Abstract
Nuclear proteins typically contain short stretches of basic amino acids (nuclear localization sequences; NLSs) that bind karyopherin α family members, directing nuclear import. Here, we identify CTNNBL1 (catenin-β-like 1), an armadillo motif-containing nuclear protein that exhibits no detectable primary sequence homology to karyopherin α, as a novel, selective NLS-binding protein. CTNNBL1 (a single-copy gene conserved from fission yeast to man) was previously found associated with Prp19-containing RNA-splicing complexes as well as with the antibody-diversifying enzyme AID. We find that CTNNBL1 association with the Prp19 complex is mediated by recognition of the NLS of the CDC5L component of the complex and show that CTNNBL1 also interacts with Prp31 (another U4/U6.U5 tri-snRNP-associated splicing factor) through its NLS. As with karyopherin αs, CTNNBL1 binds NLSs via its armadillo (ARM) domain, but displays a separate, more selective NLS binding specificity. Furthermore, the CTNNBL1/AID interaction depends on amino acids forming the AID conformational NLS with CTNNBL1-deficient cells showing a partial defect in AID nuclear accumulation. However, in further contrast to karyopherin αs, the CTNNBL1 N-terminal region itself binds karyopherin αs (rather than karyopherin β), suggesting a function divergent from canonical nuclear transport. Thus, CTNNBL1 is a novel NLS-binding protein, distinct from karyopherin αs, with the results suggesting a possible role in the selective intranuclear targeting or interactions of some splicing-associated complexes.
Keywords: Nuclear Translocation, Nuclear Transport, Protein-Protein Interactions, RNA Splicing, Spliceosome, AID, Importin, Karyopherin, NTC, Nuclear Localization Signal
Introduction
CTNNBL14 (catenin-β-like 1) was first identified as a widely expressed nuclear protein with predicted structural homology to β-catenin (1). Although speculated, based on overexpression assays, to play a role in apoptosis (1), the function of CTNNBL1 remains unknown. Protein association studies might, however, be indicative of cellular pathways in which CTNNBL1 may function. CTNNBL1 was found associated with the Prp19-CDC5L-PLRG1 RNA splicing complex (also known as the NTC or 19 complex) (2). We encountered CTNNBL1 in a screen for proteins interacting with the antibody-diversifying enzyme AID, finding that CTNNBL1-deficient chicken DT40 B cells exhibited a reduced frequency of immunoglobulin V gene conversion and somatic mutation (3).
Here, to gain possible insight into CTNNBL1 function, we have investigated the molecular basis of its interactions. We show that CTNNBL1 interaction with its known partners (the Prp19-CDC5L complex and AID) is mediated by recognition of NLS motifs. We additionally identify RNA-splicing factor Prp31 as a new CTNNBL1 interactor, with recognition also occurring through its NLS. We show that CTNNBL1 uses its central armadillo (ARM) domain to bind NLS-containing partners, analogous to the karyopherin α family of NLS-binding proteins (for review, see Refs. 4, 5). Our results reveal that CTNNBL1 differs from karyopherin αs both in its interactions with import factors as well as in its pattern of NLS preferences, indicating that it represents a novel type of NLS-binding protein.
EXPERIMENTAL PROCEDURES
Plasmids
The construction of expression plasmids is shown in supplemental Table S1. Vectors for the expression in 293T cells of Prp31-GFP and Prp31(ΔNLS)-GFP (6) were gifts from Drs. S. E. Wilkie and D. M. Hunt (University of London); the M6P8 retroviral vector was a gift from Dr. Felix Randow, and vectors for FLAG-CTNNBL1 and FLAG-APOBEC2 have been described previously (4).
Recombinant Protein Purification
Recombinant proteins were purified from Escherichia coli BL21(DE3) transformants that had been incubated at 16 °C in LB overnight following induction with 1 mm isopropyl 1-thio-β-d-galactopyranoside at an A600 of 0.6. Cells expressing GST-tagged nuclear transport factors, GST-tagged NLSs, and GST-AID were harvested by centrifugation and lysed by sonication in buffer A, B, or C respectively (supplemental Table S2). Cleared lysates were incubated with glutathione-Sepharose beads (Amersham Biosciences) to purify GST-tagged proteins. E. coli-expressing recombinant His-CTNNBL1 and His-CTNNBL1(Δ1–76) were lysed by sonication in buffer D (Table S2), and the recombinant proteins were purified by binding onto a nickel-nitrilotriacetic acid column (GE Healthcare) followed by elution with a 0–500 mm imidazole gradient. Peak fractions were further purified by anion exchange at pH 8.0 on a Q column (GE Healthcare) and subsequent gel filtration on Superdex 75 (GE Healthcare).
Monitoring Protein Interactions
All binding assays were incubated at 4 °C with end-on-end rotation for 1 h. Where required, 293T cells were transiently transfected using GeneJuice® (Merck Biosciences) according to the manufacturer's instructions and cultured to a density of 5 × 105 cells ml−1. Whole cell lysates were prepared by resuspending washed cell pellets (∼106 cells) in 0.5 ml of lysis buffer (supplemental Table S2). Cleared lysates (diluted 1:2 in lysis buffer) were incubated with antibodies against CDC5L (BD Biosciences) or GFP (Abcam) followed by precipitation with anti-mouse IgG-agarose beads (eBioscience) or protein A-Sepharose (Pharmacia Biotech). FLAG- or HA-tagged proteins were immunoprecipitated by incubation with anti-FLAG M2 agarose (Sigma) or anti-HA agarose (Roche Applied Science) and elution with 3×FLAG peptide (Sigma) or 3×HA peptide (Roche Applied Science). Proteins were visualized in Western blots using HRP-conjugated antibodies to HA (Roche Applied Science), FLAG (Sigma), GFP (Abcam), or GST (Santa Cruz Biotechnology). Endogenously expressed CDC5L and Prp19 were detected using monoclonal antibodies (BD Biosciences). Endogenous CTNNBL1 from 293T cells was detected using anti-CTNNBL1 antiserum as described previously (3) whereas His-CTNNBL1 was detected using a monoclonal anti-CTNNBL1 antibody (Abcam).
For pull downs from whole cell extracts, glutathione-Sepharose (25 μl/reaction) was preincubated with GST-tagged protein (1 h, 4 °C), washed, and then incubated with 293T cell lysate (500 μl) as described above. To detect interactions in vitro, purified GST-NLS chimeric protein (typically 5–20 μg) was incubated with 15 μg of His-tagged CTNNBL1, CTNNBL1(Δ1–76), or karyopherin α in a final volume of 30–200 μl prior to purification on glutathione-Sepharose and SDS-PAGE. In vitro interactions between GST-tagged nuclear transport factors and CTNNBL1 were assayed as above except that (because of the different yields of different recombinant GST-tagged transport factors) the GST transport factors were first purified by elution off glutathione-Sepharose and then bound back to glutathione-Sepharose (following dialysis) at a consistent stoichiometry of 250 μg of GST transport factor/100 μl of glutathione-Sepharose.
Mass Spectrometry
Whole cell lysates of 293T cells that had been transfected with either FLAG-CTNNBL1 or FLAG-APOBEC2 were immunoprecipitated using anti-FLAG M2-agarose. Following extensive washing, bound proteins were eluted using 3×FLAG peptide and separated using SDS-PAGE and stained with Coomassie. Protein-containing bands were excised from the gel and digested with trypsin (7). Peptide mixtures were separated by nanoscale liquid chromatography (LC Packings) on a reverse phase C18 column. The eluate was introduced directly into a Q-STAR hybrid tandem mass spectrometer. The peptide and ion mass data were queried against the NCBInr data base using the program MASCOT (Matrix Bioscience), and putative interactors were assigned a probability-based Mowse score as described previously (8).
Protein Localization
Transient transfectants of 293T and HeLa expressing GFP-tagged chimeric proteins were fixed with PBS/4% paraformaldehyde for 10 min, permeabilized with PBS/0.5% Triton X-100 for 10 min. Cells were counterstained with wheat germ agglutinin-Alexa Fluor 594, Hoechst 33258 (Molecular Probes), or mounting medium with DAPI (Vector Laboratories) prior to confocal microscopy. AID-HA-NLS fusion proteins were visualized in the nucleus following treatment with leptomycin B, an inhibitor of nuclear export. Following fixation and staining as described above, cells were stained with rabbit anti-HA antibody (Santa Cruz Biotechnology) followed by Alexa 568-conjugated anti-rabbit IgG antiserum (Invitrogen). Sequences encoding AID or CDC5L NLS-β-galactosidase-GFP were subcloned from the HindIII and NotI sites of vector pEGFPN1 to the same sites of the retroviral vector M6P8 and transduced into DT40 cells as described previously (9). 48 h following transduction, cells were stained with propidium iodide. GFP+ve, propidium iodide−ve cells were sorted using a Beckman Coulter MoFlo High Speed Cell Sorter, counterstained with Hoechst 33258, and visualized by live-cell confocal microscopy. All image processing and analysis were done using ImageJ software. Where required, nuclear:cytoplasmic intensity ratios of individual cells were calculated using the ImageJ line profile tool.
Isothermal Titration Calorimetry
Peptides corresponding to the sequences of SV40 NLS (PKKKRKV) and CDC5L NLS3 (KKRKRKR) (ABL Advanced Biomedical) as well as purified His-tagged CTNNBL1(Δ1–76) were dialyzed against CTNNBL1 buffer (20 mm Hepes, pH 7.5, 50 mm NaCl). Final peptide concentrations were determined by ninhydrin reaction. Binding assays were performed using an ITC200 calorimeter (MicroCal, Inc.). The cell contained 360 μl of protein solution (typically 150 μm CTNNBL1), and the syringe contained peptide solution (typically 2.8 mm). Peptide was injected into the cell in 20 injections of 2 μl (spaced every 2 min), typically to a 4–5-fold molar excess. Titration curves were fitted to the data using the manufacturer's ORIGIN software (MicroCal, Inc.).
AID Activity
To assay immunoglobulin class switching in vivo, AID-expressing pMXs-IRES-GFP based constructs were retrovirally transduced into lymphocytes from AID-deficient C57BL/6 mice and assayed by flow cytometry as described previously (3). The expression of AID in the primary B cells was monitored by Western blotting using biotinylated anti-AID N-terminal antibody (3) followed by streptavidin-HRP (Amersham Biosciences).
RESULTS
CTNNBL1 Binds the Prp19-CDC5L Complex through the CDC5L NLSs
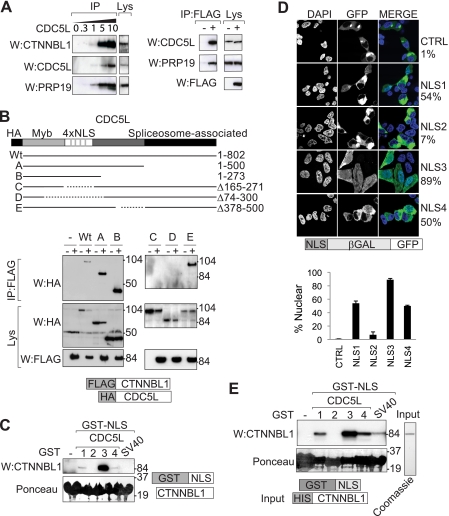
In keeping with previous proteomic studies as well as with the detection of a CTNNBL1-CDC5L association in yeast two-hybrid screens (2, 3), immunoprecipitation of FLAG-tagged CTNNBL1 from transfected 293T cells brings down several members of the Prp19-CDC5L complex (Fig. 1A). To ascertain whether there was direct interaction between CTNNBL1 and CDC5L, we exploited the fact that the CDC5L C-terminal portion is critical for CDC5L interaction with the spliceosome (10, 11). The CTNNBL1 interaction was retained by C-terminally truncated CDC5L mutants (Fig. 1B), indicating that the CTNNBL1/CDC5L interaction is likely direct or, at least, does not require other spliceosomal components. This is consistent with a recent study of the architecture of the core human Prp19-CDC5L complex and its relationship with peripheral components (12).
FIGURE 1.
CTNNBL1 associates with the NLS region of CDC5L. A, immunoprecipitation of endogenous CDC5L from 293T cells (using increasing volumes in microliters of anti-CDC5L antibody) brings down endogenous CTNNBL1 and Prp19 (left panels); immunoprecipitation of FLAG-tagged CTNNBL1 from 293T[FLAG-CTNNBL1] transfectants (+) but not control 293T cells (−) brings down endogenous CDC5L and Prp19 (right panels). IP, samples analyzed following immunoprecipitation by Western blotting (W) with anti-CTNNBL1, anti-CDC5L, anti-Prp19 or anti-FLAG antibodies as indicated. Lys, an aliquot (7%) of the total cell lysate probed as a control. B, the 4×NLS domain of CDC5L is required for interaction with CTNNBL1. Lysates of 293T cells that had been transfected with HA-tagged deletion mutants of CDC5L together with (+) or without (−) FLAG-tagged CTNNBL1 were subjected to immunoprecipitation with anti-FLAG-agarose followed by Western blotting with anti-HA antibodies. The upper panels show the Western blots of the immunoprecipitated material (IP); the lower pairs of panels show the blots of the whole cell lysates (Lys). The migration positions of prestained molecular weight marker proteins are indicated. C, purification of the endogenous CTNNBL1 from 293T cell extracts was accomplished by fishing with recombinant GST-NLS fusion proteins. Computationally predicted NLS sequences from CDC5L were expressed as GST fusion proteins in E. coli and immobilized on glutathione-Sepharose. After incubation with 293T cell lysates, total Sepharose-bound proteins were subjected to SDS-PAGE and stained with Ponceau S prior to Western blotting (W) with anti-CTNNBL1 antiserum. D, nuclear targeting activity of predicted CDC5L NLS sequences was tested using confocal microscopy to analyze the intracellular localization of NLS-β-galactosidase-GFP fusion proteins transfected into 293T cells. Nuclei were counterstained with DAPI. The percentage of fluorescent cells exhibiting clear nuclear fluorescence is indicated. 100 cells transfected with each construct were counted. The mean ± S.D. were calculated by analyzing at least 30 cells transduced with each construct from multiple independently acquired fields. E, direct interaction between GST-NLS fusion proteins and His-CTNNBL1 was assessed by incubating GST-NLS fusion proteins that had been bound to glutathione-Sepharose with purified His-CTNNBL1. Total Sepharose-bound proteins were stained with Ponceau S following SDS-PAGE prior to Western blotting (W) with anti-CTNNBL1 mAb. The purity of the recombinant His-tagged CTNNBL1 used for the binding assays is illustrated.
More extensive analysis of CDC5L truncation mutants pointed to a critical role of CDC5L region 165–271 in CTNNBL1 interaction (Fig. 1B). This region of CDC5L contains four predicted NLSs (13), raising the possibility that CTNNBL1 might bind NLSs directly, especially in view of the fact that CTNNBL1 comprises ARM motifs, a motif used by karyopherin αs to bind the NLSs of their cargoes during nuclear import.
We therefore generated GST fusion constructs in which the four putative CDC5L NLSs had been separately fused to the C terminus of GST and asked whether these fusion proteins could be used to purify endogenous CTNNBL1 from 293T cell extracts. CDC5L NLS3 was very effective at fishing the endogenous CTNNBL1 out from the 293T cell extracts (Fig. 1C), with NLS1 and NLS4 showing weaker binding and no detectable binding being obtained with either NLS2 or with the SV40 T antigen NLS.
To find out whether any of these putative NLSs actually confers nuclear-localizing activity, we fused them to a β-galactosidase/GFP chimeric protein (which is large enough to be unable to enter the nucleus by mere diffusion) and expressed them in 293T cells. NLS3 gave the strongest nuclear-localizing activity with the NLS1 and NLS4 fusions also showing substantial nuclear expression in some cells; no NLS function was evident in the chimera containing the putative NLS2 (Fig. 1D).
The interaction between CTNNBL1 and CDC5L NLS3 can be recapitulated using purified recombinant CTNNBL1 made from E. coli, revealing the interaction to be direct (Fig. 1E). Weaker interaction with CDC5L NLSs 1 and 4 as well as with the SV40 NLS is also observed in this assay with purified components.
CTNNBL1 Also Interacts with Prp31, Binding through Its NLS
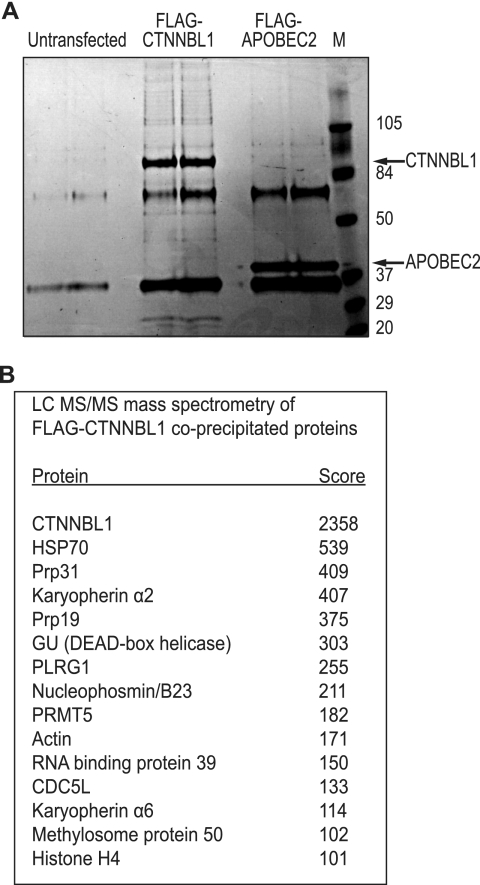
We wondered whether CTNNBL1 might interact with other nuclear partners through their NLSs. We therefore performed a mass spectrometric analysis of proteins associated with FLAG-tagged CTNNBL1 that had been immunopurified from 293T cell transfectants. Although most proteins were found only at low stoichiometry (Fig. 2A), it is notable that CDC5L, Prp19, and PLRG1 were all present among the high scoring proteins (Fig. 2B). Other high scorers include Prp31 (a splicing factor not known to associate with the CDC5L-Prp19-PLRG1 complex and which functions in the assembly of the U4/U6.U5 tri-snRNP) (14), PRMT5 and methylosome protein 50 (which form a complex implicated in protein arginine methylation), and karyopherins α2 and α6.
FIGURE 2.
Proteins associated with immunopurified FLAG-CTNNBL1. A, proteins bound to FLAG-CTNNBL1 or FLAG-APOBEC2 control were immunopurified using anti-FLAG M2-agarose, eluted with 3×FLAG peptide, separated by SDS-PAGE, and stained with Coomassie. Prestained molecular weight markers were run alongside the samples. B, proteins co-immunopurifying with FLAG-CTNNBL1 were subjected to tryptic digestion and mass spectrometric identification. All proteins identified with a Mowse score >65 and which were not also identified in the FLAG-APOBEC2 control sample (e.g. keratin; immunoglobulin fragments) are shown.
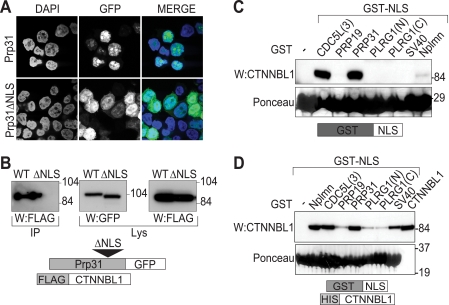
The interaction between Prp31 and CTNNBL1 could be recapitulated in transfected 293T cells (Fig. 3). Prp31 contains a well defined classical NLS (6): this NLS is required for the interaction with CTNNBL1. Furthermore, the Prp31 NLS (as a GST fusion protein) was as effective as the CDC5L NLS3 in pulling down FLAG-tagged CTNNBL1 from transfected 293T cell extracts and also interacted well with purified CTNNBL1 in vitro (Fig. 3, C and D).
FIGURE 3.
CTNNBL1 binds Prp31 through its NLS. A, selective nuclear accumulation of Prp31-GFP but not Prp31[ΔNLS]-GFP in transfected 293T cells was assessed by confocal microscopy. Nuclei were counterstained with DAPI. B, Prp31-GFP (WT) but not Prp31[ΔNLS]-GFP (ΔNLS) associates with FLAG-tagged CTNNBL1 in co-transfected 293T cells. IP, GFP-tagged Prp31 or Prp31[ΔNLS] was purified from the lysates using anti-GFP antibody and protein A-Sepharose, subjected to SDS-PAGE and Western blotted (W) with anti-FLAG antibody. Lys, samples of total cell lysates were Western blotted with ant-GFP or anti-FLAG antibodies as expression controls. C, GST-[Prp31NLS] fusion protein pulls down endogenous CTNNBL1 from a 293T whole cell lysate. The indicated GST-NLS chimeric proteins (see Fig. 4C for NLS sequences) that had been purified from E. coli and bound to glutathione-Sepharose were incubated with 293T cell lysates. Total Sepharose-bound proteins were stained with Ponceau S following SDS-PAGE prior to Western blotting (W) with anti-CTNNBL1 antiserum. D, direct interaction between GST-[Prp31NLS] (as well as other GST-NLS) fusion proteins and His-CTNNBL1 was assessed by incubating GST-NLS fusion proteins that had been bound to glutathione-Sepharose with purified His-CTNNBL1. Bound proteins were stained with Ponceau S prior to probing with anti-CTNNBL1 mAb. An aliquot of the input CTNNBL1 is shown in the right lane.
CTNNBL1 Binds NLSs via Its ARM Domain but Displays a Distinct NLS Preference Pattern from Karyopherin αs
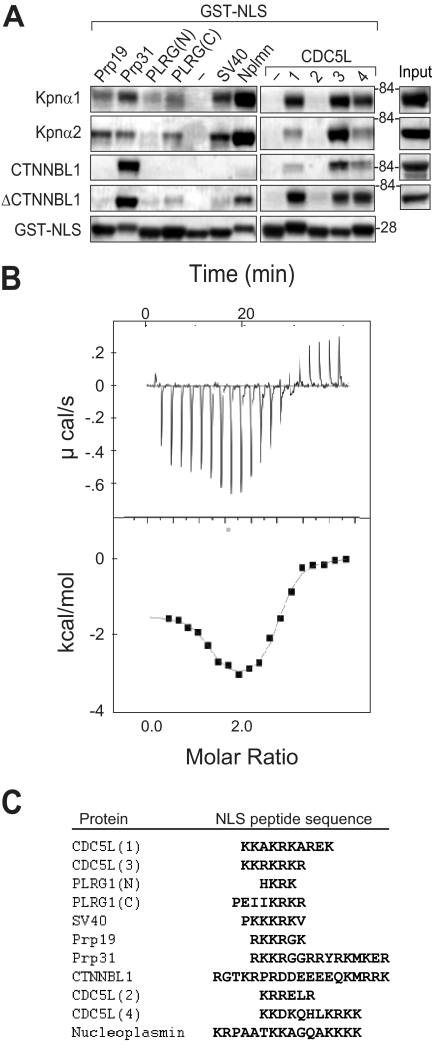
Because both CTNNBL1 and karyopherin αs bind NLSs and contain ARM repeat domains, we sought to ascertain whether CTNNBL1 and karyopherin αs exhibit similar NLS binding preferences by comparing the efficiency with which different GST-NLSs were pulled down by CTNNBL1, karyopherin α1, and karyopherin α2. The Prp31 NLS is pulled down more effectively by CTNNBL1 than by karyopherin α1/2 whereas the PLRG1 and SV40 NLSs bind better to the karyopherins; the CDC5L NLS3 is well bound by both CTNNBL1 and karyopherins (Fig. 4A). Moreover, an N-terminally truncated mutant of CTNNBL1 lacking the first 76 amino acids and consisting of the ARM domain alone (ΔCTNNBL1) was sufficient to pull down NLSs efficiently (Fig. 4A). Direct binding of the CDC5L NLS to the ARM domain of CTNNBL1 was confirmed by isothermal titration calorimetry, revealing a stoichiometry of binding of two CDC5L NLS3 peptides per ΔCTNNBL1 molecule, with the two binding sites having Kd values of 0.11 μm and 4.1 μm (Fig. 4B). This shows similarity to the binding of the SV40 T antigen NLS by karyopherin α1 (two binding sites with Kd values of 0.31 and 0.98 μm) (17). However, we could not identify specific binding of the SV40 NLS to the CTNNBL1 ARM domain by this method, consistent with the weakness of the binding in the pulldown assays (Figs. 1 and 3). The results suggest a significant difference in the fine NLS binding specificity of CTNNBL1 and karyopherin αs.
FIGURE 4.
CTNNBL1 ARM motifs bind NLSs but show a distinct specificity from karyopherin αs. A, binding of various GST-NLS fusion proteins to purified His-karyopherin α1/2, His-CTNNBL1, and His-ΔCTNNBL1 was assessed as in Fig. 3D. B, binding of CDC5L-NLS3 peptide to His-ΔCTNNBL1 was monitored by isothermal calorimetry. Kd1 = 0.11 ± 0.07 μm; n1 = 1.1; ΔH1 = −1.48 ± 0.11 kcal/mol; Kd2 = 4.1 ± 0.8 μm; n2 = 1.35; ΔH2 = −3.28 ± 0.15 kcal/mol. C, NLS sequences were assayed for binding to CTNNBL1. An alignment (using ClustalW) of the sequences of the various NLSs analyzed is shown. These NLSs in the GST fusion proteins are preceded by the sequence LYPRGSM (separating the C-terminal residue in GST from the NLS) and followed by LERPHRD*.
AID-CTNNBL1 Interaction Depends on AID Residues Critical for Nuclear Import
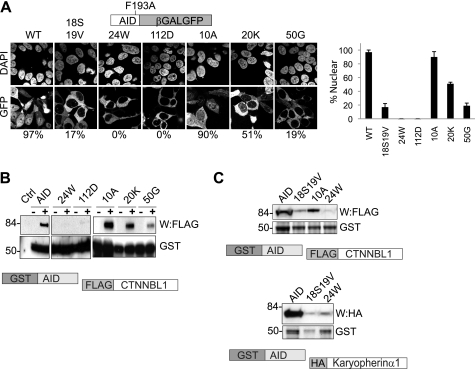
Our interest in CTNNBL1 was originally triggered by finding it associated with AID (3), a DNA deaminase that is located mainly in the cytoplasm but shuttles in and out of the nucleus. Although not containing any evident linear NLS, AID nuclear import depends on multiple basic residues distributed over the AID primary sequence (15, 16). Despite the small size of AID, its nuclear import has been shown to depend on NLS-mediated active transport, with mutations within the conformational NLS ablating the ability of AID to trigger class switch recombination in vivo (15, 16). Given that it is recognition of NLS motifs that mediates the CTNNBL1 interaction with both CDC5L and Prp31, we wondered whether CTNNBL1 recognizes a conformational NLS within AID. We made mutations at AID residues Arg19, Arg24, Arg50, and Arg112 and found (in agreement with previous observations) (15, 16) that they all interfered with nuclear import as judged by the localization of an AID-β-galactosidase-GFP fusion protein (Fig. 5A). We then asked whether these individual critical nuclear import mutations affected CTNNBL1 binding: they all perturbed it (Fig. 5, B and C). In contrast, both import and CTNNBL1 binding were retained following mutation of AID residue Lys10 or Trp20. Interestingly, in light of the finding by Patenaude et al. that AID can interact with karyopherin αs (16), we find that mutation at residue Arg24, which completely abrogates AID nuclear import (0% nuclear staining), destroys the interaction with CTNNBL1 while still allowing some (although much diminished) interaction with karyopherin α1. However, AID mutated at residues Val18 and Arg19 shows diminished nuclear accumulation (17% nuclear staining) and exhibits reduced interaction with both CTNNBL1 and karyopherin α.
FIGURE 5.
AID nuclear import mutants show diminished CTNNBL1 binding. A, subcellular distribution of chimeric AID[F193A]-β-galactosidase-GFP fusion proteins. AID[F193A]-β-galactosidase-GFP proteins with the indicated point mutations were expressed in 293T cells and visualized by confocal microscopy. Nuclei were counterstained with DAPI. The F193A mutation in the reporter protein interferes with the AID nuclear export sequence (28, 29), allowing import to be monitored. The percentage of fluorescent cells exhibiting clear nuclear fluorescence is indicated. 30 cells transfected with each construct were counted. The mean ± S.D. calculated from three independent experiments are indicated. B, diminished interaction of AID nuclear import mutants with CTNNBL1. GST-tagged AID and point mutants thereof expressed in E. coli and bound onto glutathione-Sepharose were incubated with extracts of 293T cells that had been transfected with FLAG-CTNNBL1. Following SDS-PAGE, the abundance of GST-AID proteins was demonstrated by staining with Coomassie; bound FLAG-CTNNBL1 was detected by Western blotting (W) with anti-FLAG antibody. C, R24W and V18S,R19V mutations in AID affecting binding to both CTNNBL1 (top panels) and karyopherin α1 (lower panels). Binding of GST-AID mutants to FLAG-CTNNBL1 and HA-tagged karyopherin α1 was assessed as in B, except using ant-HA antibody to detect bound HA-karyopherin α1.
CDC5L NLS3 Confers Nuclear Localization Even in the Absence of CTNNBL1
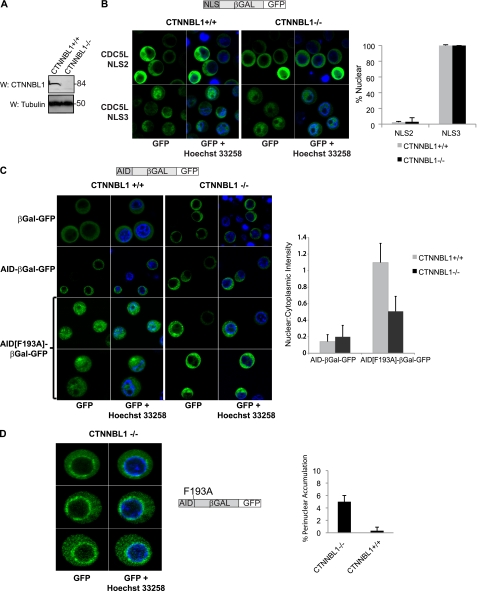
What then is the significance of NLS binding by CTNNBL1? One possibility is that CTNNBL1, although showing no primary sequence homology to karyopherin α, may nevertheless be functioning analogously in mediating nuclear import. To test whether this is the case, we used CTNNBL1+/+ and CTNNBL1−/− DT40 chicken B cells (Fig. 6A) to assay the subcellular distribution of β-galactosidase-GFP fusion proteins (which are too large to enter the nucleus by mere diffusion) that had been N-terminally tagged with CDC5L NLS3. However, we were unable to detect any difference in the nuclear localization of this fusion protein in the presence or absence of CTNNBL1 (Fig. 6B).
FIGURE 6.
Subcellular distribution of β-galactosidase-GFP fusion proteins in CTNNBL1+/+ and CTNNBL1−/− DT40 chicken B cells. A, Western blot of whole cell lysates from CTNNBL1−/− and CTNNBL1+/+ parental cells stained with anti-CTNNBL1 or anti-tubulin antibodies. B, constructs expressing β-galactosidase-GFP fusion proteins bearing an N-terminal fusion with CDC5L NLS3, which confers nuclear localization, or the putative CDC5L NLS2, which does not (Fig. 1D) retrovirally transduced into DT40 B cells. GFP-positive cells were sorted, counterstained with Hoechst 33258, and visualized using live cell confocal microscopy 48 h after transduction. The percentage of transduced cells exhibiting clear nuclear fluorescence is indicated. At least 30 cells transduced with each construct from multiple independently acquired fields were counted. C, subcellular distribution of AID and export-deficient AID[F193A]-β-galactosidase-GFP fusion proteins expressed in DT40 B cells. To quantify the heterogeneous nuclear accumulation of AID[F193A]-β-galactosidase-GFP, line profiles were obtained individually for each of at least 30 independent cells transduced with each construct, and ratios of nuclear:cytoplasmic intensity were calculated as described previously (30). The mean ± S.D. are indicated. D, representative images showing accumulation of AID[F193A]-β-galactosidase-GFP in perinuclear clumps in ∼5% of CTNNBL1−/− cells.
Nuclear Accumulation of AID Is Partially Reduced in the Absence of CTNNBL1
We next asked whether the nuclear accumulation of AID, which unlike CDC5L, is not an essential housekeeping gene, might be affected by the absence of CTNNBL1. Both β-galactosidase-GFP and AID-β-galactosidase-GFP fusion proteins remained cytosolic in CTNNBL1+/+ as well as CTNNBL1−/− DT40 cells (Fig. 6C). As expected, due to the amino acid substitution in its nuclear export sequence, the mutant AID[F193A]-β-galactosidase-GFP fusion protein is distributed throughout the nucleus and cytoplasm of CTNNBL1+/+ cells, with little accumulation in the nucleolus. However, in contrast, the localization of AID[F193A]-β-galactosidase-GFP in cells lacking CTNNBL1 appears heterogeneous. Although most of the cells display some nuclear GFP staining, the overall level of steady-state nuclear accumulation of AID[F193A]-β-galactosidase-GFP is reduced in the CTNNBL1-deficient compared with the CTNNBL1-proficient cells. The heterogeneous nuclear accumulation of AID[F193A]-β-galactosidase-GFP was quantified using the line profile tool of the ImageJ software to calculate ratios of nuclear:cytoplasmic GFP-staining intensity in individual transduced B cells (Fig. 6C). The CTNNBL1−/− cells exhibited a mean nuclear:cytoplasmic GFP intensity ratio of 0.5 (versus 1.1 in CTNNBL1+/+ cells). In addition, striking ring-like perinuclear accumulation of AID[F193A]-β-galactosidase-GFP was observed in a fraction (∼5%) of CTNNBL1−/− cells (Fig. 6D), but not in CTNNBL1+/+ cells (<0.5%) possibly reflecting a selective block in nuclear pore transit in CTNNBL1-deficient cells.
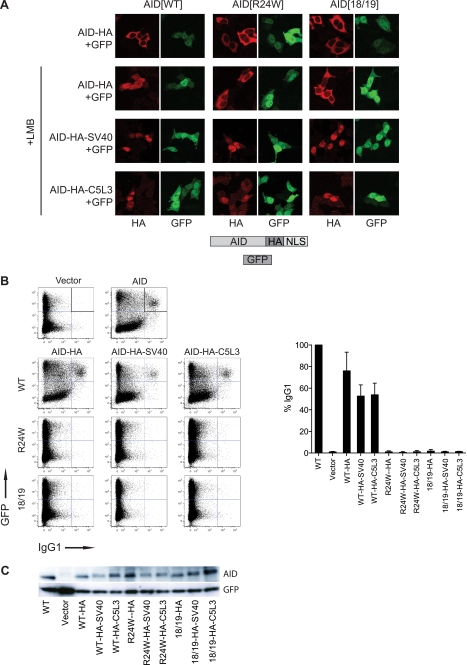
Nuclear Import of AID via Heterologous NLSs Does Not Reconstitute Antibody Class Switching
The diminished nuclear accumulation of AID[F193A]-β-galactosidase-GFP in CTNNBL1-deficient cells is consistent with the possibility that AID nuclear import/retention might be partially dependent on CTNNBL1, but with other karyopherins being able to substitute for CTNNBL1 in its absence. To test whether the mechanism of nuclear import is important in facilitating the physiological function of nuclear AID (for example by facilitating subnuclear targeting to the immunoglobulin locus), we fused an SV40 NLS (which is classically imported into the nucleus by karyopherin α but shows little binding to CTNNBL1) to the C termini of import-deficient AID mutants (R24W and V18S/R19V). The SV40 NLS fusion allowed efficient nuclear accumulation of the otherwise import-deficient AID mutants (Fig. 7A). However, these AID mutants with the heterologous SV40 NLS failed to trigger immunoglobulin class switching in AID-deficient primary mouse B cells (Fig. 7, B and C). Previous experiments have shown that the AID[V18S/R19V] mutant (although not the R24W mutant) retains DNA deaminase catalytic activity (16), suggesting that the inability of the AID[V18S/R19V]-HA-SV40 mutant to trigger class switching could be related to a defect in correct subnuclear targeting. To test whether a distinct CTNNBL1-binding NLS could rescue class switching, we fused the CDC5L NLS3 to the C termini of import-deficient AID mutants. However, although this NLS confers nuclear accumulation on the AID[V18S/R19V]-HA mutant, it does not rescue class switching (Fig. 7), leaving open the possibility that either there is something special in the AID NLS that cannot be replaced by the CDC5L NLS3 or that the V18S/R19V mutations have interfered with some critical interaction/function of AID, apart from nuclear import.
FIGURE 7.
Heterologous NLSs rescue nuclear import, but not class-switching activity, of import-deficient AID mutants. A, subcellular distribution of AID-HA-NLS fusion proteins transiently expressed in 293T cells. Cells were transfected with constructs expressing AID together with GFP (separately translated from the same transcript using an IRES), fixed in paraformaldehyde, stained with anti-HA antibody, and visualized by confocal microscopy (SV40, SV40 NLS; C5L3, CDC5L NLS3). GFP fluorescence identifies transfected cells. B, antibody class switching. B lymphocytes from AID−/− mice were activated with LPS and IL4, transduced with AID-IRES-GFP expressing retroviral vectors, and assayed for class switching from IgM to IgG1. The plots show, for each AID construct, the proportion of transduced (i.e. GFP+) B cells that have switched to IgG1 expressed as a percentage of the proportion obtained using wild-type (WT) AID. Error bars indicate the S.D. obtained in three or four experiments. C, Western blot showing the expression of AID in the transduced B lymphocytes.
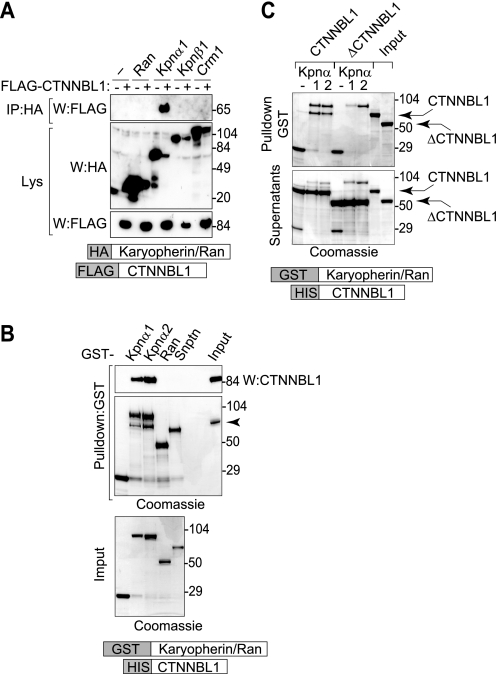
CTNNBL1 Binds Karyopherin α, but Not β, Using an N-terminal NLS
If CTNNBL1 were functioning as a karyopherin α, οne might expect CTNNBL1, like karyopherin αs, to also bind to karyopherin β1. However, no interaction between purified CTNNBL1 and purified karyopherin β1 (or Ran) could be detected. Instead, an interaction between CTNNBL1 and karyopherin αs was readily observed both in pulldowns from HEK293T cells and between recombinant proteins in vitro (Fig. 8, A and B). This interaction with karyopherin αs is consistent with the identification of karyopherin α2 and 6 in the proteomic analysis of FLAG-CTNNBL1 immunoprecipitates (Fig. 2).
FIGURE 8.
CTNNBL1 binds karyopherin α (but not β1) through its N-terminal region. A, immunoprecipitation of HA-karyopherin α1 (but not HA-tagged karyopherin β1, Crm1, or Ran) bringing down FLAG-CTNNBL1 from co-transfected 293T cell lysates. IP, samples subjected to immunoprecipitation on anti-HA-agarose were analyzed by Western blotting (W) with anti-FLAG antibody. Lys, Western blots of samples of total cell lysates were probed with anti-HA or anti-FLAG antibodies as a control. B, in vitro interaction between purified His-CTNNBL1 and purified GST-karyopherin αs. GST-tagged nuclear transport factors expressed in E. coli and bound onto glutathione-Sepharose were incubated with recombinant His-CTNNBL1. Following SDS-PAGE, bound CTNNBL1 was detected by Western blotting with anti-CTNNBL1 mAb (top panel) whereas total Sepharose-bound proteins were identified by Coomassie staining (middle panel). SDS-PAGE analysis of total input GST-fusion protein is shown in the bottom panel. The right lane in the top two panels shows the migration of purified His-CTNNBL1. Snptn, snurportin. C, the CTNNBL1 N-terminal region is required for interaction with karyopherin αs. GST-karyopherin α1/α2 bound to glutathione-Sepharose was incubated with purified His-CTNNBL1 or His-ΔCTNNBL1 (lacking the N-terminal 76 amino acids). Sepharose-bound proteins (top panel) as well as samples of the supernatants from the incubation (lower panel) were separated on SDS-PAGE and stained with Coomassie; aliquots of the input His-CTNNBL1 and His-ΔCTNNBL1 are shown in the two right lanes.
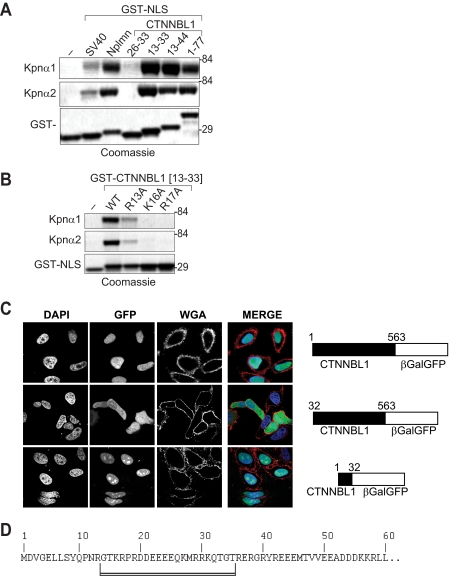
Previous results have shown that deletion of the CTNNBL1 N-terminal region causes substantial cytoplasmic accumulation (3), suggesting that this region contains a functional NLS. Indeed, immunofluorescence analysis reveals that CTNNBL1 residues 1–32 suffice to confer nuclear localization on a β-galactosidase-GFP chimeric reporter (Fig. 9C); pulldown experiments (Figs. 8C and 9A) also show that the CTNNBL1 N-terminal region mediates interaction with karyopherin α. Interestingly, although the program cNLS-Mapper suggests that residues 26–31 (KMRRK) would constitute an effective NLS, mutagenesis studies (Fig. 9, A and B) reveal that binding to karyopherin α also requires residues 16 and 17 and to a lesser extent 13 of CTNNBL1, pointing to a bipartite NLS (Fig. 9D).
FIGURE 9.
Bipartite NLS in the CTNNBL1 N-terminal region binds karyopherin α. A, direct interaction between purified karyopherin α1/2 and GST fusion proteins that contained either the indicated segments of the CTNNBL1 N-terminal region or the SV40 or nucleoplasmin (Nplmn) NLSs assessed by incubating the GST-NLS fusion proteins that had been bound to glutathione-Sepharose with purified His-tagged karyopherin. The karyopherin α and GST fusion proteins bound to the Sepharose were visualized with Coomassie following SDS-PAGE. B, mutational analysis of the interaction between karyopherin α1/2 and a GST fusion containing CTNNBL1 residues 13–33, with mutations at Arg13, Lys16, or Lys17 as indicated. C, residues 1–32 of CTNNBL1 confer nuclear localization on a CTNNBL1-β-galactosidase-GFP chimera. Nuclei were counterstained with DAPI and cell membranes with wheat germ agglutinin (WGA)-Alexa Fluor 568. D, sequence of the N-terminal region of CTNNBL1 with the bipartite NLS underlined.
DISCUSSION
The results reveal CTNNBL1 to be a novel, selective NLS-binding protein with architectural homology but no evident sequence homology to karyopherin αs. CTNNBL1 resembles karyopherin αs in using tandem ARM motifs to bind heterologous NLSs (Figs. 5 and 7). It differs in respect of the fact that (i) the CTNNBL1 N-terminal region binds karyopherin αs rather than karyopherin β1; (ii) the ARM domains of CTNNBL1 and karyopherin αs show predicted architectural homology but no primary sequence homology.
Residues critical to NLS binding by karyopherin αs are not identifiable in CTNNBL1. Although the ARM motifs of karyopherin αs contain a conserved WXXXN motif in which the tryptophan forms hydrophobic surface interactions with the long aliphatic portions of the basic side chains of the NLS cargo and the asparagine forms H bonds with the NLS backbone (18, 19), no such WXXXN motif is found within CTNNBL1. Indeed, the CTNNBL1 ARM domain contains only a single tryptophan (predicted motif 4), and this is not located in a position within CTNNBL1 ARM4 analogous to that of the WXXXN tryptophan in the karyopherin α ARM repeats. These structural differences correlate with a substantial difference in NLS binding specificity.
There are also striking parallels as well as differences when it comes to comparing the N-terminal regions (as opposed to the ARM domains) of CTNNBL1 and karyopherin αs. Karyopherin αs contain an α-helical N-terminal domain that mediates binding to the HEAT repeats of karyopherin-β and which also binds back to the karyopherin α ARM domain. CTNNBL1 also contains an evolutionarily conserved N-terminal region, but one that is distinct from that of the karyopherin αs. Rather than binding karyopherin β, this N-terminal region contains a conserved bipartite NLS that binds karyopherin αs. Thus, the CTNNBL1 interaction with the karyopherin system is the same as that of other NLS-containing import cargoes.
What insights do these results this give to CTNNBL1 function? CTNNBL1 could be an adapter for nuclear import, binding selective cargo in the cytoplasm and mediating its nuclear import through interaction with karyopherin αs and thence karyopherin β and Ran. If CTNNBL1 does fulfill such a role, it would raise questions about how cargo release and CTNNBL1 shuttling are themselves regulated. Although we observed no difference in the subcellular distribution of CDC5L NLS-tagged fusion proteins in CTNNBL1+/+ and CTNNBL1−/− cells, AID nuclear accumulation does appear to be somewhat diminished in the absence of CTNNBL1, supporting a partially redundant role for CTNNBL1 in the nuclear import or accumulation of some cargoes.
The alternative possibility is that CTNNBL1 may function solely within the nucleus. Indeed, even the classical karyopherins have roles in processes other than nuclear import/export (20, 21). Thus, CTNNBL1 might simply be an evolutionarily deviant karyopherin family member that uses the flexible ARM domain as a protein:protein recognition motif. The NLS in the CTNNBL1 N-terminal domain may therefore function merely to facilitate the import of CTNNBL1 into the nucleus.
The restricted nature of CTNNBL1 interactors identified is intriguing: the antibody mutator AID and two RNA splicing factors, CDC5L and Prp31. The snRNPs are assembled in the cytoplasm but then pass to the nucleus where they move to Cajal bodies and associate with other factors to form splicing-competent particles (22). It is likely within the Cajal body that Prp31 acts to assist formation of the U4/U6.U5 tri-snRNP particle (14, 23). CDC5L forms part of the Prp19-containing complex which ubiquitylates Prp3, thereby stabilizing the U4/U6.U5 tri-snRNP (24). Conceivably, CTNNBL1 might play a role in stabilizing interactions between snRNP components within the nucleus. The interaction with AID might similarly reflect a role in AID localization or association with spliceosomal components.
Testing these ideas will require genetic approaches. However, although CTNNBL1 deficiency yields early embryonic lethality in mice suggesting a critical role in development (A. Chandra; M. S. Neuberger and C. Rada, unpublished data), no robust phenotype (apart from a reduction in the frequency of immunoglobulin gene conversion) has been identified following CTNNBL1 gene ablation in immortalized vertebrate B cell lines or in Schizosaccharomyces pombe (3, 25).5 This is despite the fact that CTNNBL1 splice factor partners are essential genes. It might well be that even if CTNNBL1 is implicated in the trafficking or association/localization of selected RNA splicing factors, other proteins (e.g. karyopherins) might nevertheless be able to substitute for CTNNBL1 in its absence. Indeed, many organisms have multiple karyopherin α genes with different paralogs being essential in different tissues and with their deficiencies therefore leading to embryonic lethality at different stages (26, 27). A likely way forward may therefore be to identify other gene deficiencies that cause CTNNBL1, loss to yield a robust phenotype, either using a candidate approach or by a screen for synthetic lethality in fission yeast.
Supplementary Material
Acknowledgments
We thank Drs. Chris Johnson and Nicolas Basse for advice on isothermal calorimetry; Dr. Sew Peak Chew for assistance with mass spectrometry; David Owen for amino acid analysis; Maria Daly for FACS sorting; Drs. Alan Warren, Febe van Maldegem, and Anita Chandra for helpful suggestions; and Drs. Wilkie, Hunt, and Randow for the gift of plasmids.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and an additional reference.
F. van Maldegem, K. Ganesh, C. Rada, and M. S. Neuberger, unpublished data.
- CTNNBL1
- catenin-β-like 1
- ARM
- armadillo
- IRES
- internal ribosome entry site
- NLS
- nuclear localization sequence.
REFERENCES
- 1. Jabbour L., Welter J. F., Kollar J., Hering T. M. (2003) Genomics 81, 292–303 [DOI] [PubMed] [Google Scholar]
- 2. Makarova O. V., Makarov E. M., Urlaub H., Will C. L., Gentzel M., Wilm M., Lührmann R. (2004) EMBO J. 23, 2381–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Conticello S. G., Ganesh K., Xue K., Lu M., Rada C., Neuberger M. S. (2008) Mol. Cell 31, 474–484 [DOI] [PubMed] [Google Scholar]
- 4. Goldfarb D. S., Corbett A. H., Mason D. A., Harreman M. T., Adam S. A. (2004) Trends Cell Biol. 14, 505–514 [DOI] [PubMed] [Google Scholar]
- 5. Chook Y. M., Blobel G. (2001) Curr. Opin. Struct. Biol. 11, 703–715 [DOI] [PubMed] [Google Scholar]
- 6. Deery E. C., Vithana E. N., Newbold R. J., Gallon V. A., Bhattacharya S. S., Warren M. J., Hunt D. M., Wilkie S. E. (2002) Hum. Mol. Genet. 11, 3209–3219 [DOI] [PubMed] [Google Scholar]
- 7. Jensen O. N., Podtelejnikov A. V., Mann M. (1997) Anal. Chem. 69, 4741–4750 [DOI] [PubMed] [Google Scholar]
- 8. Pappin D. J., Hojrup P., Bleasby A. J. (1993) Curr. Biol. 3, 327–332 [DOI] [PubMed] [Google Scholar]
- 9. Randow F., Sale J. E. (2006) Subcell. Biochem. 40, 383–386 [DOI] [PubMed] [Google Scholar]
- 10. Tsai W. Y., Chow Y. T., Chen H. R., Huang K. T., Hong R. I., Jan S. P., Kuo N. Y., Tsao T. Y., Chen C. H., Cheng S. C. (1999) J. Biol. Chem. 274, 9455–9462 [DOI] [PubMed] [Google Scholar]
- 11. Liu L., Gräub R., Hlaing M., Epting C. L., Turck C. W., Xu X. Q., Zhang L., Bernstein H. S. (2003) Cell. Biochem. Biophys. 39, 119–132 [DOI] [PubMed] [Google Scholar]
- 12. Grote M., Wolf E., Will C. L., Lemm I., Agafonov D. E., Schomburg A., Fischle W., Urlaub H., Lührmann R. (2010) Mol. Cell. Biol. 30, 2105–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bernstein H. S., Coughlin S. R. (1997) J. Biol. Chem. 272, 5833–5837 [DOI] [PubMed] [Google Scholar]
- 14. Makarova O. V., Makarov E. M., Liu S., Vornlocher H. P., Lührmann R. (2002) EMBO J. 21, 1148–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shinkura R., Ito S., Begum N. A., Nagaoka H., Muramatsu M., Kinoshita K., Sakakibara Y., Hijikata H., Honjo T. (2004) Nat. Immunol. 5, 707–712 [DOI] [PubMed] [Google Scholar]
- 16. Patenaude A. M., Orthwein A., Hu Y., Campo V. A., Kavil B., Buschiazzo A., Di Noia J. M. (2009) Nat. Struct. Mol. Biol. 16, 517–527 [DOI] [PubMed] [Google Scholar]
- 17. Cutress M. L., Whitaker H. C., Mills I. G., Stewart M., Neal D. E. (2008) J. Cell Sci. 121, 957–968 [DOI] [PubMed] [Google Scholar]
- 18. Conti E., Uy M., Leighton L., Blobel G., Kuriyan J. (1998) Cell 94, 193–204 [DOI] [PubMed] [Google Scholar]
- 19. Fontes M. R., Teh T., Jans D., Brinkworth R. I., Kobe B. (2003) J. Biol. Chem. 278, 27981–27987 [DOI] [PubMed] [Google Scholar]
- 20. Mosammaparast N., Pemberton L. F. (2004) Trends Cell Biol. 14, 547–556 [DOI] [PubMed] [Google Scholar]
- 21. Harel A., Forbes D. J. (2004) Mol. Cell 16, 319–330 [DOI] [PubMed] [Google Scholar]
- 22. Stanek D., Neugebauer K. M. (2006) Chromosoma 115, 343–354 [DOI] [PubMed] [Google Scholar]
- 23. Schaffert N., Hossbach M., Heintzmann R., Achsel T., Lührmann R. (2004) EMBO J. 23, 3000–3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Song E. J., Werner S. L., Neubauer J., Stegmeier F., Aspden J., Rio D., Harper J. W., Elledge S. J., Kirschner M. W., Rape M. (2010) Genes Dev. 24, 1434–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Han L., Masani S., Yu K. (2010) J. Immunol. 185, 1379–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mason D. A., Fleming R. J., Goldfarb D. S. (2002) Genetics 161, 157–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yasuhara N., Shibazaki N., Tanaka S., Nagai M., Kamikawa Y., Oe S., Asally M., Kamachi Y., Kondoh H., Yoneda Y. (2007) Nat. Cell Biol. 9, 72–79 [DOI] [PubMed] [Google Scholar]
- 28. Aoufouchi S., Faili A., Zober C., D'Orlando O., Weller S., Weill J. C., Reynaud C. A. (2008) J. Exp. Med. 205, 1357–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Geisberger R., Rada C., Neuberger M. S. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 6736–6741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ordinario E. C., Yabuki M., Larson R. P., Maizels N. (2009) J. Immunol. 183, 4545–4553 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.