Abstract
Levels of many hydrophobic cellular substances are tightly regulated because of their potential cytotoxicity. These compounds tend to self-aggregate in cytoplasmic storage depots termed lipid droplets/bodies that have well defined structures that contain additional components, including cholesterol and various proteins. Hydrophobic substances in these structures become mobilized in a specific and regulated manner as dictated by cellular requirements. Retinal pigmented epithelial cells in the eye produce retinyl ester-containing lipid droplets named retinosomes. These esters are mobilized to replenish the visual chromophore, 11-cis-retinal, and their storage ensures proper visual function despite fluctuations in dietary vitamin A intake. But it remains unclear whether retinosomes are structures specific to the eye or similar to lipid droplets in other organs/tissues that contain substances other than retinyl esters. Thus, we initially investigated the production of these lipid droplets in experimental cell lines expressing lecithin:retinol acyltransferase, a key enzyme involved in formation of retinyl ester-containing retinosomes from all-trans-retinol. We found that retinosomes and oleate-derived lipid droplets form and co-localize concomitantly, indicating their intrinsic structural similarities. Next, we isolated native retinosomes from bovine retinal pigmented epithelium and found that their protein and hydrophobic small molecular constituents were similar to those of lipid droplets reported for other experimental cell lines and tissues. These unexpected findings suggest a common mechanism for lipid droplet formation that exhibits broad chemical specificity for the hydrophobic substances being stored.
Keywords: Lipid-binding Protein, Lipid Droplet, Retinal Metabolism, Retinoid, Vitamin A
Introduction
Lying behind the retina, retinal pigmented epithelial (RPE)2,3 cells have multiple functions such as nourishing and removing metabolic waste products, absorbing excess light, phagocytosis of photoreceptor fragments, and recycling the visual chromophore, 11-cis-retinal (2–6). Recycling of 11-cis-retinal starts in photoreceptors with conversion of all-trans-retinal to all-trans-retinol, which is transferred from photoreceptor cell outer segments to the RPE (7, 8). The all-trans-retinol is then esterified with a fatty acyl group at the sn-1 position of phosphatidylcholine, an enzymatic reaction catalyzed by lecithin:retinol acyltransferase (LRAT) (9–11). In the RPE, retinyl esters reportedly are substrates for retinoid isomerase, the RPE-specific 65-kDa protein (RPE65) that catalyzes conversion of retinyl esters to 11-cis-retinol (7, 12–14). Finally, 11-cis-retinol is oxidized to 11-cis-retinal and diffuses back to photoreceptors to recombine with opsins (5).
In vivo imaging of the RPE in mice revealed the presence of retinyl ester storage particles or retinosomes (15, 16). Genetic analysis indicated that their formation depends on LRAT, and several lines of evidence suggested that retinosomes located close to the RPE plasma membrane are essential particles involved in regeneration of 11-cis-retinal (5, 6, 15). Because Rpe65−/− mice cannot isomerize all-trans-retinyl esters (17), this defect leads to retention of retinoids that enlarge retinosomes (15, 16), thereby avoiding vitamin A cytotoxicity. These observations suggest that retinosomes carry out specific functions that exceed those of a mere lipid storage organelle (18). Interestingly, upon amidation, retinylamine (19), a potent inhibitor of the retinoid cycle, is stored in these structures (20, 21), as are esters of the artificial chromophore, 9-cis-retinol (22, 23). This suggests a highly specific role for retinosomes in the ocular pharmacokinetics of retinoid-based therapeutics that involves storing and releasing safe low levels of these compounds over long periods of time (24, 25). Imanishi et al. (15) found that retinosomes also are enriched in perilipin 2 (PLIN2), formerly known as adipocyte differentiation-related protein (ADRP or ADFP) (26). Mice lacking exons 2 and 3 of the gene encoding PLIN2 evidenced slowed dark adaptation, probably due to delayed clearance of all-trans-retinal and all-trans-retinol from rod photoreceptor cells (27). Indeed, two-photon microscopy (TPM) revealed aberrant trafficking of all-trans-retinyl esters in the RPE of these mice, a problem caused by prolonged maintenance of retinosomes in the dark-adapted state. These findings suggest that PLIN2 plays a unique role in vision by maintaining proper storage and trafficking of retinoids within the eye. They also imply that a common mechanism underlies the biogenesis of retinosomes and lipid droplets because PLIN2 is a prominent component of lipid droplets in other tissues (28).
Lipid droplets are ubiquitous organelles that store esterified fatty acids. Although especially prevalent in adipocytes, liver hepatocytes, and stellate cells, they are found in virtually every cell type (29–31). These droplets are surrounded by phospholipid monolayer membranes (32) and contain large amounts of proteins such as PLIN1, PLIN2, and PLIN3 (31). Based on these findings, these droplets are becoming recognized as active intracellular organelles that contribute to several processes such as lipid metabolism, intracellular trafficking, and signaling (33, 34). The current model of lipid droplet biogenesis suggests that they emerge from the endoplasmic reticulum (ER) (35, 36), but several other models of lipid droplet biogenesis have been proposed as well (37–41).
This study was designed to explore the relationship of retinosomes to lipid droplets by comparing their localization and their chemical and protein composition. Our observations provide evidence for a close relationship between retinosomes in the RPE and lipid droplets in other types of cells.
MATERIALS AND METHODS
Chemicals
All-trans-retinyl palmitate was purchased from Sigma, and all-trans-retinol was from Toronto Research Chemicals Inc., Toronto, Canada.
Immunofluorescence Microscopy and Data Analysis
Cells (NIH3T3, NIH3T3/L, NIH3T3/LR (42), and ARPE19 (42, 43)) were seeded at 1 million cells per well in multiwell cell culture plates in Dulbecco's modified Eagle's medium, pH 7.2, with 4 mm l-glutamine, 4,500 mg/liter glucose, and 110 mg/liter sodium pyruvate supplemented with 10% fetal bovine serum (FBS), 100 units/ml penicillin, and 100 units/ml streptomycin. Oleic acid was complexed with fatty acid-free bovine serum albumin at a 6:1 ratio in 0.1 m Tris-Cl, pH 8.0. After complex formation indicated by clarification, the solution was passed through a 0.2-μm filter unit (Millipore, Kankakee, IL). Cells were treated with oleic acid, 100 μm final concentration, and all-trans-retinol, 10 and 20 μm final concentration, all in N,N-dimethylformamide (Sigma). The final concentration of N,N-dimethylformamide in each cell culture dish was less than 0.9 μg/ml. After 24 h, cells were fixed with 3% formaldehyde for 30 min and immunolabeled with anti-PLIN2. Lipid droplets were stained with BODIPY493/503 (493 nm excitation/503 nm emission) (Invitrogen). After treatment with all-trans-retinol, lipid droplets were imaged by the autofluorescence of all-trans-retinol and retinyl esters as described previously (30). Images, obtained with Leica TCS SP2 fluorescence microscope (Leica Microsystems Inc., Wetzlar, Germany), were analyzed with ImageJ software (National Institutes of Health, Bethesda). Seven plates were used for each experiment, and each experiment was done independently three times. Similar results were obtained in all experiments.
Subcellular Fractionation of RPE Cell Homogenates
Bovine eyeballs were purchased from a local slaughterhouse (Mahan's Packing Co, Bristolville, OH). After enucleation, eyes were kept on ice and processed within 4 h. Vitreous fluid was removed from each eyeball, and the retina was detached. RPE cells were lightly brushed off after adding 1 ml containing 48 mm MOPS, pH 7.0, 1 mm dithiothreitol, and 245 mm sucrose to the eye cup. The collected solution, kept on ice during the entire procedure, was filtered using cotton gauze to remove larger pieces of tissue such as retinal fragments accumulated during the brushing procedure. One hundred eyeballs were used for a typical preparation. RPE cells were pelleted by centrifugation at 1,000 × g for 5 min. Cells were resuspended in 8 ml of 48 mm MOPS, pH 7.0, 1 mm dithiothreitol, and 245 mm sucrose and homogenized by 10 passes through a Dounce homogenizer with a tight plunger. The resulting suspension was centrifuged at 20,000 × g for 20 min at 4 °C to remove unbroken cells. The supernatant was collected and centrifuged at 150,000 × g for 1 h at 4 °C. Following centrifugation, the top 1 ml from each centrifuge tube, including all the white floating material, was collected. The combined suspension (totaling ∼5 ml for a typical preparation) was overlaid first with 48 mm MOPS, pH 7.0, and 150 mm sucrose (3 ml) followed by 3 ml of 48 mm MOPS, pH 7.0, in a 13-ml centrifuge tube fit to a SW41-Ti rotor (Beckman Coulter, Brea, CA). The sucrose gradient was centrifuged at 200,000 × g for 3 h at 4 °C. Seven fractions were collected from top to bottom and frozen at −20 °C until further use. After purification, lipid droplets from the seven fractions were stained with BODIPY493/503.
Subcellular Fractionation of Cell Homogenates
Cells were grown to 70% confluence in 10 10-cm diameter dishes. Growth conditions were identical to those described under “Immunofluorescence Microscopy and Data Analysis.” Cells were treated with all-trans-retinol (20 μm final concentration) for 24 h. After this incubation and removal of cell media, cells were washed three times with ice-cold phosphate-buffered saline (HyClone, Logan, UT) and filtered using a 1-μm filter. Cells were scraped and pelleted by centrifugation at 1,000 × g for 5 min. All subsequent steps followed the procedure employed under “Subcellular Fractionation of RPE Cell Homogenates.”
Assessment of Lipid Droplet Purity
SDS-PAGE was performed with 15 μl from each collected fraction by using the method of Laemmli (44). Proteins were transferred to polyvinylidene fluoride (PVDF) membranes and blocked with 5% dry milk in 137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 2 mm KH2PO4, pH 7.4 (PBS), and 0.05% Tween overnight at 4 °C. Anti-Golgi-58 antibody and antibodies against calreticulin (endoplasmic reticulum), PLIN1, retinaldehyde-binding protein (CRALBP), CGI-58, actin, cytochrome c oxygenase, peroxisome assembly factor 2 (PEX6), and PLIN3 were obtained from Abcam (Cambridge, MA). Rabbit PLIN2 C-terminal antibody was a kind gift from Dr. McManaman (45). Detection was achieved after Western blue staining (Promega, Madison, WI).
Proteomic Analysis
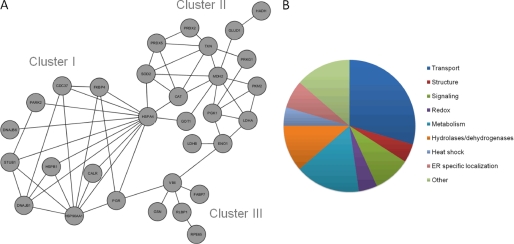
SDS-polyacrylamide gels of the seven fractions were first stained with Coomassie Brilliant Blue before destaining. Bands from each gel were cut out and washed twice for 10 min in 100 μl of 25 mm NH4HCO3 and twice in acetonitrile, 25 mm NH4HCO3, 1:3. Gel pieces were finally washed twice for 10 min with acetonitrile, 25 mm NH4HCO3, 1:1, before being dried in a SpeedVac centrifuge for 15 min. Disulfide bonds were reduced with 20 μl of 20 mm dithiothreitol for 45 min at 37 °C. Excess liquid was removed, and Cys residues were alkylated for 1 h in the dark with 30 μl of 10 mg/ml iodoacetamide dissolved in 25 mm NH4HCO3. Following alkylation, gel pieces were washed three times for 10 min each with acetonitrile, 25 mm NH4HCO3, 1:1, and dried for 20 min in a SpeedVac. Gel pieces then were rehydrated and digested with ∼13 ng/μl of sequencing grade trypsin (Promega, San Luis Obispo, CA) in 25 mm NH4HCO3 overnight at 37 °C. Resulting peptides were extracted three times with 30 μl of 5% trifluoroacetic acid in acetonitrile. Supernatants were reduced to dryness in a SpeedVac, and the resulting dried material was redissolved in 12 μl of H2O with 0.1% trifluoroacetic acid. Samples were loaded on a Dionex capillary LC system and eluted with a gradient of 0–60% acetonitrile in 0.1% formic acid over 30 min. The eluent was injected into a Fourier Transform LTQ mass spectrometer (MS) (Thermo-Finnigan, Waltham, MA) and analyzed by data-dependent methods consisting of a preliminary full scan followed by an MS/MS scan of the eight most abundant precursor ions. Peaks chosen from the MS/MS spectra with extract_msn were used to identify proteins with a Mascot search engine (version 2.2.2, Matrix Science, London, UK). Enzyme specificity was set to trypsin, and a maximum of two missed cleavages was permitted. The precursor mass tolerance was set to ±20 ppm, and the mass tolerance for fragment ions was set to ±0.8 Da. The S-carbamidomethyl Cys residue was set as a fixed modification, whereas the oxidized Met residue was set as a variable modification. The National Center for Biotechnology Information data base 20070216 (4626804 sequences, 1596079197 residues) was used for the search. The significance threshold for MS/MS searches was set to <0.05. Protein hits with three or more peptide scores of >50 were used for manual inspection with Qual Browser (version 2.07, ThermoFisher, Waltham, MA). A protein interconnectivity network (Fig. 7A) was constructed by using the on-line version of interaction databases provided by String software (46). The full protein analysis was done three times. All listed proteins were consistently observed in three preparations. The best Mascot score, sequence coverage, and number of assigned peptides are presented in the supplemental Table S1.
FIGURE 7.
Proteins recruited into retinosomes. By investigating the protein composition of purified bovine retinosomes, we found proteins involved in transport, metabolism, and structure, among others. A shows the interactive network of proteins found to be localized to retinosomes. The node labels represent gene names described in supplemental Table S1. B shows a diagram summarizing the functions of proteins localized to retinosomes. The results indicate the diversity of proteins recruited on the retinosome.
Retinyl Ester Analysis
An equal volume of n-hexane/methanol (5:1) was added to 1 ml of each collected fraction, and retinoids were extracted by vigorous shaking. Phase separation was achieved by centrifugation at 4,000 × g for 15 min at 4 °C. The organic phase was collected, dried down in a SpeedVac, and redissolved in 250 μl of n-hexane. Retinoids were separated by HPLC on a 5-μm, 250 × 4.6-mm Suplex PKB-100 column (Supelco, Bellefonte, PA). HPLC at 37 °C consisted of 14 min of isocratic elution with acetonitrile/methanol/dichloromethane/n-hexane (88:4:4:4, v/v) followed by a linear gradient to acetonitrile/methanol/dichloromethane/n-hexane (70:10:10:10, v/v) over 2 min and finishing with isocratic elution for another 14 min. Identification of retinyl palmitate and retinol was achieved by comparing sample retention times with those of authentic standards. Absorbance at 325 nm was checked for each peak. For comparison, spectra are presented after normalization of absorbance at 325 nm.
Electrospray Ionization-MS
The presence of retinyl esters in collected fractions was confirmed by the ion m/z = 269 (47) characteristic of the loss of OH group of retinol. Material collected at the retention time corresponding to m/z = 269 displayed maximal UV absorption at 325 nm. Analysis was performed with an 1100 HPLC system (Agilent Technologies, Santa Clara, CA) coupled to a Finnigan LXQ MS (Thermo Scientific, Waltham, MA) equipped with an atmospheric chemical ionization source. Retinoids were separated by isocratic elution at 37 °C with a mobile phase composed of 10% ethyl acetate in n-hexane flowing for 10 min through a 4.6 × 250-mm, 5-μm Zorbax Sil column (Agilent Technology).
Triglyceride Analysis
Triglyceride analysis was performed on each fraction collected from the lipid droplet purification. Triglycerides were quantified according to the manufacturer's instructions by a colorimetric (570 nm) method that employed the EnzyChromTM triglyceride assay kit (BioAssay Systems, Hayward, CA).
Thin Layer Chromatography (TLC)
Samples containing the lipid droplet fraction were extracted with 6 volumes of chloroform/methanol (2:1, v/v) and evaporated in a SpeedVac. Dried samples were redissolved in 200 μl of chloroform and spotted on TLC Silica 60 on aluminum (Sigma). Lipids were separated with hexane/diethyl ether/acetic acid (80:20:1, v/v) over 40 min. Lipid classes were identified by using standards obtained from Nu-Check Prep, Inc., Elysian, MN. Visualization was achieved by exposing TLC plates to iodine vapor for 10 min. Densitometry analysis was performed with ImageJ software (National Institutes of Health, Bethesda).
A2E Analysis
One and a half ml of chloroform/methanol (2:1 v/v) was added to samples that were then vigorously shaken and centrifuged at 4,000 × g for 10 min. The top fraction was collected and placed in a glass tube. The solvent was evaporated in a SpeedVac, and residues were redissolved in 200 μl of acetonitrile/water (80:20 v/v) with 0.05% trifluoroacetic acid (TFA) (solvent A). Five μl of each sample was loaded onto a C18 column (Gemini, 250 × 4.6 mm, 5 μm, Phenomenex, Torrance, CA) equilibrated with solvent A and eluted with the following gradient: 0–15 min to solvent B (0.05% TFA in acetonitrile) followed by 15–20 more min in solvent B. Absorbance at two different wavelengths, 325 and 450 nm, was monitored at a bandwidth of 10 nm. This loading provided an analysis for about one bovine eye. A2E was identified by comparing elution times of samples with those of authentic standards, and quantification was achieved by calculating areas under the peaks.
Quantification of Neutral Lipids and Phospholipids in Isolated Lipid Droplets
The lipid droplet fraction obtained from a purification procedure involving 20 eyes was mixed with 6 volumes of chloroform/methanol (2:1 v/v) at room temperature, shaken vigorously, and centrifuged at 4,000 × g for 20 min. The bottom layer was collected and evaporated in a SpeedVac, and the dried residue was immediately redissolved in 100 μl of n-hexane. HPLC was performed with a μPorasil 3.9 × 30-cm column (Waters, Franklin, MA) by a procedure described previously (48). Separation of neutral lipids and phospholipids was achieved by two different chromatographic methods. Neutral lipids were separated with linear gradients of two solutions, solvent A (100% n-hexane) and solvent B (n-hexane/isopropyl alcohol/water, 6:8:0.75 v/v), employed as follows: 0–3 min, 100% solvent A; 3–12 min, 90% solvent A and 10% solvent B; 12–30 min, 70% solvent A and 30% solvent B. For phospholipid separation, the linear gradient was as follows: 0–30 min, 100% solvent B (above) and n-hexane/isopropyl alcohol/water, 6:8:1.4, v/v. Detection was set at 206 nm and the flow rate in both cases was 1.8 ml/min. All experiments were performed in triplicate with RPE homogenates from 100 bovine eyeballs each.
TPM Imaging of the RPE
TPM imaging was performed with a Leica TCS SP2 equipped with a Ti:Sapphire laser (Chameleon XR, Coherent, Santa Clara, CA) delivering ∼140-fs pulses of 730 nm light at 90 MHz. The imaging objective was a Plan Apochromat with a 1.25 numerical aperture. After enucleation, the anterior portion of bovine eyes, including the cornea and lens, was cut out and the vitreous and retina were removed. Less than 5 h postmortem, the RPE was gently delaminated from the sclera and dissected out from the central location of the intact eye cup. Then this tissue was laid flat in the center of a glass-bottomed Matek dish, with the RPE apical side facing the coverslip. During imaging, the sample was hydrated with a solution of 9.5 mm sodium phosphate, pH 7.4, containing 137 mm NaCl and 2.7 mm KCl. The period between dissecting out the RPE and image acquisition was typically less than 30 min. All imaging was done at room temperature. The color scheme of TPM images was chosen from the standard set of Leica TCS SP2 Lookup Tables to visualize the fine details of subcellular structures. Spectral measurements initially were made from the blue to red end of the spectrum and then the reverse. Imaging was done independently four times with similar results.
Statistical Analysis
Statistical significance was determined with SigmaPlot software version 11.0 (Systat Software, Inc., Chicago) by using a one-tail type 2 t test. An asterisk denotes differences that are statistically significant (p < 0.05), and two asterisks indicate highly significant differences (p < 0.005).
RESULTS
We first investigated the formation of lipid droplets in five experimental cell lines, i.e. transformed human RPE cells called ARPE19 cells, human embryonic kidney HEK293 cells, mouse fibroblast NIH3T3 cells engineered to express LRAT (NIH/L) (42, 49), and NIH3T3 cells engineered to express both LRAT and RPE65 (NIH/RL) (42, 49). Nonengineered NIH3T3 cells served as controls for experiments involving engineered NIH3T3 cells. LRAT is required to form retinyl esters (10, 50), whereas RPE65 acts as both a retinoid isomerase (12, 14, 51) and a retinyl ester-binding protein (52). We tested whether these cells could convert all-trans-retinol to retinyl esters and use oleate to form lipid droplets. We then employed mixtures of all-trans-retinol and oleate to determine whether their products co-localized within the test cells. Finally, we repeated these experiments with native bovine RPE cells and analyzed the derived retinosomes to determine how their composition compared with that published for lipid droplets derived from other cell types.
Treatment of Cell Lines with All-trans-retinol and Oleate
We chose to expose cells to oleate because this fatty acid is converted into cytoplasmic triacylglycerol in considerably higher amounts than more toxic short chain fatty acids, such as caprylic acid (53) or palmitic acid (54), that do not readily form lipid droplets. Following incubation with either oleate or all-trans-retinol, formation of lipid droplets was observed (Fig. 1). Both control NIH3T3 cells and the NIH/L and NIH/RL cell lines engineered to produce retinyl esters formed lipid droplets following treatment with all-trans-retinol. However, fewer lipid droplets were seen in NIH3T3/RL cells as compared with NIH3T3/L cells after treatment with 10 μm all-trans-retinol. This reduction in droplets probably is due to the presence of RPE65, which uses retinyl esters as substrates. Interestingly, NIH3T3 cells that do not express LRAT and the ARPE19 cell line with dramatically reduced LRAT expression compared with primary RPE cells also formed lipid droplets following treatment with all-trans-retinol. The number of lipid droplets was observed to increase after 60 min of treatment together with a consequent reduction of cytoplasmic background staining with BODIPY493/503 (Fig. 1C). Treatment of NIH3T3 cells with all-trans-retinol alone the 1st day and a mixture of oleate and all-trans-retinol on the 2nd day showed that oleate and all-trans-retinol localized in the same lipid droplets. Fig. 2A, panel a, shows images of these cells acquired after their exposure to UV light, whereas Fig. 2A, panel b, shows staining with BODIPY493/503 that specifically targets neutral lipids. Co-localization was also observed when cells were treated with oleate the 1st day and a mixture of all-trans-retinol and oleate the 2nd day (Fig. 2B). These structures showed co-localization with PLIN2 (Fig. 2B, panel b) but not with the lysosomal marker LAMP1 (Fig. 2C, panel b).
FIGURE 1.
Fluorescence microscopy of oleic acid- and all-trans-retinol-treated cell lines. Experimental cell lines readily formed lipid droplets following treatment with oleate or all-trans-retinol. Cells were seeded at 1 million cells/plate. Three independent experiments were performed, each with seven plates. A shows lipid droplet images after treatment of ARPE19, NIH3T3, NIH3T3/L, and NIH3T3/RL cells with either oleate (100 μm) or all-trans-retinol (10 and 20 μm). B shows the lipid droplet area per cell for all five cell lines depicted in A. Quantification was accomplished with NIH Image J. software. Statistical analyses were performed with a one-tail, type 2 t test. Data shown represent means ± S.D. Their significance is denoted as follows: *, p < 0.05; **, p < 0.005. C indicates the time course of lipid droplet appearance in NIH3T3 cells following treatment with 10 μm all-trans-retinol. Arrows indicate the position of lipid droplets. The results demonstrate that lipid droplets are formed next to the plasma membranes, not only after oleate treatment but also after exposure to all-trans-retinol and retinyl esters.
FIGURE 2.
Fluorescence microscopy of NIH3T3 cells. NIH3T3 cells evidenced net formation of heterogeneous lipid droplets following incubation at 37 °C with oleate and all-trans-retinol. Cells were seeded at 1 million cells/plate. Three independent experiments were done, each with seven plates. A shows images of NIH3T3 cells after treatment with 10 μm all-trans-retinol for the first 24 h followed by a mixture of oleate (100 μm) and all-trans-retinol (10 μm) for another 24 h at 37 °C. A, panel a depicts the resulting UV-all-trans-retinol image, and panel b reveals lipid droplets stained with BODIPY493/503. The merged figure is seen in panel c. B demonstrates the fluorescence of NIH3T3 cells treated with oleate on the 1st day and a mixture of oleate and all-trans-retinol on the 2nd day, i.e. the reverse of the A protocol. B, panel a shows both UV imaging for all-trans-retinal and nuclear staining with DAPI, and panel b depicts staining with the lipid droplet marker PLIN2. Panel c displays the overlaid image of panels a and b. C exhibits NIH3T3 cells treated as described for B. C, panel a shows lipid droplets stained with BODIPY493/503, and panel b shows staining with the lysosomal marker LAMP1. Panel c is the merged image. Arrows indicate lipid droplets. The results demonstrate that all-trans-retinol and PLIN2 co-localize in lipid droplets, whereas all-trans-retinol and the lysosomal marker LAMP1 do not.
Formation of Retinyl Esters
We then tested the net production of retinyl esters following treatment with all-trans-retinol in our model cell lines. After treatment of cell lines with all-trans-retinol, retinyl esters were separated based on the length of their fatty acid chains. As expected, cell lines engineered to express LRAT, i.e. NIH/L and NIH/RL cells, produced retinyl esters (Fig. 3, C and D). The broad/merged peaks seen in Fig. 3, C and D, resulted from heterogeneity in the fatty acid composition of these esters, because our chromatographic conditions did not separate 9-cis- and 13-cis-retinyl esters, and because, for example, palmitoyl esters of these isomers eluted as sharp overlapping peaks. By contrast, a cell line such as NIH3T3 with no LRAT expression was devoid of retinyl esters (Fig. 3B). However, an increased number of lipid droplets was observed in control NIH3T3 and ARPE19 cells even in the presence of unesterified all-trans-retinol. TLC analysis of isolated lipid droplets from these cell lines showed that all-trans-retinol treatment resulted in increased triglyceride content (Fig. 3, A and B, inset). This effect was not studied further. But because of absent esterification, it could result from oxidation of retinol to retinal and then to retinoic acid, especially with increased expression of lipid-metabolizing enzymes (53). In the case of lipid droplets isolated from NIH/L and NIH/RL cell lines, treatment with all-trans-retinol did not cause a statistically significant increase in triglyceride content but rather an increase in retinyl esters as described above. These results suggest that the minimum required for lipid droplet formation is a hydrophobic substance with a propensity to aggregate, the precise chemical structure of this entity being of secondary importance.
FIGURE 3.
Retinyl ester analyses following treatment of cell cultures with 10 μm all-trans-retinol. Following treatment with 10 μm all-trans-retinol, cell homogenates were processed as described under “Materials and Methods.” The HPLCs shown are representative of triplicate experiments wherein each experiment involved 1 plate with 1 million cells. HPLCs of treated cells are depicted in solid black lines, and those of untreated cell lines are shown as dashed lines. A shows ARPE19; B shows NIH3T3; C shows NIH3T3/L, and D shows NIH3T3/RL cell chromatograms. Retinyl esters with elution times ranging from 10 to 20 min were detected only in NIH3T3/L and NIH3T3/RL cell homogenates. Insets show TLCs of triglycerides in lipid droplets isolated from untreated cells and cells treated with all-trans-retinol. The results demonstrate that the lipid droplet composition differs in the LRAT-expressing cell lines and that all-trans-retinol induces triglyceride formation in ARPE19 and NIH3T3 cell lines.
TPM Imaging of Retinosomes in Bovine RPE
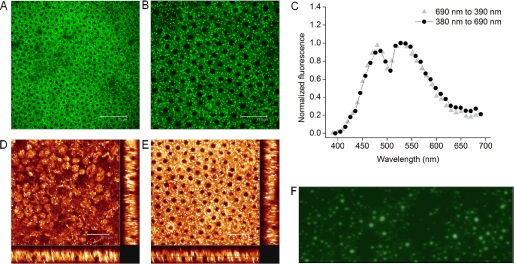
TPM is a noninvasive imaging technique that can be used to monitor retinoids within the vertebrate retina at subcellular resolution (15, 16, 20, 27, 54). This method has the great advantage of portraying endogenous retinoid fluorophores in their native environment without the need for artificial staining. Because retinosomes were discovered by applying two-photon excitation to image mouse retina and detected with TPM in human retinas (55), we first investigated if they were present in bovine eyes as well. Indeed, TPM imaging of freshly dissected, flat-mounted ex vivo bovine RPE revealed many lipid droplets visible as bright fluorescent dots located primarily near cell plasma membranes (Fig. 4A). The finer details of bovine RPE and retinosome distribution are shown in Fig. 4B. Interestingly, most bovine RPE cells contain single nuclei in contrast to mouse RPE, which predominantly house double nuclei (15). The two-photon excited emission spectrum (Fig. 4C) corresponds to that obtained from mouse RPE (16), further establishing the identity of retinosomes. A transverse view of the RPE assembled from a series of TPM optical z slice sections indicates that retinosomes extend from the apical side of the RPE toward the basal side that faces the choroid (Fig. 4, D and E). This is substantiated by supplemental movie M1, created by using 20 consecutive slice images taken every 1.85 μm along the z axis. Moreover, imaging the fraction 1 fatty layer under UV excitation revealed the characteristic autofluorescence of lipid droplets as described previously (30).
FIGURE 4.
Retinosomes in bovine RPE. A displays a large field TPM image of bovine RPE obtained from a fresh eye with the white scale bar set to 75 μm. B shows a TPM image of subcellular structures in fresh bovine RPE. Plasma membranes of cells are visible as faint black lines, and nuclei are seen as black filled circles, whereas bright fluorescent spots indicate retinosomes; scale bar, 37.5 μm. C illustrates the normalized fluorescence spectrum obtained as a function of wavelength upon scanning back from 690 to 390 nm (▴) and forward from 380 to 690 nm (●). D, main box displays an en face TPM image of the apical side of the RPE. The two transverse images, one shown at the bottom and one at the right edge, were assembled from a series of z slice images. Scale bar was set to 37.5 μm. E, main box displays an en face TPM image of the RPE with light focused close to the center of the nuclei; the two transverse images were obtained as described in D. The scale bar was set to 37.5 μm. F shows a microscopic fluorescent image of the fraction 1 fatty layer stained with BODIPY493/503. Pictures are representative snapshots of experiments performed in triplicate. The results demonstrate that bovine RPE cells contain bright fluorescent retinosomes, and the lipid droplets purified from the REP homogenates are similar to those imaged in vivo.
Isolation of Lipid Droplets from Bovine RPE
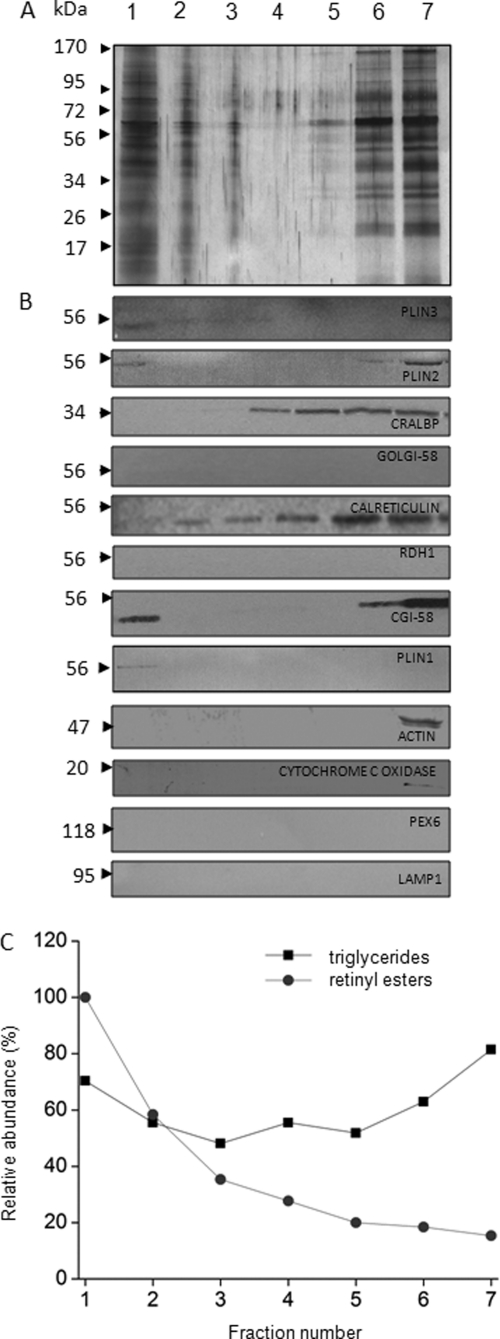
To analyze the composition of retinosomes, we designed a purification method similar to that used for lipid droplets and tested the purity of the preparation by using known markers of cellular structures (28, 33, 56). Analysis of RPE homogenates after sucrose gradient centrifugation (see “Materials and Methods”) indicated that the top lowest density fraction (fraction 1) stained with BODIPY493/503 (Fig. 4F) indicating the presence of lipid droplets. Moreover, and as expected, fraction 1 contained higher levels of retinyl esters heterogeneously acylated with fatty acids (mostly palmitate) than any other fraction (Fig. 5A). The presence of retinyl esters in fraction 1 was confirmed by MS analysis (Fig. 5, B and C). To assess the purity of the lipid droplet preparation, we tested these fractions with lipid droplet markers such as PLIN1, PLIN2, PLIN3, and CGI-58. Fraction 1 not only contained all these lipid droplet markers (Fig. 6B) but also had the highest amounts of retinyl esters (Fig. 6C), whereas triglycerides were spread throughout the gradient (Fig. 6C). Markers for cytoplasm (actin), mitochondria (cytochrome c oxygenase), and peroxisomes (PEX6) were absent from fraction 1 (Fig. 6B). Fractions 6 and 7 contained higher amounts of triglycerides as compared with fractions 2–4 (Fig. 6C), and these fractions also contained other lipid droplet markers (Fig. 6A, lanes 6 and 7). Possibly, these fractions held ER or other organelles such as peroxisomes that accounted for their increased density during centrifugation.
FIGURE 5.
Analysis of retinosomes from bovine RPE. Shown here are chromatograms of in vivo purified retinosomes with absorbance measured at 325 nm together with the retinyl ester content of the collected fractions 1–7. These fractions were obtained by sucrose gradient centrifugation of bovine RPE homogenates and analyzed by MS. Chromatograms shown are representative of triplicate experiments. A, features an HPLC analysis of retinyl esters from the seven fractions. B indicates the relative absorbance at 326 nm as a function of elution time (min), and C shows the representative ion m/z = 269 characteristic of retinol and retinyl esters. The mass spectrometry analysis was performed on fraction 1. The results demonstrate that lipid droplets isolated from RPE homogenates contain retinyl esters.
FIGURE 6.
Purification of retinosomes from bovine RPE. The purity of retinosomes obtained from bovine RPE was assessed based on both the detection of lipid droplet proteins in the top sucrose gradient fraction (fraction 1) and the absence of contaminating proteins such as calreticulin, CRALBP, actin, cytochrome c oxidase, PEX6, LAMP1, RDH1, and Golgi-58 from this fraction. Yet another criterion was the increased level of retinyl esters found in the top fraction. Images are representative of experimental triplicates. A shows SDS-PAGE of fractions collected after sucrose step centrifugation (Coomassie Blue stain). Fractions are labeled 1–7, and the lane with molecular markers indicates the molecular mass in kDa. B displays immunoblots of these collected fractions. Fraction 1 contained a notable level of CGI-58 with lesser amounts of PLIN3, PLIN2 and PLIN1. C shows the relative abundance of retinyl esters (●) and triglycerides (■) in the collected fractions. Levels of retinyl esters were highest in fraction 1, and triglycerides were broadly distributed. The results demonstrate that the protein composition of fraction 1 containing lipid droplets is different from the other fractions, and some of these proteins were previously identified in lipid droplet preparations from a variety of cell lines. Lipid droplets isolated from RPE homogenates show that retinyl esters are not the only components but that triglycerides are present as well.
Lipid Droplet Proteomics
We next performed a proteomic analysis of fraction 1 (supplemental Table S1). We found proteins implicated in lipid metabolism and trafficking (Fig. 7) and identified structural proteins and proteins involved in redox processes. Importantly, we identified proteins such as the RAB proteins, PLIN3, CGI-58, and PLIN1, previously shown by others to be recruited to lipid droplets (36, 38). RPE-specific proteins such as CRALBP and RPE65 also were present in fraction 1.
Lipid Droplet Metabolome
Because purified lipid droplet fraction 1 from bovine RPE contained both retinyl esters and triglycerides (Figs. 4C and 5A), we quantified the amounts of triglycerides/phospholipids and esters present (30). The main components of retinosomes shown in Table 1 were retinoids, but cholesterol was documented at levels similar to those described for liver stellate cells. Because retinosomes do not co-localize with lipofuscin (15), it was anticipated that the main fluorescent component of these structures, namely A2E, would not be present. Indeed, we did not detect A2E in purified fraction 1 by the HPLC analytic procedure described under “Materials and Methods.”
TABLE 1.
Analyses of bovine retinosomes and liver lipid droplet-containing fractions
Analyses were carried out on fraction 1 and performed as described under “Quantification of Neutral Lipids and Phospholipids in Isolated Lipid Droplets.” Values represent normalized mean values of experiments done in triplicate.
| Lipid droplets from bovine RPE | Lipid droplets from rat liver stellate cellsa | |
|---|---|---|
| % | % | |
| Retinyl esters | 42.6 | 39.5 |
| Retinol | 13.1 | |
| Phospholipid | 2.5 | 31.7 |
| Free fatty acids | 12.7 | |
| Cholesterol esters | 10.4 | 20.1 |
| Cholesterol | 6.5 | |
| Triglycerides | 12.2 | 8.7 |
a Values are from a study performed by Moriwaki et al. (48) with stellate cells isolated from rat liver.
DISCUSSION
Because of their physicochemical properties in aqueous environments, many hydrophobic substances tend to self-aggregate. Their storage deposits in the cytoplasm, termed lipid droplets or lipid bodies, have well defined structures and contain several other components, including cholesterol and various proteins. Specific lipid droplet-like storage particles in the eye, called retinosomes, accumulate retinyl esters that are the essential components of the visual (retinoid) cycle (8). In this study, we documented formation of lipid droplets in experimental cell lines and found that retinosomes and oleate-derived lipid droplets and their metabolic products, such as fatty acid esters and triglycerides, form together and co-localize (Fig. 2B), indicating their intrinsic structural similarity.
Homogeneity of Lipid Droplets
Using five cell types to test the heterogeneity of lipid droplets formed following exposure to oleate and all-trans-retinol, we observed that, irrespective of the precursors used to generate triglycerides or esters, these products were distributed throughout lipid droplets rather than recruited to different compartments (Fig. 2). Previous studies with hepatocytes also found that treatment with labeled fatty acid analogues resulted in incorporation of the resulting esters into the cores of lipid droplets (57, 58). Interestingly, however, a few studies indicated heterogeneity within single lipid droplets. For example, 3T3-L1 adipocytes treated with palmitate and linoleate formed heterogeneous lipid droplets (59, 60), and more importantly, different regions of a droplet contained different ratios of its components. At the resolution employed in our experiments, the lipid core of retinosomes appears to be homogeneous.
Protein Networks in Lipid Droplets, Protein Content of Retinosomes and Other Lipid Droplets
SDS-PAGE analysis of sucrose gradient fractions from our RPE homogenates showed that the retinosome fraction (fraction 1) contained a specific set of proteins identified under “Results.” These proteins had a different migration pattern than those found in fractions 2–7 (Fig. 6A). This interesting result prompted an analysis of the proteins in fraction 1 by a well established proteomic approach. Most proteins identified were found to be involved in transport, metabolism, or signaling (Fig. 7B). Several were shown to be components of lipid droplets by other investigators. For example, proteins such as PLIN2, PLIN3 (28, 61), CGI-58 (62), and PLIN1 (61) that we detected by immunoblotting were found to be recruited to lipid droplets by a variety of cell lines. Several structural proteins (33, 63), chaperone proteins (64), and redox enzymes (65) were also noted to be prominent constituents of lipid droplets. Moreover, we also corroborated the presence of caveolin-1, a major player in regulating lipid droplet phospholipid composition (66, 67), and sterol carrier protein-2, implicated in cholesterol efflux from lipid droplets (68).
The possibility that contaminating proteins are present in our preparations cannot be overlooked. Three ways the contaminating proteins could fractionate with lipid droplets should be considered. First, such proteins could be introduced during the sucrose gradient purification. Based on their buoyancy, lipid droplets must travel against the gravitational field induced by centrifugation. Second, contaminating proteins might be present that are broadly distributed throughout all fractions of the gradient. Third, certain proteins freed from their native compartments could selectively stick to lipid droplets through hydrophobic interactions. Although the markers used in this study cannot establish the absolute purity of the lipid droplet preparation, they do rule out the possibility of gross contamination by cellular components such as the Golgi, ER, peroxisomes, lysosomes, mitochondria, and cytosol. Thus proteins such as RPE65, calreticulin, malate dehydrogenase, plasma retinol-binding protein, retinaldehyde-binding protein, hemoglobin, and cellular retinoic acid-binding protein 2 identified by our proteomics study probably were recruited by one of the three mechanisms described above because their presence in the lipid droplet fraction was not confirmed by other methods. However, the cytosolic marker lactate dehydrogenase was previously found to co-fractionate with lipid droplets (69), and small amounts of abundant proteins could also be associated with retinosomes (for example, see Refs. 70, 71). Polytropic proteins identified by our studies are not likely to be recruited to the lipid bilayer based on the currently accepted model. Proteins are expected to bind to lipid droplets via lipid anchors, amphipathic helices, or hairpin-like topologies. In contrast to the currently accepted model for protein recruitment to lipid droplets, the double membrane spanning protein DGAT2 was also shown to be localized to ER and lipid droplets (72, 73). Overall, our results strengthen the argument that retinosomes in RPE cells perform functions similar to those of lipid droplets in other types of cells.
The generated protein interaction network grouped our identified proteins into three major clusters (Fig. 7A). Important nodes connecting these clusters were HSPA4 (a heat shock protein) and VIM (a structural protein). Cluster I contained heat shock proteins; cluster II mainly included dehydrogenases and transferases, whereas cluster III contained structural proteins and RPE-specific proteins. The last set of proteins is needed to form retinosome structures and carry out their various functions. Most identified proteins have transport functions (Fig. 7B). Proteins involved in metabolism made up another category. Lipid droplets are highly dynamic and can interact with other cellular organelles such as the endoplasmic reticulum (74, 75), ribosomes (41), endosomes (76), mitochondria (77), and peroxisomes (78).
Chemical Makeup of Retinosomes and Lipid Droplets from Other Tissues
We found that purified retinosomes from bovine RPE contain not only retinyl esters (esters of the visual chromophore), but triglycerides, cholesterol, and cholesterol esters as well (Fig. 6C and Table 1). Quantities of these compounds were found to be similar to those reported for lipid droplets isolated from rat liver stellate cells by Moriwaki et al. (48) but differed from values obtained for lipid droplets isolated from bovine heart where the major constituents were triglycerides (79). Moreover, macrophage foam cells (80) and oviduct epithelial cells (81) contain high amounts of cholesterol esters and low amounts of triglycerides. The higher amounts of cholesterol in foam and oviduct epithelial cells were attributed to the specific needs of these specialized cells. In addition, Bartz et al. (82) showed that some cell lines, but not all, contain significant amounts of ether lipids. The same study presented a detailed analysis of phospholipid composition for the first time and concluded that lipid droplet phospholipid composition totals ∼160 molecular species.
Together, these findings suggest that the lipid droplet is a rather dynamic tissue-specific organelle that changes its composition in response to fatty acid, cholesterol, and all-trans-retinol availability, variations in protein expression, and other signals. Moreover, our results also show a clear relationship between retinosomes and lipid droplets from other tissues.
In summary, many hydrophobic substances are critical components of cellular compartments and vital intermediates in various metabolic transformations. Across phyla, these compounds have a propensity to aggregate in aqueous micro-environments. This study suggests a close relationship between retinosomes of the eye and lipid-derived droplets in other tissues. Almost every cell contains a chemically diverse group of lipid droplets. These structures are typically present in low quantities with the exception of a few specialized cells, such as lipocytes, hepatocytes, or stellate cells. Moreover, most studies of these droplets are restricted to either mice with knockouts of key components of these structures or to experimental cell lines maintained under artificial conditions. Thus, elucidating the mechanisms of lipid droplet formation, action, and utilization in native systems still remains an important scientific challenge. Retinosomes are formed in the RPE of the vertebrate retina, where they localize to the rims of these hexagonal cells. Because retinoids are naturally fluorescent, the study of retinosomes could provide important insights into the formation and metabolism of lipid droplets in general, especially because these structures are accessible for real time TPM imaging in vivo (16, 54).
Supplementary Material
Acknowledgments
We acknowledge Grzegorz Bereta for providing the NIH3T3/L and NIH3T3/RL cell lines and David Peck, Susan Farr, and Satsumi Ross and Drs. Marcin Golczak and Philip Kiser for helping with the eye preparations. The help of Dr. Benlian Wang from the Case Western Reserve Proteomics department is greatly appreciated during the initial studies involving protein identification. We are grateful to James L. McManaman (University of Colorado) for generously providing us with anti-PLIN2 antibody. We also thank Drs. Leslie T. Webster, Jr., Johannes von Lintig, Yoshikazu Imanishi, Brian Kevany and Debarshi Mustafi for critical reading of this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants EY009339, EY021126, EY020715, and P30 EY11373. This work was also supported by TECH 09-004 from the State of Ohio Department of Development and Third Frontier Commission, European Life Scientist Organization sponsored by the Klaus Tschira Foundation, Heidelberg, Germany.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Movie 1.
“Retinal pigmented epithelium” wherein pigmented modifies epithelium and specifies a particular type of epithelium should be used. Historically, “retina pigment epithelium” was used by Uhlenhuth (1). In PubMed, the number of citations for the following terms is as follows: retina pigment epithelium, 8,244; retinal pigment epithelium, 11,670; retina pigmented epithelium, 1,123; and retinal pigmented epithelium, 1,590.
- RPE
- retinal pigmented epithelium
- A2E
- pyridinium bis-retinoid formed from two molecules of retinal and one molecule of ethanolamine
- LRAT
- lecithin:retinol acyltransferase
- TPM
- two-photon microscopy
- ER
- endoplasmic reticulum.
REFERENCES
- 1. Uhlenhuth E. (1916) J. Exp. Med. 24, 689–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kevany B. M., Palczewski K. (2010) Physiology 25, 8–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bok D. (1993) J. Cell Sci. Suppl. 17, 189–195 [DOI] [PubMed] [Google Scholar]
- 4. Flannery J. G., O'Day W., Pfeffer B. A., Horwitz J., Bok D. (1990) Exp. Eye Res. 51, 717–728 [DOI] [PubMed] [Google Scholar]
- 5. von Lintig J., Kiser P. D., Golczak M., Palczewski K. (2010) Trends Biochem. Sci. 35, 400–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Palczewski K. (2006) Annu. Rev. Biochem. 75, 743–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Travis G. H., Golczak M., Moise A. R., Palczewski K. (2007) Annu. Rev. Pharmacol. Toxicol. 47, 469–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. McBee J. K., Palczewski K., Baehr W., Pepperberg D. R. (2001) Prog. Retin. Eye Res. 20, 469–529 [DOI] [PubMed] [Google Scholar]
- 9. Saari J. C., Bredberg D. L. (1988) J. Biol. Chem. 263, 8084–8090 [PubMed] [Google Scholar]
- 10. Batten M. L., Imanishi Y., Maeda T., Tu D. C., Moise A. R., Bronson D., Possin D., Van Gelder R. N., Baehr W., Palczewski K. (2004) J. Biol. Chem. 279, 10422–10432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Moise A. R., Golczak M., Imanishi Y., Palczewski K. (2007) J. Biol. Chem. 282, 2081–2090 [DOI] [PubMed] [Google Scholar]
- 12. Jin M., Li S., Moghrabi W. N., Sun H., Travis G. H. (2005) Cell 122, 449–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Moiseyev G., Crouch R. K., Goletz P., Oatis J., Jr., Redmond T. M., Ma J. X. (2003) Biochemistry 42, 2229–2238 [DOI] [PubMed] [Google Scholar]
- 14. Moiseyev G., Chen Y., Takahashi Y., Wu B. X., Ma J. X. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 12413–12418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Imanishi Y., Batten M. L., Piston D. W., Baehr W., Palczewski K. (2004) J. Cell Biol. 164, 373–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Palczewska G., Maeda T., Imanishi Y., Sun W., Chen Y., Williams D. R., Piston D. W., Maeda A., Palczewski K. (2010) Nat. Med. 16, 1444–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Redmond T. M., Yu S., Lee E., Bok D., Hamasaki D., Chen N., Goletz P., Ma J. X., Crouch R. K., Pfeifer K. (1998) Nat. Genet. 20, 344–351 [DOI] [PubMed] [Google Scholar]
- 18. Imanishi Y., Gerke V., Palczewski K. (2004) J. Cell Biol. 166, 447–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Golczak M., Kuksa V., Maeda T., Moise A. R., Palczewski K. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 8162–8167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Golczak M., Imanishi Y., Kuksa V., Maeda T., Kubota R., Palczewski K. (2005) J. Biol. Chem. 280, 42263–42273 [DOI] [PubMed] [Google Scholar]
- 21. Maeda A., Maeda T., Imanishi Y., Golczak M., Moise A. R., Palczewski K. (2006) Biochemistry 45, 4210–4219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Van Hooser J. P., Aleman T. S., He Y. G., Cideciyan A. V., Kuksa V., Pittler S. J., Stone E. M., Jacobson S. G., Palczewski K. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 8623–8628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Van Hooser J. P., Liang Y., Maeda T., Kuksa V., Jang G. F., He Y. G., Rieke F., Fong H. K., Detwiler P. B., Palczewski K. (2002) J. Biol. Chem. 277, 19173–19182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moise A. R., Noy N., Palczewski K., Blaner W. S. (2007) Biochemistry 46, 4449–4458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Palczewski K. (2010) Trends Pharmacol. Sci. 31, 284–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kimmel A. R., Brasaemle D. L., McAndrews-Hill M., Sztalryd C., Londos C. (2010) J. Lipid Res. 51, 468–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Imanishi Y., Sun W., Maeda T., Maeda A., Palczewski K. (2008) J. Biol. Chem. 283, 25091–25102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brasaemle D. L., Barber T., Wolins N. E., Serrero G., Blanchette-Mackie E. J., Londos C. (1997) J. Lipid Res. 38, 2249–2263 [PubMed] [Google Scholar]
- 29. Beller M., Thiel K., Thul P. J., Jäckle H. (2010) FEBS Lett. 584, 2176–2182 [DOI] [PubMed] [Google Scholar]
- 30. Blaner W. S., O'Byrne S. M., Wongsiriroj N., Kluwe J., D'Ambrosio D. M., Jiang H., Schwabe R. F., Hillman E. M., Piantedosi R., Libien J. (2009) Biochim. Biophys. Acta 1791, 467–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Londos C., Brasaemle D. L., Schultz C. J., Adler-Wailes D. C., Levin D. M., Kimmel A. R., Rondinone C. M. (1999) Ann. N. Y. Acad. Sci. 892, 155–168 [DOI] [PubMed] [Google Scholar]
- 32. Tauchi-Sato K., Ozeki S., Houjou T., Taguchi R., Fujimoto T. (2002) J. Biol. Chem. 277, 44507–44512 [DOI] [PubMed] [Google Scholar]
- 33. Brasaemle D. L., Dolios G., Shapiro L., Wang R. (2004) J. Biol. Chem. 279, 46835–46842 [DOI] [PubMed] [Google Scholar]
- 34. Cermelli S., Guo Y., Gross S. P., Welte M. A. (2006) Curr. Biol. 16, 1783–1795 [DOI] [PubMed] [Google Scholar]
- 35. Brown D. A. (2001) Curr. Biol. 11, R446–R449 [DOI] [PubMed] [Google Scholar]
- 36. Martin S., Parton R. G. (2006) Nat. Rev. Mol. Cell Biol. 7, 373–378 [DOI] [PubMed] [Google Scholar]
- 37. Bozza P. T., Magalhães K. G., Weller P. F. (2009) Biochim. Biophys. Acta 1791, 540–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Guo Y., Cordes K. R., Farese R. V., Jr., Walther T. C. (2009) J. Cell Sci. 122, 749–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ploegh H. L. (2007) Nature 448, 435–438 [DOI] [PubMed] [Google Scholar]
- 40. Robenek M. J., Severs N. J., Schlattmann K., Plenz G., Zimmer K. P., Troyer D., Robenek H. (2004) FASEB J. 18, 866–868 [DOI] [PubMed] [Google Scholar]
- 41. Wan H. C., Melo R. C., Jin Z., Dvorak A. M., Weller P. F. (2007) FASEB J. 21, 167–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Golczak M., Maeda A., Bereta G., Maeda T., Kiser P. D., Hunzelmann S., von Lintig J., Blaner W. S., Palczewski K. (2008) J. Biol. Chem. 283, 9543–9554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dunn K. C., Aotaki-Keen A. E., Putkey F. R., Hjelmeland L. M. (1996) Exp. Eye Res. 62, 155–169 [DOI] [PubMed] [Google Scholar]
- 44. Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 45. Russell T. D., Palmer C. A., Orlicky D. J., Bales E. S., Chang B. H., Chan L., McManaman J. L. (2008) J. Lipid Res. 49, 206–216 [DOI] [PubMed] [Google Scholar]
- 46. Jensen L. J., Kuhn M., Stark M., Chaffron S., Creevey C., Muller J., Doerks T., Julien P., Roth A., Simonovic M., Bork P., von Mering C. (2009) Nucleic Acids Res. 37, D412–D416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wingerath T., Kirsch D., Spengler B., Kaufmann R., Stahl W. (1997) Anal. Chem. 69, 3855–3860 [DOI] [PubMed] [Google Scholar]
- 48. Moriwaki H., Blaner W. S., Piantedosi R., Goodman D. S. (1988) J. Lipid Res. 29, 1523–1534 [PubMed] [Google Scholar]
- 49. Bereta G., Kiser P. D., Golczak M., Sun W., Heon E., Saperstein D. A., Palczewski K. (2008) Biochemistry 47, 9856–9865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Batten M. L., Imanishi Y., Tu D. C., Doan T., Zhu L., Pang J., Glushakova L., Moise A. R., Baehr W., Van Gelder R. N., Hauswirth W. W., Rieke F., Palczewski K. (2005) PLoS Med. 2, e333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Redmond T. M., Poliakov E., Yu S., Tsai J. Y., Lu Z., Gentleman S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13658–13663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mata N. L., Moghrabi W. N., Lee J. S., Bui T. V., Radu R. A., Horwitz J., Travis G. H. (2004) J. Biol. Chem. 279, 635–643 [DOI] [PubMed] [Google Scholar]
- 53. Wongsiriroj N., Piantedosi R., Palczewski K., Goldberg I. J., Johnston T. P., Li E., Blaner W. S. (2008) J. Biol. Chem. 283, 13510–13519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Imanishi Y., Palczewski K. (2010) Methods Mol. Biol. 652, 247–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Guo X., Ruiz A., Rando R. R., Bok D., Gudas L. J. (2000) Carcinogenesis 21, 1925–1933 [DOI] [PubMed] [Google Scholar]
- 56. Brasaemle D. L., Rubin B., Harten I. A., Gruia-Gray J., Kimmel A. R., Londos C. (2000) J. Biol. Chem. 275, 38486–38493 [DOI] [PubMed] [Google Scholar]
- 57. Kasurinen J. (1992) Biochem. Biophys. Res. Commun. 187, 1594–1601 [DOI] [PubMed] [Google Scholar]
- 58. Wang H., Wei E., Quiroga A. D., Sun X., Touret N., Lehner R. (2010) Mol. Biol. Cell 21, 1991–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rinia H. A., Burger K. N., Bonn M., Müller M. (2008) Biophys. J. 95, 4908–4914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Xie X. S., Yu J., Yang W. Y. (2006) Science 312, 228–230 [DOI] [PubMed] [Google Scholar]
- 61. Londos C., Brasaemle D. L., Schultz C. J., Segrest J. P., Kimmel A. R. (1999) Semin. Cell Dev. Biol. 10, 51–58 [DOI] [PubMed] [Google Scholar]
- 62. Yamaguchi T., Osumi T. (2009) Biochim. Biophys. Acta 1791, 519–523 [DOI] [PubMed] [Google Scholar]
- 63. Lieber J. G., Evans R. M. (1996) J. Cell Sci. 109, 3047–3058 [DOI] [PubMed] [Google Scholar]
- 64. Jiang H., He J., Pu S., Tang C., Xu G. (2007) Biochim. Biophys. Acta 1771, 66–74 [DOI] [PubMed] [Google Scholar]
- 65. Goodman J. M. (2009) J. Lipid. Res. 50, 2148–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Blouin C. M., Le Lay S., Eberl A., Köfeler H. C., Guerrera I. C., Klein C., Le Liepvre X., Lasnier F., Bourron O., Gautier J. F., Ferré P., Hajduch E., Dugail I. (2010) J. Lipid. Res. 51, 945–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Pol A., Martin S., Fernandez M. A., Ferguson C., Carozzi A., Luetterforst R., Enrich C., Parton R. G. (2004) Mol. Biol. Cell 15, 99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Atshaves B. P., Starodub O., McIntosh A., Petrescu A., Roths J. B., Kier A. B., Schroeder F. (2000) J. Biol. Chem. 275, 36852–36861 [DOI] [PubMed] [Google Scholar]
- 69. Wolins N. E., Rubin B., Brasaemle D. L. (2001) J. Biol. Chem. 276, 5101–5108 [DOI] [PubMed] [Google Scholar]
- 70. Kiser P. D., Palczewski K. (2010) Prog. Retin. Eye Res. 29, 428–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Golczak M., Kiser P. D., Lodowski D. T., Maeda A., Palczewski K. (2010) J. Biol. Chem. 285, 9667–9682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kuerschner L., Moessinger C., Thiele C. (2008) Traffic 9, 338–352 [DOI] [PubMed] [Google Scholar]
- 73. Stone S. J., Levin M. C., Zhou P., Han J., Walther T. C., Farese R. V., Jr. (2009) J. Biol. Chem. 284, 5352–5361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wu C. C., Howell K. E., Neville M. C., Yates J. R., 3rd, McManaman J. L. (2000) Electrophoresis 21, 3470–3482 [DOI] [PubMed] [Google Scholar]
- 75. Cushman S. W. (1970) J. Cell Biol. 46, 326–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Liu P., Bartz R., Zehmer J. K., Ying Y. S., Zhu M., Serrero G., Anderson R. G. (2007) Biochim. Biophys. Acta 1773, 784–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Jägerström S., Polesie S., Wickström Y., Johansson B. R., Schröder H. D., Højlund K., Boström P. (2009) Cell Biol. Int. 33, 934–940 [DOI] [PubMed] [Google Scholar]
- 78. Binns D., Januszewski T., Chen Y., Hill J., Markin V. S., Zhao Y., Gilpin C., Chapman K. D., Anderson R. G., Goodman J. M. (2006) J. Cell Biol. 173, 719–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Christiansen K., Jensen P. K. (1972) Biochim. Biophys. Acta 260, 449–459 [DOI] [PubMed] [Google Scholar]
- 80. McGookey D. J., Anderson R. G. (1983) J. Cell Biol. 97, 1156–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Henault M. A., Killian G. J. (1993) Anat. Rec. 237, 466–474 [DOI] [PubMed] [Google Scholar]
- 82. Bartz R., Li W. H., Venables B., Zehmer J. K., Roth M. R., Welti R., Anderson R. G., Liu P., Chapman K. D. (2007) J. Lipid Res. 48, 837–847 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.