Abstract
Autosomal recessive and autosomal dominant forms of Weill-Marchesani syndrome, an inherited connective tissue disorder, are caused by mutations in ADAMTS10 (encoding a secreted metalloprotease) and FBN1 (encoding fibrillin-1, which forms tissue microfibrils), respectively, yet they are clinically indistinguishable. This genetic connection prompted investigation of a potential functional relationship between ADAMTS10 and fibrillin-1. Specifically, fibrillin-1 was investigated as a potential ADAMTS10 binding partner and substrate, and the role of ADAMTS10 in influencing microfibril biogenesis was addressed. Using ligand affinity blotting and surface plasmon resonance, recombinant ADAMTS10 was found to bind to fibrillin-1 with a high degree of specificity and with high affinity. Two sites of ADAMTS10 binding to fibrillin-1 were identified, one toward the N terminus and another in the C-terminal half of fibrillin-1. Confocal microscopy and immunoelectron microscopy localized ADAMTS10 to fibrillin-1-containing microfibrils in human tissues. Furin-activated ADAMTS10 could cleave fibrillin-1, but innate resistance of ADAMTS10 zymogen to propeptide excision by furin was observed, suggesting that, unless activated, ADAMTS10 is an inefficient fibrillinase. To investigate the role of ADAMTS10 in microfibril biogenesis, fetal bovine nuchal ligament cells were cultured in the presence or absence of ADAMTS10. Exogenously added ADAMTS10 led to accelerated fibrillin-1 microfibril biogenesis. Conversely, fibroblasts obtained from a Weill-Marchesani syndrome patient with ADAMTS10 mutations deposited fibrillin-1 microfibrils sparsely compared with unaffected control cells. Taken together, these findings suggest that ADAMTS10 participates in microfibril biogenesis rather than in fibrillin-1 turnover.
Keywords: ADAM ADAMTS, Connective Tissue, Convertases, Electron Microscopy (EM), Fibrillin
Introduction
Inherited connective tissue disorders that result from mutations of genes encoding extracellular matrix (ECM)3 components or their modifying enzymes are potentially insightful in identifying structural and regulatory roles of ECM constituents or their supramolecular assemblies. A classic example of such an ECM component is fibrillin-1, a large, secreted glycoprotein, which is a major component of 8–12-nm-diameter tissue microfibrils (1). Bundles of microfibrils, containing fibrillin-1, fibrillin-2, and fibrillin-3 (in humans), are ubiquitous in the body but are especially abundant in tissues containing elastin (2). The structural role of microfibrils includes maintenance of the ocular lens in its appropriate position in the eye via the zonule of Zinn (the suspensory ligament of the lens), which comprises primarily fibrillin-1 microfibrils (2, 3). Inadequacy of the zonule or traumatic breakage of zonule fibers can result in subluxation or dislocation of the lens (ectopia lentis).
Dominantly inherited FBN1 mutations lead to diverse connective tissue anomalies, the most common of which is Marfan syndrome (MFS) (4). Major features of MFS include musculoskeletal anomalies, aortic aneurysms, and ectopia lentis. Several features of MFS appear to be a consequence of FBN1 haploinsufficiency (5, 6), although interference of mutant fibrillin-1 with microfibril assembly may also occur. Recent evidence strongly suggests that aortic dissection, pulmonary anomalies, excess skeletal growth, and muscular weakness in MFS may be a consequence of TGFβ dysregulation (7, 8). One mechanism potentially underlying this role is that fibrillin-1 interacts with latent TGFβ-binding protein-1 (9), which mediates efficient tissue sequestration and activation of TGFβ (10). Thus, microfibrils may act as a repository for TGFβ. Fibrillin-1 also binds directly to bone morphogenetic proteins (11).
FBN1 mutations were identified in autosomal dominant Weill-Marchesani syndrome (WMS) (6, 12). WMS is a rare eye and connective tissue disorder characterized by anomalies of the anterior chamber of the eye, including ectopia lentis, short stature, short distal extremities, thick skin, and stiff joints. Autosomal recessive WMS is caused by ADAMTS10 mutations (13–15). WMS resulting from either FBN1 or ADAMTS10 mutations is clinically indistinguishable (16), which suggests that ADAMTS10 and fibrillin-1 may interact and/or act cooperatively in a common pathway. Several clinical manifestations of WMS, such as short stature, thick skin, stiff joints, and short hands and feet, contrast with the presentation of MFS, although ectopia lentis is present in both disorders (16). A typical clinical presentation of WMS consists of repeated attacks of acute congestive glaucoma starting in early childhood. In addition to closed angle glaucoma resulting from a shallow anterior chamber in WMS, the small, spherical lens is untethered by zonules and is thus susceptible to anterior dislocation through the pupil where it blocks the flow of aqueous humor. Despite surgical intervention, vision is usually severely compromised in affected individuals. The occurrence of ectopia lentis in WMS together with dysgenesis of the zonule raises the possibility that ADAMTS10 may be involved in zonule formation or maintenance and perhaps specifically in biogenesis of fibrillin-1 microfibrils. Widespread expression of Adamts10 mRNA in mesenchymal tissues during mouse embryogenesis was shown using in situ hybridization, and continuing expression was noted in adult tissues by Northern analysis (17). Fibrillin-1 is also widely expressed in tissues.
ADAMTS10 belongs to a superfamily of secreted proteins containing both ADAMTS proteases and ADAMTS-like proteins (18). Some ADAMTS proteases, such as procollagen aminopropeptidases (ADAMTS2, ADAMTS3, and ADAMTS14) and ADAMTS13, which is required for maturation of functional von Willebrand factor, are highly specialized (18). A major cluster of ADAMTS proteases (e.g. ADAMTS1, ADAMTS4, ADAMTS5, ADAMTS9, and ADAMTS20) has the ability to cleave large aggregating proteoglycans, which is not shared by ADAMTS10 (17). Instead, ADAMTS10 together with its homolog, ADAMTS6, represents a distinct gene duplication event in mammals, suggestive of a specialized function. The genetics of WMS (17) suggests a function that could be related to fibrillin-1. Therefore, in the present work, we investigated interactions between ADAMTS10 and fibrillin-1. The findings identify ADAMTS10 as a novel potential participant in the biology of fibrillin microfibrils and provide new insights on WMS.
EXPERIMENTAL PROCEDURES
Reagents used were from Sigma-Aldrich unless otherwise specified.
Expression Plasmids, Site-directed Mutagenesis, and Transfection
The plasmid for expression of full-length human ADAMTS10 with C-terminal tandem myc + His6 tags was described previously (17). Site-directed mutagenesis of this plasmid was undertaken using the QuikChange site-directed mutagenesis kit (Stratagene, Thousand Oaks, CA) to generate the following ADAMTS10 mutants: ADAMTS10 G230R,L231R (designated ADAMTS10-RRKR), ADAMTS10 E393A (designated ADAMTS10-EA), and ADAMTS10-RRKR E393A (designated ADAMTS10-RRKR-EA). These mutants were transfected into HEK293F cells using FuGENE 6 (Roche Applied Science) followed by selection with G418 (1 mg/ml) for isolation of stably transfected clones. Protein expression was confirmed by Western blot analysis of conditioned medium using anti-myc monoclonal antibody 9E10 (Invitrogen). To determine the role of furin in processing ADAMTS10 propeptide, full-length ADAMTS10 was transfected into furin-deficient CHO-RPE.40 cells (19) and compared with transfection into CHO-K1 cells as a control.
Recombinant Proteins and Antibodies
Wild-type ADAMTS10 and ADAMTS10-RRKR were purified from the conditioned medium of stably transfected HEK293F cells using Ni2+-Sepharose affinity chromatography essentially as described previously (14) and analyzed by Western blotting and Coomassie staining under reducing and non-reducing conditions. Recombinant human fibrillin-1 peptides were described previously (9, 11). hFib1–49 is a new, tandem myc- and His6-tagged human fibrillin-1 peptide with essentially similar domain structure as rF11. It was generated by cloning a SpeI-XhoI fragment from a full-length fibrillin-1 plasmid (provided by Dr. Penny Handford, Oxford University) into pcDNA3.1 myc-His A+ (Stratagene). Anti-ADAMTS10 monoclonal antibodies (mAbs) were generated to the peptide RGKFYKWKTYRG (a sequence within the cysteine-rich domain), which was conjugated to bovine serum albumin for hybridoma production in mice. Hybridoma supernatant and rabbit sera were screened using ELISA against the respective immobilized peptides. Of the various hybridomas isolated, clone 287-3E2 (subclass γ1/κ) was used for obtaining monoclonal antibody for subsequent applications. ADAMTS10 polyclonal antibody was generated by injecting rabbits with the peptide CSEFDSIPFRGKFYKWKTYR, which was conjugated to keyhole limpet hemocyanin via the N-terminal Cys (Alpha Diagnostic International, San Antonio, TX). Immune sera were tested by Western blotting against ADAMTS10-conditioned medium and were affinity-purified against the immobilized peptide immunogen. The characterization of an affinity-purified rabbit polyclonal antibody (pAb 10041) to the human ADAMTS10 propeptide (residues 153–182) was described previously (14). Antibody 287-3E2 was tested for application in immunofluorescence microscopy by staining HEK293F cells that were stably transfected or transiently transfected with a myc-tagged ADAMTS10 expression plasmid or the empty vector. The cells were fixed and permeabilized prior to immunofluorescence microscopy essentially as described previously (20).
Analytical Gel Filtration Chromatography of ADAMTS10-RRKR
290 ml of conditioned 293 serum-free medium (SFM) II were collected from HEK293F cells stably transfected with ADAMTS10-RRKR and concentrated to 1.2 ml using Amicon Ultra centrifugal filter devices with a 10-kDa-cutoff membrane (Millipore). Three gel filtration molecular mass standards, apoferritin (443 kDa), alcohol dehydrogenase (150 kDa), and carbonic anhydrase (29 kDa), were added to the concentrated ADAMTS10-RRKR sample at a final concentration of 0.5, 1, and 0.5 mg/ml, respectively. 1.0 ml of the sample was loaded to a Superose 6 10/30 column (Amersham Biosciences), which was connected to a fast protein liquid chromatography system (BioLogic HR chromatography system, Bio-Rad). The sample was eluted with PBS at 0.3 ml/min, and 1-ml fractions were collected. The collected fractions were analyzed by Western blot using anti-myc and affinity-purified anti-ADAMTS10 propeptide pAb 10041 antibody under reducing conditions.
Electrophoresis, Western Blot, and Ligand Affinity Blot Analysis
400 ng each of purified ADAMTS10 and other ADAMTS proteins (human ADAMTS1 was kindly provided by Dr. Luisa Iruela-Arispe, bovine ADAMTS2 was provided by Dr. Alain Colige, human ADAMTS4 and ADAMTS5 were provided by Wyeth Pharmaceuticals, and human ADAMTS13 was provided by Drs. Elaine Majerus and J. Evan Sadler) were mixed with an equal volume of 2× sample loading buffer and electrophoresed under non-reducing conditions on a 6% Tris-HCl gel. Proteins were then transferred to PVDF membrane and blocked for 1 h at room temperature in 5% (w/v) milk in Tris-buffered saline (TBS; 10 mm Tris-HCl, pH 7.2, 0.15 m NaCl). Blots were then incubated overnight with fibrillin-1 peptide rF6 or rF11 diluted in 2.5% milk in TBS at a concentration of 50 μg/ml. The following day, blots were washed three times (5 min each) with TBST (TBS containing 0.05% Tween 20) prior to incubation with mAb 69 (to detect rF6) or mAb 201 (to detect rF11) diluted in 2.5% (w/v) milk in TBS for 3 h at room temperature. Blots were washed with TBST as before and incubated with goat anti-mouse HRP diluted in 2.5% (w/v) milk in TBS for 1 h at room temperature. Washing with TBST was done before the blots were developed using the enhanced chemiluminescence Western blotting detection reagent (GE Healthcare). Signal was detected by exposure to radiography film (Denville Scientific, Metuchen, NJ).
Surface Plasmon Resonance (SPR) Analysis
Purified ADAMTS10 in 10 mm MES buffer, pH 6.0 was immobilized for SPR analysis on a BIAcore CM5 sensor chip (research grade) with the amine coupling kit following the manufacturer's instructions (chip and kit were from GE Healthcare). 1506 resonance units were coupled to the chip for analysis in a BIAcore 3000 instrument (GE Healthcare). The kinetic analysis was performed at 25 °C in 10 mm Hepes buffer, pH 7.4 with 0.15 m NaCl, 2 mm CaCl2, and 0.005% (v/v) surfactant P20 at a flow rate of 20 μl/min. The purified analytes were diluted in the above buffer at different concentrations and injected through an uncoupled control flow cell as well as the flow cell coupled to ADAMTS10. The sample injection time was 3–6 min and was followed by a pause of 10 min for dissociation. 1 m NaCl with 2–10% (v/v) acetonitrile was used for regeneration after each injection at a flow rate of 50 μl/min for 30–60 s. The stabilization time following each regeneration was 2 min. All the data were corrected with reference to the background binding in the control flow cell. The kinetic constants were calculated assuming a 1:1 (Langmuir) binding model using the BIAevaluation software (version 4.0.1; GE Healthcare). In reciprocal SPR experiments, ADAMTS10 was used as the analyte with rFib1–49 as the ligand bound to the chip.
Fibrillin-1 Cleavage Analysis
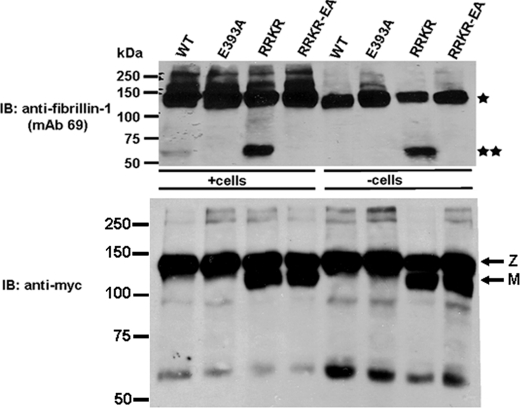
HEK293F clones expressing ADAMTS10, ADAMTS10-EA, ADAMTS10-RRKR, and ADAMTS10-RRKR-EA were plated at equal density in a 6-well plate. When cells reached confluence (2–3 days later), the medium was collected, and cells were washed with serum-free DMEM and detached mechanically by streaming 1 ml of serum-free DMEM across the monolayer. Cells were placed in separate 1.5-ml tubes and centrifuged for 5 min at 1500 rpm. The supernatant was removed, and the cells were washed five times with 1 ml of serum-free DMEM and resuspended in 90 μl of 293 SFM II and 10 μl of 10× proteinase inhibitor mixture (without EDTA; Roche Diagnostics). rF6 (500 ng) was added to the cell suspension, and the tubes were incubated at 37 °C for 4 h (designated as +cells in Fig. 6). The supernatant was collected and electrophoresed under reducing conditions on a 6% Tris-HCl gel. Western blotting was performed using the fibrillin-1 antibody mAb 69. These peptides were also incubated with the conditioned cell-free medium of transfected cells (designated as −cells in Fig. 6).
FIGURE 6.
Furin-processed ADAMTS10 can cleave fibrillin-1. ADAMTS10 proteolytic activity toward fibrillin-1 was determined in HEK293F cells stably transfected with wild-type ADAMTS10 (WT) or the indicated mutants. The transfected cells (+cells) or their conditioned medium (−cells) as indicated was incubated with peptide rF6 (asterisk). ADAMTS10 (WT) cleaves rF6 to generate a 60-kDa peptide (top panel, double asterisk), but this cleavage is much more robust with ADAMTS10-RRKR. When rF6 digests were done using ADAMTS10-conditioned cell-free medium, cleavage by wild-type ADAMTS10 was undetectable. Mutation of the ADAMTS10 catalytic Glu393 residue (to Ala) in each construct abrogated cleavage and is indicative of specific cleavage by ADAMTS10 in this experiment. The lower panel shows the ADAMTS10 molecular species detected by Western blotting with anti-myc. IB, immunoblot; Z, zymogen; M, mature form.
Tissue Specimens and Confocal Microscopy
Human skin was obtained from a 1-year-old patient undergoing surgery with Institutional Review Board approval (Oregon Health Sciences Center), snap frozen in dry ice-chilled hexanes, stored at −80 °C until use, and then embedded in Tissue-Tek OCTTM (Thermofischer Scientific, Waltham, MA) for sectioning. 25-μm-thick sections were obtained on a cryostat, collected on glass microscope slides, then dried overnight, and fixed for 20 min at −20 °C in 100% acetone. After rinsing three times in PBS, sections were incubated with ADAMTS10 monoclonal antibody 287-3E2 (diluted 1:50) and fibrillin-1 polyclonal antibody 9543 (diluted 1:50) for 2 h at room temperature and then rinsed thrice in PBS prior to incubation with Alexa Fluor 488-conjugated goat anti-mouse antibody and Alexa Fluor 633-conjugated goat anti-rabbit for 1 h (antibodies were from Invitrogen). Stained sections of skin were mounted in 90% glycerol and observed using a 63× water-corrected Plan-Apochromat lens with a Leica TCS SP2 confocal microscope (Leica Microsystems, Wetzlar, Germany) operated with simultaneous excitation at 488 and 633 nm and with collection windows adjusted to 494–529 (fibrillin) and 639–739 nm (ADAMTS10). 39 exposures were collected at “Z” heights separated by 222 nm to produce the Z series (shown in supplemental Video File 1). As negative controls, skin sections were incubated with the respective secondary antibodies alone.
Transmission Immunoelectron Microscopy
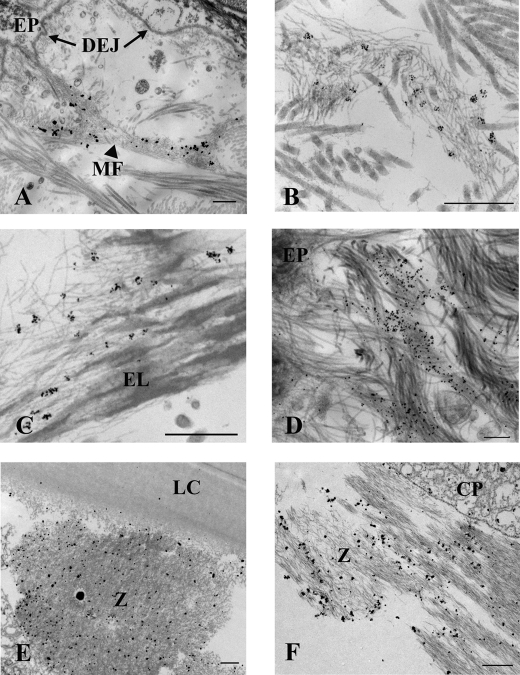
En bloc immunohistochemistry was performed as described previously (21) with some modification. Skin from the same 1-year-old individual used for confocal evaluation and skin from a 2.5-year-old patient were cut into ∼1 × 2-mm pieces (including the epithelium) and immersed at 4 °C overnight in primary antibody (monoclonal antibody 287-3E2 to ADAMTS10) diluted 1:5 in SFM, then rinsed in medium, and incubated further in goat anti-rabbit 1-nm colloidal gold conjugate overnight at 4 °C (Electron Microscopy Sciences, Hatfield, PA). The samples were rinsed in SFM and “gold-enhanced” in GoldEnhance EM (Nanoprobes, Stony Brook, NY) to better visualize the 1-nm gold particles. Gold enhancement proceeded for 5 min for the 1-year-old skin samples imaged in Fig. 4, A and D, and for the eye tissue imaged in Fig. 4, E and F, and for 4.5 min for the 2.5-year-old skin samples imaged in Fig. 4, B and C. Additional skin samples from the same 2.5-year-old patient were treated identically except a 5-nm gold secondary conjugate was substituted for the 1-nm conjugate, and the enhancement protocol was eliminated (because 5-nm gold particles are easily imaged in the transmission electron microscope). Immunoelectron microscopy of fibrillin-1 was done using polyclonal antibody 9543, which was raised against a peptide representing the N-terminal half of fibrillin-1 (rF11) (22) and 10-nm colloidal gold conjugate. The samples were then rinsed in SFM; fixed in 1.5% glutaraldehyde, 1.5% paraformaldehyde containing 0.05% tannic acid; osmicated in 1% OsO4, then dehydrated, and embedded in Spurr's epoxy. Intact human lens and ciliary body obtained from a 35-year-old individual (see above) were immunolabeled using an identical protocol as for the 1-year-old skin. Ultrathin sections were observed using an FEI Tecnai G2 microscope operated at 120 kV.
FIGURE 4.
Immunoelectron microscopy illustrates that ADAMTS10 is specifically bound to tissue microfibrils in skin and zonules. ADAMTS10 was localized en bloc using mAb 287-3E2 followed by 1-nm gold secondary conjugate and then gold-enhanced to allow visualization at low magnification. Labeling of microfibril bundles (MF) is most intense close to the dermal-epidermal junction; however, labeling is absent immediately adjacent to the epithelium (EP) and the basement membrane at the dermal-epidermal junction (DEJ; A, arrows). Immunogold particles seen at higher magnification following slightly less time in gold enhancement solution demonstrate that ADAMTS10 localizes to microfibrils in small clumps represented by a cluster of gold particles (B; image taken near the dermal-epidermal junction) also seen among elastin-associated microfibrils in the shallow reticular dermis (C). An image collected at 0° tilt through an exceptionally thick section (300 nm; D) of immunolabeled skin shows several labeled microfibril bundles intersecting the lamina densa of the dermal-epidermal junction best appreciated in the aligned tilt series (supplemental Video File 2). Ciliary zonules (Z) intersecting both the lens capsule (LC; E) and ciliary process (CP; F) are well labeled with antibody to ADAMTS10. Scale bars for A–D, 500 nm; scale bars for E and F, 1 μm.
Analysis of Microfibril Biogenesis
Fetal bovine nuchal ligament (fBNL) cells (<10 passages) were seeded on cover slips and cultured with DMEM-F-12 medium supplemented with 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin. At 80% confluence, medium conditioned by HEK293F cells stably expressing wild-type ADAMTS10 or stably transfected with the empty pcDNA3.1 expression vector (as control) was transferred to fBNL cultures. The pH of the conditioned medium (DMEM-F-12 medium supplemented with 10% FBS) was adjusted to 7.0, and it was supplemented with d-glucose prior to adding to fBNL cells at a ratio of ⅔ conditioned medium and ⅓ fresh medium. The fBNL cells were cultured in this medium for 3 or 5 days with new fresh/conditioned medium mixture provided to cells every 48 h. Cells were fixed for 7 min with ice-cold methanol, blocked with 5% normal goat serum in PBS, and incubated with anti-fibrillin-1 monoclonal antibody (MAB1919, Millipore Corp., Billerica, MA) at 4 °C overnight to detect fibrillin-1 made by the fBNL cells. Goat anti-mouse Alexa Fluor 488 (Invitrogen) was used as a secondary antibody at 6 μg/ml. Nuclei were stained with DAPI.
Dermal fibroblasts were obtained from a skin biopsy of an 84-year-old case of recessive WMS in whom ADAMTS10 mutations were recently identified (14) as well as from unaffected individuals (ages, 97 and 74 years). Cells (passages 2–4) from these donors were cultured to equivalent density for 5 days and stained for the presence of fibrillin-1 microfibrils using polyclonal antibody 9543 and monoclonal antibody 69 with essentially similar results obtained using both antibodies.
Real Time Quantitative PCR
fBNL cells cultured for 3 days in the presence of vector-conditioned medium or ADAMTS10-conditioned medium as described above were trypsinized and washed with PBS. Subsequently, RNA was extracted using the RNeasy kit (Qiagen, Valencia, CA). Using random primers, 450 ng of RNA was reversed transcribed (Superscript III kit, Invitrogen), and real time PCR was performed using the Power SYBR Green PCR Mastermix and the 7500 System (Applied Biosystems, Carlsbad, CA). After verifying that the PCR efficiencies were 90–100%, relative quantification was performed using GAPDH RNA as an endogenous control. The following primers were used: bovine FBN1: forward primer, GTGAAAACACCAAAGGCTCATTTAT; reverse primer, ATACAGCATGTCTGTCGCAGTTG; amplicon, 129 bp; bovine GAPDH: forward primer, CCTGCCCGTTCGACAGATAG; reverse primer, GCGACGATGTCCACTTTGC; amplicon, 150 bp. Optimal concentrations were 50 nm for the FBN1 primers and 75 nm for the GAPDH primers. Data are from three independent experiments; for each experiment, quantitative PCR was performed twice.
RESULTS
ADAMTS10 Binds Specifically to Fibrillin-1
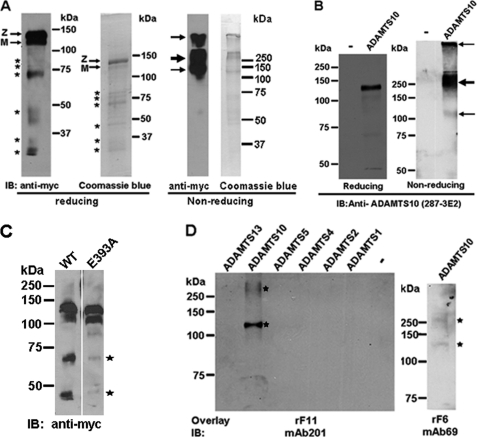
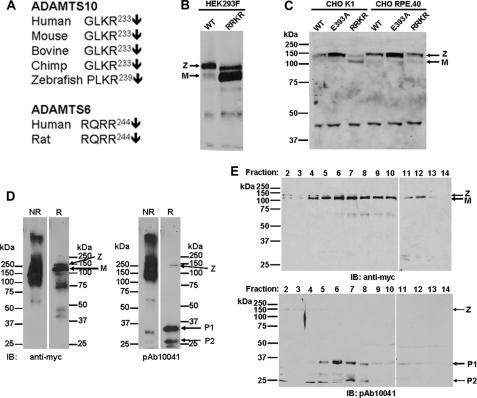
Recombinant ADAMTS10 was substantially purified from the conditioned medium of stably transfected HEK293F cells. Analysis of purified ADAMTS10 under reducing conditions by Western blotting with antibody to the C-terminal myc tag or Coomassie Blue staining (Fig. 1A) demonstrated a major species of 130 kDa representing ADAMTS10 zymogen, i.e. including the N-terminal propeptide with a lower proportion of the 120-kDa furin-processed form lacking the propeptide as described previously (17). Although the predicted molecular mass of ADAMTS10 zymogen is 110 kDa, it typically migrates more slowly than expected because of N-glycosylation (17). Additional immunoreactive species that were visible with Coomassie Blue staining and on Western blotting are likely derived from ADAMTS10 on the basis of their binding to the anti-myc monoclonal antibody and are presumed to be of proteolytic origin. However, as seen in Fig. 1A, not all Coomassie Blue-stained fragments reacted with anti-myc antibody because some of these presumably represent N-terminal ADAMTS10 fragments lacking the C-terminal myc tag. Under non-reducing conditions, purified ADAMTS10 migrated very differently (Fig. 1A). A major band of ∼250 kDa was seen on both Coomassie Blue staining and anti-myc Western blotting, whereas the 120-kDa species representing presumed monomeric ADAMTS10 appeared to be at lower levels. In addition, some ADAMTS10 was clearly retained in the stacking gel. The cause of this aberrant migration is not known, but we conjecture that ADAMTS10 may form homophilic complexes or heterophilic complexes, such as with the nonspecific protease inhibitor α2-macroglobulin, which has been shown previously to bind it (17). Conditioned medium from HEK293F cells expressing ADAMTS10 was used for evaluation of the ADAMTS10 monoclonal and polyclonal antibodies. In ADAMTS10-transfected but not vector-transfected cells, the anti-ADAMTS10 antibodies detected the expected 130-kDa ADAMTS10 zymogen and the 120-kDa furin-processed form (Fig. 1B shows the monoclonal antibody; data for the ADAMTS10 polyclonal antibody were similar because their epitopes overlap and are not shown). Under non-reducing conditions, a major species migrating at 250 kDa and a minor species at 120 kDa were detected by the ADAMTS10 mAb (Fig. 1B). Immunoreactive ADAMTS10 was also visible high in the stacking gel. These data are essentially similar to the major species observed with anti-myc with differences in smaller species that reflect the different locations of the epitopes for 287-3E2 and anti-myc. To determine whether ADAMTS10 fragmentation could occur by autoproteolysis, we compared the medium from HEK293F cells expressing either ADAMTS10 or ADAMTS10 E393A (mutation of the catalytic Glu residue is a standard inactivating mutation in the metalloprotease field) by Western blotting with anti-myc. Indeed, the 70- and 45-kDa species seen in wild-type ADAMTS10 were considerably attenuated in ADAMTS10 E393A (Fig. 1C). We conclude that ADAMTS10 fragmentation in the conditioned medium and in the purified protein was autocatalytic, although it remains possible that other proteases could also contribute.
FIGURE 1.
ADAMTS10 binds to fibrillin-1. A, analysis of purified recombinant ADAMTS10 by Western blot analysis (under reducing and non-reducing conditions as indicated) with anti-myc antibody or Coomassie Blue staining. Under reducing conditions, the major ADAMTS10 species migrates at 130 kDa, consistent with it being the unprocessed zymogen (Z). A 120-kDa band corresponding to mature (M) ADAMTS10 is present. Additional, smaller immunoreactive bands seen (asterisks) appear to be derived from ADAMTS10 on the basis of their binding to anti-myc. Under non-reducing conditions, the major species detected is at 250 kDa (bold arrow) with minor species seen at 120 kDa and within the stacking gel (fine arrows). B, characterization of monoclonal antibody 287-3E2 by Western blot analysis of conditioned medium from ADAMTS10-transfected HEK293F cells. Transfection with the pcDNA vector alone was used as the control (−). Under reducing conditions, the expected 130-kDa zymogen (Z; a major species) and the 120-kDa mature form (M; a minor species) are specifically seen only in the medium of ADAMTS10-transfected cells. Under non-reducing conditions, most of the ADAMTS10 migrates with an apparent molecular mass of 250 kDa (bold arrow). Some is trapped in the stacking gel, and a small amount with an apparent molecular mass of 120 kDa is also seen (fine arrows). C, wild type (WT) or ADAMTS10 E393A was transiently transfected in parallel into HEK293F cells, and the collected medium was electrophoresed under reducing conditions followed by immunoblotting with anti-myc. Note that molecular species corresponding to proteolytic fragments in the WT sample (asterisks) are considerably weaker in the E393A medium. D, left-hand panel, 400 ng each of ADAMTS10 and the indicated ADAMTS proteases were electrophoresed under non-reducing conditions. Following electroblotting, membranes were incubated with fibrillin-1 peptide corresponding to the N-terminal half (rF11) followed by detection with the indicated fibrillin-1 antibody. The peptide bound only to ADAMTS10 (asterisks). Right-hand panel, ADAMTS10 was electrophoresed as above for investigation of interaction with the C-terminal half of fibrillin-1 (rF6) followed by detection using mAb 69. The ADAMTS10 species detected were similar to those that bound rF11 (asterisks). IB, immunoblot.
For the initial analysis of ADAMTS10-fibrillin-1 interactions, we used a blot overlay assay in which ADAMTS10 and selected ADAMTS proteases were electrophoresed by non-reducing SDS-PAGE, transferred to a membrane, and probed with recombinant fibrillin-1 peptides comprising the N-terminal half. The fibrillin-1 peptide rF11 (representing the N-terminal half) bound to ADAMTS10 but not to equivalent amounts of recombinant ADAMTS1, ADAMTS2, ADAMTS4, ADAMTS5, and ADAMTS13 in the blot (Fig. 1D). In a separate experiment, we found that rF6 (C-terminal half of fibrillin-1) also bound ADAMTS10 (Fig. 1D). The relative binding affinity of these peptides could not be compared because different antibodies were used for detection of each. Both fibrillin peptides bound to the ∼120- and ∼250-kDa ADAMTS10 species seen under non-reducing conditions, although binding of rF11 to the ∼120-kDa species was clearly stronger than to the 250-kDa species (Fig. 1D).
Identification of Binding Sites in N-terminal and C-terminal Halves of Fibrillin-1
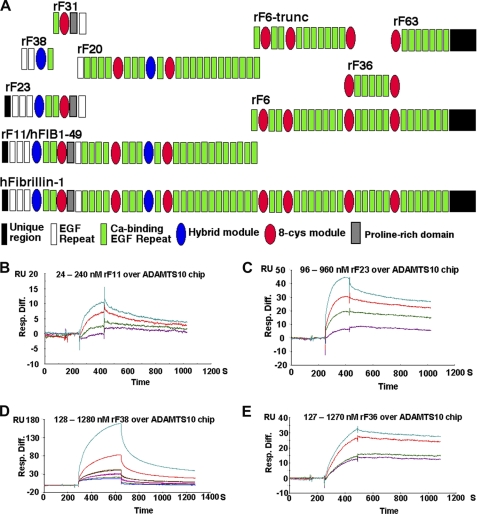
To define the binding sites of ADAMTS10 on fibrillin-1 and to determine binding kinetics, we used SPR. Specifically, the ability of recombinant fibrillin-1 peptide rF11, peptide rF6, or smaller fragments of these (Fig. 2A) to bind to ADAMTS10 was determined using full-length ADAMTS10 as the bound ligand (Fig. 2, B–E). rF11 bound to ADAMTS10 with a characteristic binding curve (sensorgram) indicative of a specific interaction (Fig. 2B) and with a KD of 28 nm. A peptide containing only the N-terminal one-third of rF11, rF23, bound with a similar dissociation constant (45 nm), suggesting that it fully contained the ADAMTS10 binding site identified on rF11 (Fig. 2C and Table 1). Consistent with this finding, a peptide representing the C-terminal portion of rF11, rF20, which did not overlap with rF23, did not provide a specific binding curve (data not shown). The sensorgram for a construct contained within rF23 (rF38) suggested that it bound specifically to ADAMTS10, but the calculated binding constant was lower (3 μm), suggesting that it did not contain the complete binding site within rF23 (Fig. 2D and Table 1). Peptide rF31, representing the region of rF23 complementary to rF38, did not bind to ADAMTS10 (Table 1). In a reverse assay, we used a new fibrillin-1 peptide, hFib1–49, which is similar to rF11, as the bound ligand and ADAMTS10 as an analyte, which confirmed the interactions observed using ADAMTS10 as the ligand (data not shown). These data suggested that an ADAMTS10 binding site was present within the N-terminal fibrillin-1 fragment represented by rF23. To further delineate the binding site in the C-terminal half of fibrillin-1 suggested by the blot overlay assay with peptide rF6, we undertook SPR analysis using peptides representing subfragments of the C-terminal half. Among these, peptide rF36 bound to ADAMTS10 with a comparable affinity as rF23 (Fig. 2E and Table 1), whereas peptides rF63 and rF6-trunc did not bind (Table 1). These observations strongly suggest the existence of at least two independent ADAMTS10 binding sites of comparable affinity on fibrillin-1.
FIGURE 2.
SPR analysis of interactions of full-length ADAMTS10 with fibrillin-1. A, depiction of the domain structure of fibrillin-1 and the peptides used in SPR analysis. B–E, the indicated fibrillin-1 peptides were used as the analyte and flowed over a chip coupled with ADAMTS10 as the ligand. The observed SPR change is indicated in resonance units (RU). Injections utilized a series of molar concentrations for each analyte (the concentration range used is indicated above each set of sensorgrams). Table 1 lists the specific numerical values for binding obtained from these and additional experiments of which the illustrated data are representative. Resp. Diff., response difference.
TABLE 1.
Summary of ADAMTS10 BIAcore kinetic data
| Analytes | Interaction | ka | kd | KA | KD | χ2 | Rmax |
|---|---|---|---|---|---|---|---|
| 1/m × s | 1/s | 1/m | nm | RUa | |||
| Fibrillin-1: rF11 | Yes | 6.01 × 104 | 16.9 × 10−4 | 35.5 × 106 | 28 | 0.055 | 11 |
| rF23 | Yes | 2.18 × 104 | 9.77 × 10−4 | 22.3 × 106 | 45 | 0.994 | 35 |
| rF20 | No | —b | — | — | — | — | — |
| rF38 | Yes | 0.16 × 104 | 46.8 × 10−4 | 0.33 × 106 | 3330 | 0.501 | 180 |
| rF31 | No | — | — | — | — | — | — |
| rF6 | Yes | — | — | — | — | — | — |
| rF6-trunc | No | — | — | — | — | — | — |
| rF36 | Yes | 1.27 × 104 | 9.21 × 10−4 | 13.8 × 106 | 72 | 0.586 | 32 |
| rF63 | No | — | — | — | — | — | — |
a Response units.
b —, no data.
Intriguingly, rF23 and rF6 contain the domains affected by previously identified in-frame deletions in fibrillin-1 that result in WMS (12).4 We conjecture that these deletions may affect the binding of ADAMTS10 to fibrillin-1. To further validate the specificity of the ADAMTS10 binding to fibrillin-1, SPR was used to evaluate binding of ADAMTS10 to several other proteins known to interact with fibrillin-1, namely latent TGFβ-binding protein-1, fibulin-1, fibulin-2, and fibronectin. None of them bound to ADAMTS10 (data not shown).
ADAMTS10 Is Associated with Microfibril Bundles in Situ
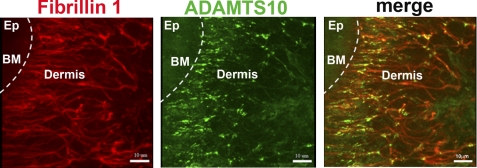
Because these data strongly suggested that fibrillin-1 bound specifically and directly to ADAMTS10 and because the genetic evidence suggested a functional association between these two proteins, we asked whether ADAMTS10 was localized in tissue microfibrils. We generated a new mouse monoclonal antibody to ADAMTS10, 287-3E2 (Fig. 1B), and used it for immunofluorescence and immunoelectron microscopy. When sections of skin were co-immunostained, examined by fluorescence microscopy using antibodies to fibrillin-1 and ADAMTS10, and observed by confocal microscopy, the pattern of ADAMTS10 immunostaining overlapped substantially with fibrillin-1 immunostaining (Fig. 3 and supplemental Video File 1). Control data not shown demonstrated the specificity of immunostaining. There was a region of very intense localization of ADAMTS10 to the narrow, branched microfibril bundles present in the superficial (papillary) dermis located very close to the dermal-epidermal junction (Fig. 3 and supplemental Video File 1). Less intense co-localization of fibrillin-1 and ADAMTS10 was also seen throughout the papillary dermis and into the shallow reticular dermis. Images collected of control sections using the same instrument settings eliminated the possibility that the patterns represented elastin autofluorescence.
FIGURE 3.
Co-localization of ADAMTS10 with fibrillin-1 in human skin. A, immunofluorescent visualization of the distribution of fibrillin-1 (red) and ADAMTS10 (green) in human skin. The locations of the epidermis (Ep), dermis, and basement membrane (BM; broken white line) are shown. Note the substantial overlap between ADAMTS10 and fibrillin microfibrils in the papillary dermis (closest to the basement membrane) with relatively lower ADAMTS10 staining intensity in the further removed reticular dermis (center and right of each panel). Scale bar, 10 μm. Negative controls indicating antibody specificity are shown in the supplemental data.
Immunoelectron microscopy of human skin clearly identified binding of the ADAMTS10 antibody to microfibril bundles (Fig. 4, A–D, and supplemental Video File 2), whereas no association was seen with collagen fibrils, basement membrane, and other structures (Fig. 4A and supplemental Video File 2). Immunoelectron microscopy of fibrillin-1 (data not shown) was consistent with previous work showing that it localizes to all microfibrils in the dermis, including the complete extent of those intersecting the lamina densa and reaching into the papillary dermis at the dermal-epidermal junction. ADAMTS10 also localized to these microfibril bundles; however, it was conspicuously absent from a region within approximately 1 μm from the epithelial basement membrane lamina densa. Microfibril bundles extending into the deeper papillary dermis and those associated with small elastic fibers present in the papillary dermis and the reticular layer were also labeled with antibody to ADAMTS10. The larger elastin fibers of the deep reticular layer were only occasionally labeled. Interestingly, in these en bloc immunogold experiments, gold conjugates larger than 1 nm did not localize ADAMTS10 to any microfibrils in skin. This is in contrast to the localization of primary antibodies to fibrillin, which consistently demonstrated a periodic labeling pattern using immunogold particles ranging in size from 1 up to 15 nm (data not shown). This indicated that the epitope for ADAMTS10 was more cryptic than the epitope for fibrillin.
Immunoelectron microscopy of human lens and zonule demonstrated ADAMTS10 antibody binding specifically to microfibrils along the length of the ciliary zonules, including those regions adjacent to the lens capsule (Fig. 4D) and those adjacent to the ciliary processes (Fig. 4E). Taken together, these co-localization data show that ADAMTS10 is present in microfibril bundles in vivo and provide additional evidence supporting a specific interaction of ADAMTS10 with fibillin-1.
Furin-processed ADAMTS10 Cleaves Fibrillin-1
All ADAMTS proteases contain a consensus furin recognition and cleavage sequence (Arg-Xaa-(Arg/Lys)-Arg↓) at the junction of the propeptide and catalytic domain. However, human ADAMTS10 has the sequence Gly-Leu-Lys-Arg (17) at this site and lacks the Arg residue at the P4 position that is required for optimal furin cleavage (23). Indeed, an Arg residue is lacking at the P4 position in ADAMTS10 from other species as well in contrast to its closest relative, ADAMTS6 (24), which has an optimal furin-processing site (Fig. 5A). A low level but variable amount of activated ADAMTS10 was observed (e.g. Figs. 1A and 5B) following ADAMTS10 transfection into a variety of cell types (HEK293F, COS-1, and CHO-K1), and this mature ADAMTS10 from HEK293F cells was shown previously to be appropriately processed at the Arg233-Ser234 peptide bond by N-terminal sequencing (17). This suggested that ADAMTS10 is indeed processed by furin or related proprotein convertases but with very low efficiency. Because HEK293F cells robustly express furin (25), incomplete processing of ADAMTS10 suggested innate resistance of ADAMTS10 to furin, although we cannot presently exclude the possibility that ADAMTS10 zymogen may be processed to maturity by enzymes other than proprotein convertases.
FIGURE 5.
Wild-type ADAMTS10 is normally resistant to furin processing but can be substantially processed to mature form by optimization of furin recognition sequence. A, sequence alignment of the putative furin cleavage site of ADAMTS10 and its closest homolog, ADAMTS6, from the indicated species. Note that the cleavage site of ADAMTS6 matches the desired RX(K/R)R consensus but that ADAMTS10 consistently lacks the P4 Arg residue. B, site-directed mutagenesis of ADAMTS10 was used to convert the wild-type (WT) G230L231KR sequence to R230R231KR (RRKR), and both forms were analyzed by Western blot of the conditioned medium of transfected HEK293F cells. Note that the major species detected in WT ADAMTS10 is the zymogen (Z), whereas the mature form (M) constitutes the major species following ADAMTS10-RRKR transfection. C, analysis of the indicated ADAMTS10 constructs in medium of transiently transfected CHO-K1 cells or furin-deficient CHO-RPE.40 cells by reducing SDS-PAGE and Western blotting with anti-myc. Note that the prevalent mature form (M) of the ADAMTS10-RRKR mutant is decreased in intensity relative to the zymogen (Z) in CHO-RPE.40, cells indicating that it was the consequence of furin processing. In contrast, there is little change in amount of the mature and zymogen forms observed in wild-type ADAMTS10 (WT) or ADAMTS10 E393A in either cell line. D, comparison of ADAMTS10-RRKR migration under reducing (R) and non-reducing (NR) conditions using anti-myc or the anti-propeptide antibody pAb 10041. Note the similar migration pattern observed with both antibodies under non-reducing conditions with the exception of some cleaved propeptide observed with pAb 10041. In contrast, reducing Western blots with anti-myc show a prevalent 120-kDa band corresponding to mature ADAMTS10 (M) with a low prevalence of zymogen (Z), and pAb 10041 showed prominent species of 35 (P1) and 27 kDa (P2) arising from the processed ADAMTS10 propeptide. E, Western blots following reducing SDS-PAGE of fractions collected from analytical gel filtration chromatography of ADAMTS10-RRKR. Note that cleaved ADAMTS10 propeptide species P1 and P2 co-elute with the mature ADAMTS10 in fractions 4–12. IB, immunoblot.
To investigate a putative proteolytic activity of this “activated” fraction of ADAMTS10, we generated a mutant in which the sequence G230L231KR was altered to R230R231KR. Expression of this mutant in HEK293F cells provided mature ADAMTS10 as the majority species in the medium (Fig. 5B). That the observed 120-kDa major species in ADAMTS10-RRKR was a result of furin processing was supported by a comparison of the expressed protein in furin-deficient CHO-RPE.40 and wild-type CHO-K1 cells. In these cells, both wild-type ADAMTS10 and ADAMTS10 E393A were processed poorly. Although ADAMTS10-RRKR was processed substantially in CHO-K1 cells, lower levels of the mature form were seen in CHO-RPE.40 medium (Fig. 5C). This suggested that the mature species of ADAMTS10-RRKR indeed resulted from furin activity; minimal processing seen in the other two constructs resulted not from furin activity but from another processing enzyme present in CHO-RPE.40 cells (Fig. 5C). We further demonstrated the significant extent of ADAMTS-RRKR processing in HEK293F cells by Western blotting using anti-myc and a previously described polyclonal antibody to the ADAMTS10 propeptide. Anti-myc Western blotting (Fig. 5D, left-hand panels) demonstrated significant zymogen conversion to the mature form under reducing conditions as well as ADAMTS10 proteolytic fragments. Under non-reducing conditions, ADAMTS10-RRKR migrated similarly to wild-type ADAMTS10 (Fig. 5D). Western blotting with the anti-propeptide antibody (pAb 10041) showed two major species of 35 and 27 kDa corresponding to the N-glycosylated and unglycosylated forms of the propeptide, respectively. In this blot, the intact zymogen constituted a very minor reactive species. However, under non-reducing conditions, the visualized species were very similar to those seen with anti-myc with the exception of a small amount of free propeptide (Fig. 5D). This suggested the possibility that the ADAMTS10 propeptide remained associated with the mature enzyme after furin processing. To extend this analysis further, we performed analytical gel filtration chromatography of ADAMTS10-RRKR. The analysis of eluted fractions using anti-myc and the propeptide antibody in Western blot analysis under reducing conditions clearly demonstrated that the fractions that contained mature ADAMTS10 also contained the cleaved propeptide (Fig. 5E). Taken together, the data in Fig. 5, D and E, strongly suggested that following furin cleavage the cleaved propeptide remains associated with the mature enzyme.
When fibrillin peptide rF6 was incubated with cells expressing wild-type ADAMTS10, a small amount of rF6 processing to a smaller, 60-kDa fragment was seen (Fig. 6); incubation of rF6 with cell-free medium from these cells did not generate this species (Fig. 6). ADAMTS10-RRKR-transfected cells as well as the conditioned medium of these cells generated the 60-kDa fibrillin-1 fragment more robustly than wild-type ADAMTS10 (Fig. 6). The corresponding catalytically inactive mutants ADAMTS10-EA and ADAMTS10-RRKR-EA did not generate the 60-kDa species, suggesting that it arose from specific proteolytic activity of ADAMTS10 or ADAMTS10-RRKR (Fig. 6). In contrast to rF6, the N-terminal fibrillin-1 peptide rF11 was not cleaved by ADAMTS10 (data not shown). These data together with the observation that ADAMTS10 binds to α2-macroglobulin argue that mature ADAMTS10, if produced physiologically, is an active protease that can cleave the C-terminal half of fibrillin-1. However, wild-type ADAMTS10 zymogen is innately resistant to furin processing, arguing that fibrillin-1 turnover may not be its major function or that it deliberately evolved to cleave fibrillin-1 inefficiently.
ADAMTS10 Enables Biogenesis of Fibrillin-1-containing Microfibrils in Cultured Cells
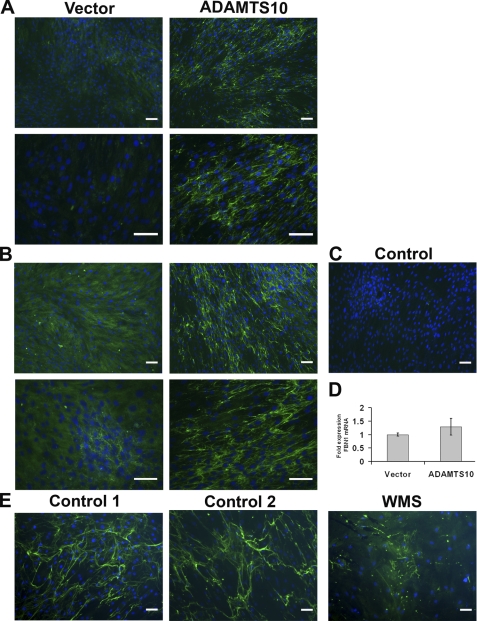
In WMS, ectopia lentis appears to result from the absence or dysgenesis of the zonule. In MFS, the zonule is present but is believed to be mechanically deficient with ectopia lentis occurring after the zonule fibers break and as patients get older. This clinical observation stimulated consideration of whether ADAMTS10 participated in the biogenesis of microfibrils. An in vitro biogenesis assay was established using fBNL cells, which deposit fibrillin-1 into their ECM upon culture to confluence. We investigated the potential role of ADAMTS10 by incubating the cultures with conditioned medium containing ADAMTS10 or with vector-conditioned medium as a control. These experiments demonstrated accelerated fibrillin-1 microfibril biogenesis in the ECM of confluent cells in the presence of ADAMTS10 at 3 (Fig. 7A) and 5 days (Fig. 7B) compared with vector-conditioned medium. A negative control for immunostaining, i.e. deletion of fibrillin-1 monoclonal antibody from the staining protocol, resulted in a total lack of fluorescence (Fig. 7C), indicating that the detected structures were specifically fibrillin-1-reactive. Accelerated microfibril biogenesis was not a consequence of increased production of fibrillin-1 as evidenced by the lack of a statistically significant change in the level of FBN1 mRNA after addition of exogenous ADAMTS10 (Fig. 7D). This observation suggested that ADAMTS10 could accelerate fibrillin microfibril biogenesis by fBNL.
FIGURE 7.
ADAMTS10 accelerates microfibril biogenesis in cultured fibroblasts, and WMS fibroblasts assemble a sparse microfibril network compared with normal skin fibroblasts. A and B, panels show fibrillin-1 immunofluorescence (green) in cultures of bovine fetal nuchal ligament cells as described under “Experimental Procedures.” Two fields are shown from cultures grown for 3 (A) or 5 days (B) at two different magnifications and are representative of multiple (n = 10) independent experiments. The cells treated with ADAMTS10-conditioned medium consistently show more fibrillin-1 microfibrils than cells treated with vector-transfected conditioned medium. In the negative control panel (C), the anti-fibrillin-1 antibody was omitted, and this preparation showed no green fluorescence. D, quantitative RT-PCR of FBN1 mRNA from fBNL fibroblasts treated with ADAMTS10-conditioned medium or vector-conditioned medium (n = 3) shows that there was no significant difference. Error bars represent S.D. E, skin fibroblasts from two control male subjects and an 86-year-old man with WMS were grown to confluence and stained for fibrillin-1 microfibrils (green) using monoclonal antibody 69. Note the robust microfibril production in cells from the control subjects, whereas fewer microfibrils were present in the WMS fibroblasts after 5 days. Scale bar, 50 μm.
We compared the deposition of fibrillin-1 in skin fibroblasts obtained from a case of WMS harboring two ADAMTS mutations, A25T and Q318Stop, which we recently characterized at the genetic and molecular level (14). In contrast to skin fibroblasts from two unaffected individuals of comparable age grown at a similar relative density, the patient fibroblasts consistently deposited less fibrillin-1 in the ECM (Fig. 7E). Together, these experiments supported the possibility that ADAMTS10 accelerated assembly of fibrillin microfibrils as a principal function.
DISCUSSION
The similar phenotypes resulting from ADAMTS10 and FBN1 mutations in recessive and dominant WMS, respectively, argue that ADAMTS10 and fibrillin-1 are functionally related; i.e. they might work in the same pathway or perhaps interact directly. The present studies, showing that ADAMTS10 not only directly bound to fibrillin-1 but also localized to fibrillin-1 microfibril bundles in situ, support this hypothesis. Our confocal and immuno-EM data suggest that ADAMTS10 is highly concentrated in a region close to the epithelium, a region of potential specific importance to the assembly of epithelial cell-derived fibrillin-1. Localization of ADAMTS10 to ciliary zonules, which are defective, leading to ectopia lentis in WMS patients, suggests a critical role in the maintenance or assembly of zonular microfibrils. Two distinct regions of fibrillin-1 that bound to full-length ADAMTS10 were identified, one in the N-terminal half and another in the C-terminal half. Preliminary data5 suggest that the recombinant ADAMTS10 ancillary domain (i.e. lacking the propeptide and catalytic domain) also binds fibrillin-1. Although the SPR sensorgrams obtained with specific fibrillin-1 fragments had the characteristic curves typical of specific binding interactions, the observed increases in resonance units following binding were relatively low. This phenomenon could have resulted from the fact that in solution, without prior reduction, the monomeric form of ADAMTS10 appeared as a minority, and instead, an unexpectedly large species (i.e. a complex, either homophilic or with a residual serum component such as α2-macroglobulin) was seen. Therefore, the ADAMTS10 available for fibrillin-1 binding could have been low. ADAMTS10 did not bind other fibrillin-1-interacting molecules, specifically latent TGFβ-binding protein-1, fibronectin, fibulin-1, and fibulin-2. Thus, direct binding of ADAMTS10 to fibrillin-1 is likely to occur in vivo and may underlie the observed genetic relationships.
To determine the consequences of the interaction of ADAMTS10 with fibrillin-1, we initially investigated whether fibrillin-1 was a substrate for ADAMTS10. However, the ADAMTS10 zymogen was found to be innately resistant to activation by furin because only a relatively small proportion of furin-processed, mature ADAMTS10 was present in three different cell lines upon transfection. This innate resistance was attributed to the lack of an optimal consensus sequence for processing by furin. The sequence conservation of ADAMTS10 to exclude the required P4 Arg residue in several species strongly suggests that ADAMTS10 may have evolved specifically not to be efficiently activated by furin and/or related proprotein convertases. Indeed, sensitivity to cleavage by furin was substantially increased by mutagenesis to create a consensus site for furin processing, and this mutant was inefficiently cleaved in furin-deficient CHO-RPE.40 cells. Alternatively, it is a possibility that ADAMTS10 could be activated by a proprotein convertase-independent mechanism.
Previously, ADAMTS9 zymogen was shown to have activity against its substrate versican (26), which provided a precedent for the possibility that ADAMTS10 zymogen could have proteolytic activity. However, wild-type ADAMTS10, comprising substantially the zymogen, did not cleave fibrillin-1 peptide rF6 efficiently. Next, the quantitatively minor mature form of ADAMTS10 was tested for its ability to cleave fibrillin-1. This form was generated by site-directed mutagenesis of the furin-processing site to optimize maturation; the activated ADAMTS10 generated from the mutant is essentially representative of the bioactivity of the small proportion of wild-type ADAMTS10 that is furin-processed. ADAMTS10-RRKR demonstrated processing of fibrillin-1 fragment rF6 but not rF11. In ADAMTS10-RRKR, we observed that following propeptide processing the cleaved propeptide remained bound to the mature enzyme. Because ADAMTS10-RRKR is more proteolytically active than wild-type ADAMTS10, we speculate that the cleaved propeptide is bound to the mature protease in a different conformation than when contiguous with it. We conclude that the ADAMTS10 zymogen does not cleave fibrillin-1 but that the quantitatively minor activated species may do so. The potential significance of fibrillin-1 processing by ADAMTS10 is therefore intimately tied to whether or not the ADAMTS10 propeptide is processed. At the present time, we have not been able to detect ADAMTS10 in tissue extracts to determine whether the propeptide is excised in vivo. Therefore, future studies will address this question and investigate other possible mechanisms by which ADAMTS10 propeptide could be excised. The possibility that ADAMTS10 cleaves another fibrillin-1 associated molecule, such as a fibrillin-2, or a microfibril-associated protein or proteoglycan remains to be addressed.
Because ectopia lentis is a major feature of WMS and clinical observations suggest an absence of zonular fibers in WMS eyes (as opposed to stretched or broken fibers in MFS), we next considered the possibility that ADAMTS10 promoted microfibril assembly. Zonule assembly is poorly understood but is believed to occur following synthesis of fibrillin-1 by non-pigmented ciliary body epithelial cells (27). The lens and ciliary body are closely apposed during eye development and subsequently separate so that zonule formation occurs within a narrow extracellular space that expands as development proceeds. Acceleration of fibrillin-1 assembly may be crucial during this developmental process.
To test a potential role for ADAMTS10 in this process, we used a previously described assay (28) in which confluent fibroblasts from various sources such as skin and nuchal ligament laid down a network of fibrillin-1 microfibrils in their extracellular matrix. This assay suggested a role for ADAMTS10 in fibrillin-1 assembly. To complement this assay, we asked whether skin fibroblasts from a WMS patient could assemble a robust fibrillin network. These cells assembled a sparse fibrillin network in ECM compared with wild-type cells of a comparable age. We propose that ADAMTS10 may normally facilitate fibrillin-1 microfibril biogenesis and that this function may be lost in WMS. It is important to point out that the microfibril biogenesis assay was done in the presence of 10% serum (i.e. in the presence of abundant α2-macroglobulin) so that any possible proteolytic activity of ADAMTS10 was eliminated. Therefore, the observed effects are independent of any potential proteolytic activity of ADAMTS10.
In addition to ADAMTS proteases, the ADAMTS superfamily contains ADAMTS-like proteins, which lack the propeptide, metalloprotease, and disintegrin-like domains and do not have proteolytic activity (29). Instead, they appear to be secreted glycoproteins closely related to ADAMTS ancillary domains and reside in extracellular matrix. Of relevance to WMS, ADAMTSL2 mutations lead to a WMS-like condition named geleophysic dysplasia (30). Furthermore, ADAMTSL4 mutations occur in recessively inherited isolated ectopia lentis (31, 32), and ADAMTS17 mutations result in ectopia lentis in dogs (33) and a WMS-like syndrome in humans (15). Recently, it was shown that ADAMTSL6 was capable of enhancing fibrillin-1 assembly into ECM (34).
Together, the present work and the recent publications on ADAMTS17 and ADAMTS-like proteins suggest that in addition to evolutionarily related clusters of ADAMTS proteases that mediate procollagen processing and proteoglycan turnover (29) the ADAMTS superfamily contains a group of proteins that are highly relevant to disorders related to fibrillin-1. By virtue of its resistance to furin processing and strong affinity for fibrillin-1 as well as a comparable functional effect as ADAMTSL6, ADAMTS10 appears to function as an ADAMTS-like protein. We speculate that ADAMTSL2, ADAMTSL4, ADAMTSL6, and ADAMTS17 may act via the same mechanism as ADAMTS10 or within a novel shared pathway relevant to the biology of fibrillin-1 and thus could hypothetically enhance the action of ADAMTS10 or work cooperatively with it. Thus, members of the ADAMTS cluster of fibrillin-1-related proteins may work cooperatively to influence microfibril biogenesis, a model similar to the novel cooperative model recently established for ADAMTS proteases in versican turnover (35, 36).
In conclusion, the present work substantially clarifies the relationship between ADAMTS10 and fibrillin-1 that was suggested by WMS genetics, and will enable further understanding of the roles of ADAMTS proteins in microfibril biology.
Supplementary Material
Acknowledgments
We thank Daiichi Fine Chemical Co. Ltd. for generating ADAMTS10 monoclonal antibody.
This work was supported, in whole or in part, by National Institutes of Health Grant AR53890 from the NIAMS and Grant EY21151 (to S. S. A.) and Postdoctoral Training Grants T32 AR050959 (to W. E. K.; training program in orthopedics; principal investigator, Joseph P. Iannotti) and T32 HL007914-08 (to H. L. B.; training program in vascular biology and pathology; principal investigator, Edward Plow). This work was also supported by a grant from the National Marfan Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Video Files 1 and 2.
G. Sengle and L. Y. Sakai, manuscript in preparation.
G. Sengle and L. Y. Sakai, unpublished data.
- ECM
- extracellular matrix
- WMS
- Weill-Marchesani syndrome
- MFS
- Marfan syndrome
- ADAMTS
- a disintegrin-like and metalloproteinase with thrombospondin motifs
- pAb
- polyclonal antibody
- SFM
- serum-free Dulbecco's medium
- SPR
- surface plasmon resonance
- fBNL
- fetal bovine nuchal ligament.
REFERENCES
- 1. Sakai L. Y., Keene D. R., Engvall E. (1986) J. Cell Biol. 103, 2499–2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ramirez F., Sakai L. Y., Dietz H. C., Rifkin D. B. (2004) Physiol. Genomics 19, 151–154 [DOI] [PubMed] [Google Scholar]
- 3. Cain S. A., Morgan A., Sherratt M. J., Ball S. G., Shuttleworth C. A., Kielty C. M. (2006) Proteomics 6, 111–122 [DOI] [PubMed] [Google Scholar]
- 4. Pyeritz R. E. (2000) Annu. Rev. Med. 51, 481–510 [DOI] [PubMed] [Google Scholar]
- 5. Judge D. P., Biery N. J., Keene D. R., Geubtner J., Myers L., Huso D. L., Sakai L. Y., Dietz H. C. (2004) J. Clin. Investig. 114, 172–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Robinson P. N., Arteaga-Solis E., Baldock C., Collod-Béroud G., Booms P., De Paepe A., Dietz H. C., Guo G., Handford P. A., Judge D. P., Kielty C. M., Loeys B., Milewicz D. M., Ney A., Ramirez F., Reinhardt D. P., Tiedemann K., Whiteman P., Godfrey M. (2006) J. Med. Genet. 43, 769–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Habashi J. P., Judge D. P., Holm T. M., Cohn R. D., Loeys B. L., Cooper T. K., Myers L., Klein E. C., Liu G., Calvi C., Podowski M., Neptune E. R., Halushka M. K., Bedja D., Gabrielson K., Rifkin D. B., Carta L., Ramirez F., Huso D. L., Dietz H. C. (2006) Science 312, 117–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ng C. M., Cheng A., Myers L. A., Martinez-Murillo F., Jie C., Bedja D., Gabrielson K. L., Hausladen J. M., Mecham R. P., Judge D. P., Dietz H. C. (2004) J. Clin. Investig. 114, 1586–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Isogai Z., Ono R. N., Ushiro S., Keene D. R., Chen Y., Mazzieri R., Charbonneau N. L., Reinhardt D. P., Rifkin D. B., Sakai L. Y. (2003) J. Biol. Chem. 278, 2750–2757 [DOI] [PubMed] [Google Scholar]
- 10. Rifkin D. B. (2005) J. Biol. Chem. 280, 7409–7412 [DOI] [PubMed] [Google Scholar]
- 11. Sengle G., Charbonneau N. L., Ono R. N., Sasaki T., Alvarez J., Keene D. R., Bächinger H. P., Sakai L. Y. (2008) J. Biol. Chem. 283, 13874–13888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Faivre L., Gorlin R. J., Wirtz M. K., Godfrey M., Dagoneau N., Samples J. R., Le Merrer M., Collod-Beroud G., Boileau C., Munnich A., Cormier-Daire V. (2003) J. Med. Genet. 40, 34–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dagoneau N., Benoist-Lasselin C., Huber C., Faivre L., Mégarbané A., Alswaid A., Dollfus H., Alembik Y., Munnich A., Legeai-Mallet L., Cormier-Daire V. (2004) Am. J. Hum. Genet. 75, 801–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kutz W. E., Wang L. W., Dagoneau N., Odrcic K. J., Cormier-Daire V., Traboulsi E. I., Apte S. S. (2008) Hum. Mutat. 29, 1425–1434 [DOI] [PubMed] [Google Scholar]
- 15. Morales J., Al-Sharif L., Khalil D. S., Shinwari J. M., Bavi P., Al-Mahrouqi R. A., Al-Rajhi A., Alkuraya F. S., Meyer B. F., Al Tassan N. (2009) Am. J. Hum. Genet. 85, 558–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Faivre L., Dollfus H., Lyonnet S., Alembik Y., Mégarbané A., Samples J., Gorlin R. J., Alswaid A., Feingold J., Le Merrer M., Munnich A., Cormier-Daire V. (2003) Am. J. Med. Genet. 123A, 204–207 [DOI] [PubMed] [Google Scholar]
- 17. Somerville R. P., Jungers K. A., Apte S. S. (2004) J. Biol. Chem. 279, 51208–51217 [DOI] [PubMed] [Google Scholar]
- 18. Apte S. S. (2004) Int. J. Biochem. Cell Biol. 36, 981–985 [DOI] [PubMed] [Google Scholar]
- 19. Spence M. J., Sucic J. F., Foley B. T., Moehring T. J. (1995) Somat. Cell Mol. Genet. 21, 1–18 [DOI] [PubMed] [Google Scholar]
- 20. Hall N. G., Klenotic P., Anand-Apte B., Apte S. S. (2003) Matrix Biol. 22, 501–510 [DOI] [PubMed] [Google Scholar]
- 21. Rousselle P., Lunstrum G. P., Keene D. R., Burgeson R. E. (1991) J. Cell Biol. 114, 567–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Reinhardt D. P., Keene D. R., Corson G. M., Pöschl E., Bächinger H. P., Gambee J. E., Sakai L. Y. (1996) J. Mol. Biol. 258, 104–116 [DOI] [PubMed] [Google Scholar]
- 23. Nakayama K. (1997) Biochem. J. 327, 625–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hurskainen T. L., Hirohata S., Seldin M. F., Apte S. S. (1999) J. Biol. Chem. 274, 25555–25563 [DOI] [PubMed] [Google Scholar]
- 25. Koo B. H., Apte S. S. (2010) J. Biol. Chem. 285, 197–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Koo B. H., Longpré J. M., Somerville R. P., Alexander J. P., Leduc R., Apte S. S. (2007) J. Biol. Chem. 282, 16146–16154 [DOI] [PubMed] [Google Scholar]
- 27. Hanssen E., Franc S., Garrone R. (2001) Matrix Biol. 20, 77–85 [DOI] [PubMed] [Google Scholar]
- 28. Hollister D. W., Godfrey M., Sakai L. Y., Pyeritz R. E. (1990) N. Engl. J. Med. 323, 152–159 [DOI] [PubMed] [Google Scholar]
- 29. Apte S. S. (2009) J. Biol. Chem. 284, 31493–31497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Le Goff C., Morice-Picard F., Dagoneau N., Wang L. W., Perrot C., Crow Y. J., Bauer F., Flori E., Prost-Squarcioni C., Krakow D., Ge G., Greenspan D. S., Bonnet D., Le Merrer M., Munnich A., Apte S. S., Cormier-Daire V. (2008) Nat. Genet. 40, 1119–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Greene V. B., Stoetzel C., Pelletier V., Perdomo-Trujillo Y., Liebermann L., Marion V., De Korvin H., Boileau C., Dufier J. L., Dollfus H. (2010) Ophthalmic Genet. 31, 47–51 [DOI] [PubMed] [Google Scholar]
- 32. Ahram D., Sato T. S., Kohilan A., Tayeh M., Chen S., Leal S., Al-Salem M., El-Shanti H. (2009) Am. J. Hum. Genet. 84, 274–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Farias F. H., Johnson G. S., Taylor J. F., Giuliano E., Katz M. L., Sanders D. N., Schnabel R. D., McKay S. D., Khan S., Gharahkhani P., O'Leary C. A., Pettitt L., Forman O. P., Boursnell M., McLaughlin B., Ahonen S., Lohi H., Hernandez-Merino E., Gould D. J., Sargan D. R., Mellersh C. S. (2010) Invest. Ophthalmol. Vis. Sci. 51, 4716–4721 [DOI] [PubMed] [Google Scholar]
- 34. Tsutsui K., Manabe R., Yamada T., Nakano I., Oguri Y., Keene D. R., Sengle G., Sakai L. Y., Sekiguchi K. (2010) J. Biol. Chem. 285, 4870–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McCulloch D. R., Nelson C. M., Dixon L. J., Silver D. L., Wylie J. D., Lindner V., Sasaki T., Cooley M. A., Argraves W. S., Apte S. S. (2009) Dev. Cell 17, 687–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Le Goff C., Somerville R. P., Kesteloot F., Powell K., Birk D. E., Colige A. C., Apte S. S. (2006) Development 133, 1587–1596 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.