 to the sides of actin filaments, whereas high concentrations block Arp2/3 from binding. Given that we observed activation only at low concentrations of coronin, recruitment to the filament may explain activation. Crn1-UCC and Crn1-WDU, mutants defective in actin filament binding, did not activate the complex (Figs. 2B and 3, D and F), providing further support for this mechanism. Importantly, the Crn1 construct lacking the unique region (Crn1-WD-CC) bound actin filaments with affinity similar to the wild-type Crn1 (Fig. 3, E and F) but did not activate Arp2/3 complex or recruit it to filaments (Figs. 2B and 3B). A double mutation in the CA region (Crn1-ALEE-AKK) also prevented activation and recruitment (Figs. 2D and 3G). This led us to hypothesize that interaction of Crn1 with both actin filaments and Arp2/3 complex is required for recruitment and activation.
The Crn1 CA Region Binds Preferentially to the Arp2 Site in Arp2/3 Complex
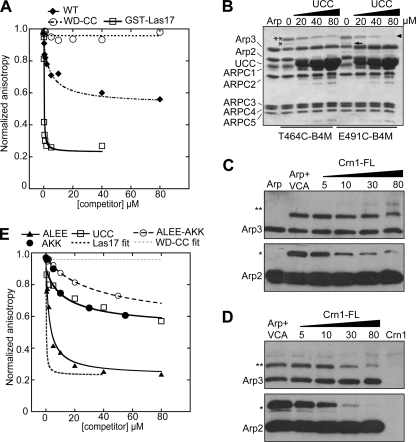
The CA region from WASp/Scar proteins interacts directly with Arp2/3 complex (29, 30), so we reasoned that Crn1 might bind to the complex through its CA sequence. In a fluorescence anisotropy competition binding assay, titration of a complex of rhodamine-labeled VCA (Rh-VCA) and Arp2/3 complex with Crn1 decreased the anisotropy, suggesting that Crn1 competes with Rh-VCA for binding to Arp2/3 complex. The anisotropy did not decrease to the level of Rh-VCA alone even at the highest Crn1 concentration (80 μm), so coronin did not completely displace VCA (Fig. 4A).
FIGURE 4.
Crn1 interacts directly with Arp2/3 complex through its CA region and competes with VCA. A and E, fluorescence anisotropy competition assay to measure interaction between Arp2/3 complex and Crn1 constructs. Increasing concentrations of Crn1, Crn1-WD-CC, Crn1-ALEE, Crn1-AKK, Crn1-ALEE-AKK, Crn1-UCC, or GST-Las17-VCA were equilibrated with 60 nm ScArp2/3 complex and 50 nm SpWsp1-Rh-VCA, and the anisotropy was measured. Data were fit as described under “Experimental Procedures.” The KD for the Crn1-ALEE-AKK was estimated by assuming saturation at a normalized anisotropy value of 0.6, as observed for wild-type Crn1. B, Coomassie Brilliant Blue-stained gel of 5.0 μm Bos taurus Arp2/3 complex cross-linked to 10 μm T464C-B4M or E491C-B4M in the presence of increasing concentrations of Crn1-UCC. Cross-linked bands are labeled as follows: two asterisks, Arp3xT464C-B4M-VCA; single asterisks, Arp2xT464C-B4M-VCA; arrowhead, Arp3xE491C-B4M-VCA; arrow, Arp2xE491C-B4M-VCA. Supplemental Fig. S5 shows that Crn1 activates B. taurus Arp2/3 complex. C and D, Western blots of cross-linking reactions with T464C-VCA (C) or E491C-VCA (D). Conditions were identical to B, except that increasing concentrations of Crn1 (full length (FL)) were added to the reaction. The top row shows anti-Arp3 blots, and the bottom row shows anti-Arp2 blots.
Multiple lines of evidence suggest that there are two binding sites for VCA on Arp2/3 complex (39, 40). We reasoned that like cortactin, a type II NPF that interacts with Arp2/3 complex through an N-terminal acidic domain (6), Crn1 may bind preferentially to one of these two sites, explaining the failure to compete all Rh-VCA off the complex. To test this, we labeled an engineered cysteine in the C or A regions of N-WASp-VCA (T464C or E491C) with benzophenone-4-maleimide and used UV radiation to cross-link the benzophenone moiety to interacting residues on the surface of Arp2/3 complex. Cross-linking resulted in two high molecular weight bands for each VCA construct (Fig. 4B). Western blotting (Fig. 4, C and D) and mass spectrometry identified the top band as Arp3-VCA and the lower band as Arp2-VCA. These data indicate that there are two binding sites for CA on Arp2/3 complex, consistent with the results of Padrick et al. (40). The addition of full-length Crn1 to the reaction decreased cross-linking to both Arp2 and Arp3, indicating that Crn1 can block both sites (Fig. 4, C and D). However, the Arp2 site was blocked at lower concentrations than the Arp3 site, suggesting that coronin competes more strongly with VCA at the Arp2 site than at the Arp3 site.
We next used the competition assay to map Crn1 domain requirements for binding to the complex. The WD repeat domain was not required for competition with VCA because Crn1-UCC displaced VCA in both the anisotropy and the cross-linking experiments (Fig. 4, B and E). In contrast, deletion of the entire unique domain (Crn1-WD-CC) abolished competition with VCA (Fig. 4A). The double mutant Crn1-ALEE-AKK bound to the complex at least 8-fold more weakly than Crn1 and did not completely compete Rh-VCA from the complex at 50 μm (Fig. 4E), suggesting that the unique region CA is critical for the interaction with Arp2/3 complex. Surprisingly, mutation of either the A or the C motifs individually did not prevent competition with VCA (Fig. 4E). The acidic region mutant, Crn1-AKK, bound only slightly worse than Crn1 (apparent KD ∼2 μm, when compared with 0.6 ± 0.2 μm for Crn1) and, like Crn1, did not completely displace VCA. The C region mutant, Crn1-ALEE, bound more tightly than wild-type coronin (apparent KD = 0.3 ± 0.05 μm) and decreased the anisotropy to the same value as unbound Rh-VCA alone, suggesting that the C-region mutant competes with both VCA binding sites. We do not currently understand why this mutant binds more tightly than wild-type Crn1, but one possibility is that the wild-type C sequence inhibits the A region from binding to one of the two CA binding sites on Arp2/3 complex. High resolution x-ray crystal structures of CA bound to each site on Arp2/3 complex will likely be required to resolve this question.
Trimerization of Crn1 Is Required for Activation
Oligomerization of WASp/SCAR proteins greatly enhances their potency (39, 41, 42), so we asked whether oligomerization of Crn1 influences its activation potential. We used analytical ultracentrifugation to demonstrate that Crn1 trimerizes (Fig. 5). Deletion of the coiled-coil domain (Crn1-WDU), which is known to oligomerize other coronins (43), abolished activation by Crn1 (Fig. 2B). To determine the role of the coiled coil in activation, we replaced the coiled-coil domain with a 27-amino acid trimerization domain, called foldon, from bacteriophage T4 fibritin (44). This construct, Crn1-foldon, activated Arp2/3 complex and recruited it to actin filaments similarly to wild-type coronin (Figs. 2B and 3B), suggesting that oligomerization is the only required function of the coiled coil in activating the complex. Multivalent binding of the WD repeat domains to actin filaments has been proposed previously (45) and explains the increased affinity of Crn1-foldon for actin filaments when compared with Crn1-WDU (Fig. 3F).
FIGURE 5.
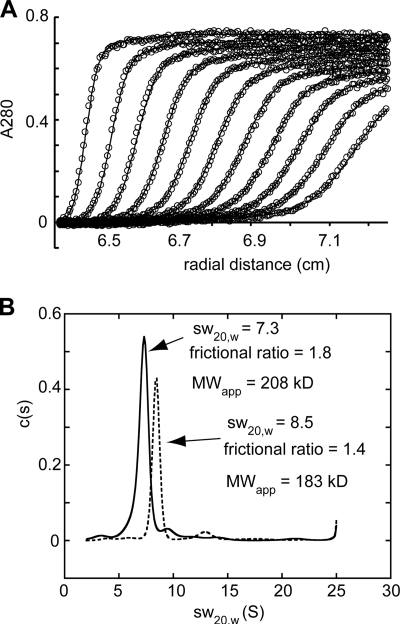
Oligomerization state of Crn1 and Crn1-WD-CC. A, raw data (open circles) for sedimentation velocity analytical ultracentrifugation experiment containing 9.0 μm full-length Crn1 with superimposed fit using a continuous c(S) distribution model. B, sedimentation coefficient distribution for analytical ultracentrifugation runs containing either 9.0 μm Crn1 (solid line, monomer molecular mass = 72.7 kDa) or 9.0 μm Crn1 (WD-CC) (dashed line, monomer molecular mass = 55.3 kDa). The calculated apparent molecular weights (MWapp) are indicated. The major species in the analysis of Crn1 had a sedimentation coefficient of 7.3, a frictional ratio of 1.8, and an apparent molecular mass of 208 kDa, indicating that Crn1 forms an extended trimer. The major species in the analysis of Crn1-WD-CC had a sedimentation coefficient of 8.5 and a frictional ratio of 1.4, yielding an apparent molecular mass of 183 kDa, consistent with a more compact trimer or tetramer.
Crn1 Acts Synergistically with Las17
Activation of the complex by Crn1 was enhanced in the presence of the VCA region of Las17, suggesting that these activators act synergistically. However, because Crn1 shows activation of the complex on its own, it is possible that they act additively rather than synergistically. To test this, we ran actin assembly assays with a range of ratios of Crn1 to VCA, keeping the total activator concentration at 60 nm. Given that Crn1 on its own activates Arp2/3 complex more weakly than VCA, maximal activation at 100% VCA would argue for an additive effect, whereas maximum activation at lower ratios of VCA to Crn1 would indicate that activation is synergistic. We found maximal activation when the molar ratio of Crn1:VCA was 1:1, demonstrating that Crn1 and VCA act synergistically (supplemental Fig. S2).
DISCUSSION
Crn1 Is an Arp2/3 Complex Activator with Similarities to Both Cortactin and WASp/Scar Proteins
We show here that Crn1 activates Arp2/3 complex both on its own and synergistically with Las17-VCA. Activation requires all three regions of Crn1: the N-terminal WD repeat domain, the central unique domain, and the C-terminal coiled-coil domain. Within the unique region, we discovered a CA sequence, a conserved feature of the WASp/Scar family proteins. Like WASp/Scar CA, Crn1 CA is required for activation and interaction with the complex. However, we discovered important mechanistic differences between Crn1- and WASp-mediated activation. First, Crn1 does not contain a V sequence, an actin monomer binding region found N-terminal to CA in WASp/Scar proteins (29, 34, 35). Not surprisingly, we found that Crn1 failed to bind actin monomers, so we concluded that Crn1 does not activate the complex by recruiting actin monomers to the branch junction, a mechanism proposed for the VCA region of WASp/Scar (29, 34). Second, our anisotropy and cross-linking assays demonstrate that Crn1 CA has distinct binding preferences for Arp2/3 complex when compared with WASp/Scar CA because it preferentially competes with one of two WASp/Scar binding sites. In these respects, Crn1 is similar to cortactin, the prototypical type II NPF from higher eukaryotes that, like Crn1, binds to only one of two activator binding sites on the complex and contains an F-actin binding domain instead of an actin monomer binding domain (5, 6). Although functionally this classifies Crn1 as a type II NPF, we note that our data already point to important mechanistic differences between Crn1 and cortactin because 1) cortactin contains an A but not a C sequence and 2) cortactin binds to the Arp3 subunit, whereas Crn1 preferentially binds Arp2. Dissection of these differences will likely provide valuable insight into the molecular mechanism of Arp2/3 complex activation.
The Role of Filament Recruitment in Crn1-mediated Activation of Arp2/3 Complex
Previous experiments showed that Crn1 recruits Arp2/3 complex to actin filaments, but the functional relevance of this observation was unknown (18). Here we showed that only low concentrations of Crn1 could increase copelleting of Arp2/3 with actin filaments or activate the complex in actin polymerization assays, so recruitment and activation are correlated. Deletion of the coiled-coil domain or the WD repeat domain decreased actin filament binding by at least 200- and 1500-fold, respectively, and completely eliminated activation, indicating that actin filament binding is required for activation. Mutants with reduced (or no) binding to Arp2/3 (Crn1-WD-CC and Crn1-ALEE-AKK) failed to recruit or activate the complex, so direct binding to both Arp2/3 complex and actin filaments is required. Therefore, these data are consistent with a tethering recruitment activation model, in which Crn1 simultaneously binds an actin filament and Arp2/3 complex. The low binding affinity and unusually slow on-rate of Arp2/3 complex for filaments suggests that recruitment is a plausible mechanism (3, 38). The tethering recruitment model predicts simultaneous interaction of Arp2/3 complex with Crn1 and Las17. Although this is supported by our anisotropy and cross-linking data, we cannot rule out the possibility that Crn1 and Las17 interact sequentially to activate the complex. This remains an open question for both Crn1 and cortactin and will require further study (6, 46). The stoichiometry of Crn1 to Arp2/3 complex on filaments also remains an open question. The data in Fig. 3B show that in the absence of Crn1, approximately one molecule of Arp2/3 complex is bound per 10 actin subunits in a filament. The addition of coronin results in maximal recruitment of one additional complex when ∼0.83 coronin trimers (or 2.5 Crn1 monomers) are bound per 10 actin subunits. Although this suggests that on average one coronin trimer recruits slightly more than one complex, further experiments will be required to unequivocally determine the stoichiometry.
Indirect Inhibition Explains the Dual Modes of Regulation Observed for Crn1
Our data showed a dramatic concentration dependence in the effect of Crn1 on Arp2/3 complex activity. Activation was maximal at about 100 nm and decreased at higher concentrations. Low micromolar concentrations completely inhibited the complex, as observed previously (18). Humphries et al. (18) reported that a construct of Crn1 containing both the unique and the coiled-coil regions is sufficient to bind and inhibit Arp2/3 complex. This led to the proposal of a direct inhibition model, in which binding of the UCC region locks Arp2/3 complex into an inactive conformation (18, 32). Our competition binding assay showed that deletion of the unique region abolished binding between Crn1 and Arp2/3 complex, but this construct, Crn1-WD-CC, strongly inhibited the complex, arguing against direct inhibition as the main mode of down-regulation. In addition, we show that mutations that reduced or eliminated actin filament binding (Crn1-WDU, Crn1-UCC) caused drastic reductions in inhibition (Fig. 2B, supplemental Fig. S3) and that high concentrations of Crn1 reduce Arp2/3 binding to actin filaments (Fig. 3B). Therefore, we propose an alternative mechanism in which coronin inhibits nucleation by saturating actin filaments and blocking Arp2/3 complex binding sites (Fig. 6). The three-dimensional reconstruction of coronin-saturated F-actin is consistent with this mechanism because it shows that Arp2/3 complex and coronin binding sites on F-actin overlap (27, 47) (supplemental Fig. S4). In addition, previous experiments showed that inhibition was relieved by adding preformed filaments (18), which we propose is due to the presence of free side binding sites on the added filaments. We note that in our assays, 4.0 μm Crn1 or Crn1-foldon was sufficient to completely inhibit Arp2/3 complex but did not completely block copelleting of the complex with actin filaments (Fig. 3B). In contrast, 4.0 μm Crn1-WD-CC or Crn1-ALEE-AKK, constructs with impaired Arp2/3 binding, strongly blocked copelleting of the complex at 4.0 μm. These data suggest that Crn1 constructs that bind Arp2/3 can tether the complex to actin filaments even when they are saturated with Crn1 and are consistent with a tethering recruitment mechanism of activation.
FIGURE 6.
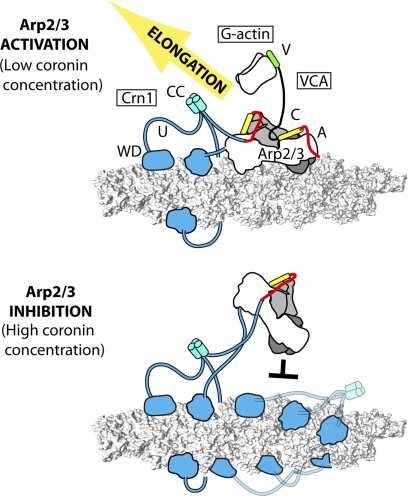
Proposed mechanism for dual modes of regulation of Arp2/3 complex by Crn1. Coronin trimers (cyan) bind actin filaments (gray surface representation) through their WD repeat domains and Arp2/3 complex (Arp3 is dark gray; Arp2 is light gray; other subunits are white) through the unique region CA. The unique CA interacts with Arp2, leaving a second activator binding site on Arp3 open for VCA-containing NPF. Synergistic activation may occur through simultaneous recruitment of the complex to filaments and actin monomers to the complex.
An indirect mechanism of inhibition explains the observed switch between activating and inhibiting modes of coronin. We propose that the switch is controlled by the ability of Crn1 to either recruit Arp2/3 to filaments or block filament binding sites for the complex. At low Crn1 concentrations, side binding sites adjacent to the bound coronin are free, and the complex can be recruited to these sites, increasing the apparent binding affinity of the complex for filaments (Fig. 6). At high Crn1 concentrations, Arp2/3 complex binding sites are blocked, so nucleation is inhibited. We note that three other actin-binding proteins, caldesmon, epithelial protein lost in neoplasm (EPLIN), and tropomyosin (10, 48, 49), have been shown to inhibit nucleation by blocking filament binding sites. Understanding how these proteins contribute to the architecture of actin networks in vivo is an important challenge in the field.
Regulation of Arp2/3 complex by Coronin in Vivo
We showed that activation of Arp2/3 complex by Crn1 is important for proper actin patch dynamics in budding yeast, and our data support a model in which Crn1 activates the complex by recruiting it to the sides of filaments. These observations are consistent with mathematical modeling of actin patch assembly in fission yeast, which demonstrated that Arp2/3 complex must bind to the sides of actin filaments faster in vivo than observed in vitro to account for the rate of patch assembly (33, 50). Therefore, type II NPFs like Crn1 may play a general role in accelerating Arp2/3 complex-mediated branching in actin patch assembly. Of the six known activators of Arp2/3 complex in budding yeast (Las17, Abp1, Myo3 and Myo5, Pan1, and Crn1), only Las17 is a prototypical type I NPF with an actin monomer-recruiting V region (51). Understanding how these regulators coordinate to control Arp2/3 complex activity in vivo will provide valuable insight into Arp2/3-mediated actin assembly in actin patches and other branched actin networks.
Our in vitro data show that the role of Crn1 in regulating Arp2/3 complex switches depending on its concentration. In endocytic patches, Crn1 reaches an average peak concentration of 93 molecules in budding yeast and 400–500 molecules in fission yeast when compared with 5000–7000 molecules of actin (19, 33). Crn1 recruitment is delayed when compared with actin, and its concentration peaks about 4 s later than the core actin module (19, 33). Therefore, Crn1 is present at low concentrations during patch assembly, consistent with our proposed activation mechanism.
At peak concentrations, budding yeast actin patches contain an average of 5100 molecules of actin, greater than 50 times the amount of Crn1 at its peak (19). This suggests that Crn1 on its own does not saturate filaments to the extent required to block binding of the complex during actin patch lifetimes. However, we cannot rule out cooperation of Crn1 with other actin filament-binding proteins to inhibit branching nucleation. Although this leaves open the question of a role for Crn1 inhibition of Arp2/3 complex in yeast actin patches, other actin filament networks appear to require inhibition of Arp2/3 complex by coronin for proper dynamics (7).
Implications for Understanding Other Coronin Family Proteins
We examined other coronin sequences to determine whether activation of Arp2/3 complex is unique to budding yeast Crn1 or a general feature of coronin family proteins. Humans have seven coronins, divided into three classes (52). Class 1 and class 2 coronins (Coro1A-1C, Coro6, Coro2A-B) have the same domain organization as Crn1, whereas the sole member of third class, Coro7/Pod-1, has two WD repeat domains and no predicted coiled coil. We did not find a CA sequence in any of the class 1 or class 2 coronins, making it unlikely that that they activate Arp2/3 complex. However, we and others have noted the presence of an acidic region conserved among diverse species in the C terminus of Coro7 (52). Our analysis suggests that Coro7 proteins also have a C sequence N-terminal to the acidic region (Fig. 2C). Therefore, Coro7/Pod-1 proteins may be functionally similar to budding yeast Crn1. Interestingly, like budding yeast Crn1, Coro1B interacts directly with both actin filaments and Arp2/3 complex (9). However, instead of activating the complex, Coro1B has been reported to target Arp2/3 complex at existing branch junctions to disassemble the branch (23). In addition, multiple laboratories have shown that some coronins play a role in regulating the disassembly of actin filaments through the actin depolymerization factor cofilin (17, 19, 53). Clearly, additional in vitro and in vivo experiments are required to understand the molecular basis of actin cytoskeletal regulation by different classes of coronins.
Supplementary Material
Acknowledgments
We thank John Cooper and Brian Galletta for providing patch tracking software and Vladamir Sirotkin and Ken Prehoda for critically reading the manuscript. We also thank Stephan Weitzel for help with analytical ultracentrifugal experiments.
This work was supported, in whole or in part, by National Institutes of Health Grant (to B. J. N).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures, Figs. S1–S5, and Table 1.
- NPF
- nucleation-promoting factor
- WASp
- Wiskott-Aldrich Syndrome Protein
- Rh
- rhodamine
- N-WASp
- neuronal WASp
- Scar
- Suppressor of cAMP receptor.
REFERENCES
- 1. Pollard T. D., Cooper J. A. (2009) Science 326, 1208–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Goley E. D., Welch M. D. (2006) Nat. Rev. Mol. Cell Biol. 7, 713–726 [DOI] [PubMed] [Google Scholar]
- 3. Goley E. D., Rammohan A., Znameroski E. A., Firat-Karalar E. N., Sept D., Welch M. D. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 8159–8164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Padrick S. B., Rosen M. K. (2010) Annu. Rev. Biochem. 79, 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Uruno T., Liu J., Zhang P., Fan Yx., Egile C., Li R., Mueller S. C., Zhan X. (2001) Nat. Cell Biol. 3, 259–266 [DOI] [PubMed] [Google Scholar]
- 6. Weaver A. M., Heuser J. E., Karginov A. V., Lee W. L., Parsons J. T., Cooper J. A. (2002) Curr. Biol. 12, 1270–1278 [DOI] [PubMed] [Google Scholar]
- 7. Shiow L. R., Roadcap D. W., Paris K., Watson S. R., Grigorova I. L., Lebet T., An J., Xu Y., Jenne C. N., Föger N., Sorensen R. U., Goodnow C. C., Bear J. E., Puck J. M., Cyster J. G. (2008) Nat. Immunol. 9, 1307–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cai L., Holoweckyj N., Schaller M. D., Bear J. E. (2005) J. Biol. Chem. 280, 31913–31923 [DOI] [PubMed] [Google Scholar]
- 9. Cai L., Marshall T. W., Uetrecht A. C., Schafer D. A., Bear J. E. (2007) Cell 128, 915–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Blanchoin L., Pollard T. D., Hitchcock-DeGregori S. E. (2001) Curr. Biol. 11, 1300–1304 [DOI] [PubMed] [Google Scholar]
- 11. Iwasa J. H., Mullins R. D. (2007) Curr. Biol. 17, 395–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Engqvist-Goldstein A. E., Drubin D. G. (2003) Annu. Rev. Cell Dev. Biol. 19, 287–332 [DOI] [PubMed] [Google Scholar]
- 13. Galletta B. J., Chuang D. Y., Cooper J. A. (2008) PLoS Biol. 6, e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. D'Agostino J. L., Goode B. L. (2005) Genetics 171, 35–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kaksonen M., Sun Y., Drubin D. G. (2003) Cell 115, 475–487 [DOI] [PubMed] [Google Scholar]
- 16. Evangelista M., Klebl B. M., Tong A. H., Webb B. A., Leeuw T., Leberer E., Whiteway M., Thomas D. Y., Boone C. (2000) J. Cell Biol. 148, 353–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gandhi M., Achard V., Blanchoin L., Goode B. L. (2009) Mol. Cell 34, 364–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Humphries C. L., Balcer H. I., D'Agostino J. L., Winsor B., Drubin D. G., Barnes G., Andrews B. J., Goode B. L. (2002) J. Cell Biol. 159, 993–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lin M. C., Galletta B. J., Sept D., Cooper J. A. (2010) J. Cell Sci. 123, 1329–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Toshima J. Y., Toshima J., Kaksonen M., Martin A. C., King D. S., Drubin D. G. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 5793–5798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kaksonen M., Toret C. P., Drubin D. G. (2005) Cell 123, 305–320 [DOI] [PubMed] [Google Scholar]
- 22. Nolen B. J., Pollard T. D. (2008) J. Biol. Chem. 283, 26490–26498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cai L., Makhov A. M., Schafer D. A., Bear J. E. (2008) Cell 134, 828–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Goode B. L., Wong J. J., Butty A. C., Peter M., McCormack A. L., Yates J. R., Drubin D. G., Barnes G. (1999) J. Cell Biol. 144, 83–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Goode B. L., Rodal A. A., Barnes G., Drubin D. G. (2001) J. Cell Biol. 153, 627–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gandhi M., Jangi M., Goode B. L. (2010) J. Biol. Chem. 285, 34899–34908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Galkin V. E., Orlova A., Brieher W., Kueh H. Y., Mitchison T. J., Egelman E. H. (2008) J. Mol. Biol. 376, 607–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Higgs H. N., Pollard T. D. (2001) Annu. Rev. Biochem. 70, 649–676 [DOI] [PubMed] [Google Scholar]
- 29. Marchand J. B., Kaiser D. A., Pollard T. D., Higgs H. N. (2001) Nat. Cell Biol. 3, 76–82 [DOI] [PubMed] [Google Scholar]
- 30. Panchal S. C., Kaiser D. A., Torres E., Pollard T. D., Rosen M. K. (2003) Nat. Struct. Biol. 10, 591–598 [DOI] [PubMed] [Google Scholar]
- 31. Goley E. D., Rodenbusch S. E., Martin A. C., Welch M. D. (2004) Mol. Cell 16, 269–279 [DOI] [PubMed] [Google Scholar]
- 32. Rodal A. A., Sokolova O., Robins D. B., Daugherty K. M., Hippenmeyer S., Riezman H., Grigorieff N., Goode B. L. (2005) Nat. Struct. Mol. Biol. 12, 26–31 [DOI] [PubMed] [Google Scholar]
- 33. Sirotkin V., Berro J., Macmillan K., Zhao L., Pollard T. D. (2010) Mol. Biol. Cell 21, 2894–2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chereau D., Kerff F., Graceffa P., Grabarek Z., Langsetmo K., Dominguez R. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 16644–16649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dayel M. J., Mullins R. D. (2004) PLoS Biol. 2, E91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Higgs H. N., Blanchoin L., Pollard T. D. (1999) Biochemistry 38, 15212–15222 [DOI] [PubMed] [Google Scholar]
- 37. Achard V., Martiel J. L., Michelot A., Guérin C., Reymann A. C., Blanchoin L., Boujemaa-Paterski R. (2010) Curr. Biol. 20, 423–428 [DOI] [PubMed] [Google Scholar]
- 38. Beltzner C. C., Pollard T. D. (2008) J. Biol. Chem. 283, 7135–7144 [DOI] [PubMed] [Google Scholar]
- 39. Padrick S. B., Cheng H. C., Ismail A. M., Panchal S. C., Doolittle L. K., Kim S., Skehan B. M., Umetani J., Brautigam C. A., Leong J. M., Rosen M. K. (2008) Mol. Cell 32, 426–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Padrick S. B., Deka R. K., Chuang J. L., Wynn R. M., Chuang D. T., Norgard M. V., Rosen M. K., Brautigam C. A. (2010) Anal. Biochem. 407, 89–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Footer M. J., Lyo J. K., Theriot J. A. (2008) J. Biol. Chem. 283, 23852–23862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Higgs H. N., Pollard T. D. (2000) J. Cell Biol. 150, 1311–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kammerer R. A., Kostrewa D., Progias P., Honnappa S., Avila D., Lustig A., Winkler F. K., Pieters J., Steinmetz M. O. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13891–13896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Frank S., Kammerer R. A., Mechling D., Schulthess T., Landwehr R., Bann J., Guo Y., Lustig A., Bächinger H. P., Engel J. (2001) J. Mol. Biol. 308, 1081–1089 [DOI] [PubMed] [Google Scholar]
- 45. Cai L., Makhov A. M., Bear J. E. (2007) J. Cell Sci. 120, 1779–1790 [DOI] [PubMed] [Google Scholar]
- 46. Uruno T., Liu J., Li Y., Smith N., Zhan X. (2003) J. Biol. Chem. 278, 26086–26093 [DOI] [PubMed] [Google Scholar]
- 47. Rouiller I., Xu X. P., Amann K. J., Egile C., Nickell S., Nicastro D., Li R., Pollard T. D., Volkmann N., Hanein D. (2008) J. Cell Biol. 180, 887–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Maul R. S., Song Y., Amann K. J., Gerbin S. C., Pollard T. D., Chang D. D. (2003) J. Cell Biol. 160, 399–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yamakita Y., Oosawa F., Yamashiro S., Matsumura F. (2003) J. Biol. Chem. 278, 17937–17944 [DOI] [PubMed] [Google Scholar]
- 50. Berro J., Sirotkin V., Pollard T. D. (2010) Mol. Biol. Cell 21, 2905–2915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Weaver A. M., Young M. E., Lee W. L., Cooper J. A. (2003) Curr. Opin. Cell Biol. 15, 23–30 [DOI] [PubMed] [Google Scholar]
- 52. Uetrecht A. C., Bear J. E. (2006) Trends Cell Biol. 16, 421–426 [DOI] [PubMed] [Google Scholar]
- 53. Brieher W. M., Kueh H. Y., Ballif B. A., Mitchison T. J. (2006) J. Cell Biol. 175, 315–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rodal A. A., Manning A. L., Goode B. L., Drubin D. G. (2003) Curr. Biol. 13, 1000–1008 [DOI] [PubMed] [Google Scholar]
- 55. Wen K. K., Rubenstein P. A. (2005) J. Biol. Chem. 280, 24168–24174 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.