Abstract
The repair of DNA double-strand breaks (DSBs) by homologous recombination (HR) requires processing of broken ends. For repair to commence, the DSB must first be resected to generate a 3'-single-stranded DNA (ssDNA) overhang, which becomes a substrate for the DNA strand exchange protein, Rad511. Genetic studies have implicated a multitude of proteins in the process, including helicases, nucleases, and topoisomerases2–4. Here we have biochemically reconstituted elements of the resection process and reveal that it requires the nuclease, Dna2, the RecQ-family helicase, Sgs1, and the ssDNA-binding protein, Replication protein-A (RPA). We establish that Dna2, Sgs1, and RPA comprise a minimal protein complex capable of DNA resection in vitro. Sgs1 helicase unwinds the DNA to produce an intermediate that is digested by Dna2, and RPA stimulates DNA unwinding by Sgs1 in a species-specific manner. Interestingly, RPA is also required both to direct Dna2 nucleolytic activity to the 5'-terminated strand of the DNA break and to inhibit 3'→5' degradation by Dna2, actions which generate and protect the 3'-ssDNA overhang, respectively. In addition to this core machinery, we establish that both the topoisomerase 3 (Top3) and Rmi1 complex and the Mre11-Rad50-Xrs2 complex (MRX) play important roles as stimulatory components. Stimulation of end resection by the Top3-Rmi1 heterodimer and the MRX proteins is via complex formation with Sgs15,6 that unexpectedly stimulates DNA unwinding. We suggest that Top3-Rmi1 and MRX are important for recruitment of the Sgs1-Dna2 complex to DSBs. Our experiments provide a mechanistic framework for understanding initial steps of recombinational DNA repair in eukaryotes.
Keywords: Sgs1, Dna2, Top3, Rmi1, replication protein-A, DNA helicase, double-strand break resection, recombination, topoisomerase
Recent genetic studies in Saccharomyces cerevisiae identified two independent pathways capable of rapid and extensive resection of DNA DSBs: one catalyzed by the 5'→3' dsDNA exonuclease, Exo17, and a second requiring the nuclease/helicase, Dna28,9, and the 3' →5' helicase, Sgs12,4,5. In addition, the MRX-Sae2 complex (MRXS) mediates a short-range resection2,4. In cells deleted for RAD50 or MRE11, the long-range resection by Dna2/Sgs1 or Exo1 occurs at the same rate, but only after an initial delay, demonstrating that the early function of MRXS can be bypassed when chemically clean breaks are generated by HO-endonuclease2,4,5. Work using point mutations in the nuclease site of MRE11 showed that processing of HO-induced breaks is not defective10 and that Dna2 nuclease can replace Mre11-dependent nuclease activity in DSB repair11, suggesting MRX has a function in end resection independent of nuclease activity10,11.
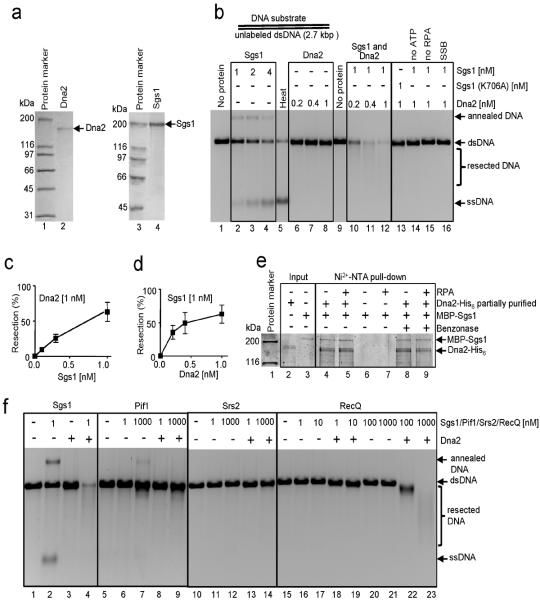
To define the roles of Dna2, Sgs1, and other proteins in this intricate in vivo process, we examined DNA resection in vitro by reconstituting a core reaction using purified proteins (Fig. 1a). Full-length Sgs1 is a vigorous DNA helicase, as recently reported12, and can fully unwind the 2.7 kb linear dsDNA substrate at nanomolar concentrations (Fig. 1b, lanes 2–4). RPA is essential for resection because it is needed for Sgs1 unwinding at these concentrations (lane 15), and could not be replaced by Escherichia coli ssDNA-binding protein (SSB) (lane 16)12; this stimulatory effect of RPA on Sgs1 is the consequence of species-specific interaction between Sgs1 and RPA13 superimposed on a non-specific stimulation due to ssDNA binding12. Dna2 showed no detectable nuclease or helicase activity on the dsDNA, as expected due to its weak unwinding capability14 (lanes 6–8). However, in the presence of Sgs1, Dna2 degraded the DNA (lanes 10–12), showing that up to 2.7 kb can be readily processed. Consistent with previous findings4,9, degradation was not observed with nuclease-dead Dna2 (K677R), whereas helicase-dead Dna2 (K1080E) supported resection, albeit with a lower efficiency (Supplementary Fig. 2). A DNA end was required for Dna2-dependent degradation, as no DNA cleavage was observed on a covalently-closed circular dsDNA, with or without Sgs1, even if it contained a 450 nucleotide (nt) `bubble' of non-complementary ssDNA (Supplementary Fig. 3). Processing of linear dsDNA (Fig. 1b) required Sgs1 helicase activity (lane 13) and ATP (lane 14). The amount of DNA resected depended on both Sgs1 and Dna2 concentrations (Fig. 1c,d). Thus, Sgs1, Dna2, and RPA comprise a minimal set of proteins required for DNA end resection.
Figure 1. Sgs1 and Dna2 resect DNA in a reaction dependent on yeast RPA.
a, Purified Dna2 (80 ng) and Sgs1 (880 ng) stained with Coomassie Brilliant Blue. b, Linear pUC19 dsDNA incubated with Sgs1 and/or Dna2, and RPA (3 μM); “SSB”: SSB substituted for RPA; “Heat”: heat-denatured dsDNA; ”annealed DNA”: partial unwinding and annealing of DNA30. c, and d, Quantification of experiments as shown in b. e, Dna2 and Sgs1 physically interact in the absence or presence of RPA (lanes 4 and 5). f, Resection by Dna2 (1 nM) is specific for yeast Sgs1 helicase; RPA is 3 μM.
To determine whether Dna2 and Sgs1 interact functionally, we replaced Sgs1 with other helicases. Neither S. cerevisiae Pif1 nor Srs2 could replace Sgs1, even when at a 1000-fold higher concentration than Sgs1 (Fig. 1f). E. coli RecQ could partially replace Sgs1, but a 1000-fold higher concentration (1 μM) was needed to equivalently degrade the substrate (Fig. 1f, lane 23). Collectively, these results imply a specific interaction between Sgs1 and Dna2. To establish whether Dna2 and Sgs1 physically interact, we tested whether partially purified His6-tagged Dna215 could pull-down MBP-tagged Sgs112 (Fig. 1e). The results show that recombinant Sgs1 and Dna2 do directly interact, independently of DNA, and that RPA neither blocked nor is required for the interaction. Thus, resection catalyzed by Dna2-Sgs1-RPA is likely a concerted process where nucleolytic cleavage occurs concomitantly with DNA unwinding by Sgs1.
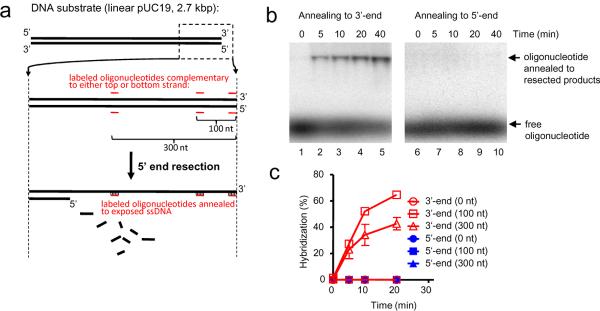
Resection of a DSB in mitotically growing cells is dependent on the nuclease activity of Dna22,4,11 and is largely, but not absolutely, limited to the 5'-strand16. This directionality is essential to form 3'-ending ssDNA, which is a primer for DNA synthesis from the joint molecule intermediate17. However, it was not clear how this specificity is achieved in the Sgs1/Dna2 pathway, because Dna2 degrades both 5'- and 3'-terminated ssDNA14. To determine which strands are resected in our reconstituted system, we designed a set of 32P-labeled oligonucleotides that are complementary to either the 5'- or 3'-terminated strands at positions that are directly adjacent to, 100 nt, or 300 nt from the DNA end (Fig. 2a). These oligonucleotides were used as hybridization probes to determine the DNA strand, and length, exposed by resection. The probes for the 100 nt and 300 nt positions annealed exclusively to the 3'-terminated strand indicating that extensive resection is limited to the 5'-strand, leaving the 3'-end largely intact (Fig. 2b). Resection required Dna2 because, without it, both the 3'- and 5'-terminated strands are intact and unpaired, as expected from simple unwinding by Sgs1 helicase (Supplementary Fig. 4a). Unexpectedly, but consistent with the ability of Dna2 to degrade 5'- and 3'-strands, we did not detect hybridization using probes for sequences adjacent to the DNA end (Fig. 2c) suggesting that, while extensive degradation occurs only on the 5'-strand, both strands are degraded in the vicinity of the DNA break. Loss of ssDNA at the 3'-terminus was confirmed independently in assays using dsDNA 32P-labeled at the 3'-end, where the combined action of Dna2-Sgs1-RPA resulted in a rapid loss of signal and the appearance of rapidly migrating degradation products (Supplementary Fig. 4b). Kinetic analysis of resection at the 0, 100 nt, and 300 nt sites is shown in Fig. 2c; resection to 100 nt is slightly faster than to 300 nt, as expected. We note that, because the enzyme concentrations and incubation time are limiting, resection originating from the opposite DNA end is unlikely to reach the end analyzed by hybridization and, therefore, does not affect our analysis (Supplementary Figure 4c,d). In summary, with the exception of the region directly next to the end, resection of DNA by Dna2-Sgs1-RPA is limited to the 5'-terminated DNA strand.
Figure 2. Sgs1, Dna2, and RPA preferentially resect the 5'-terminated strand of a DNA break.
a, Assay. b, Annealing of oligonucleotide to resection products (0.5 nM Sgs1, 0.5 nM Dna2, and 3 μM RPA) using probe complementary to either top (3'-) or bottom (5'-) strand, 300 nucleotides from end. c, Quantification of DNA end resection at various distances from DSB, based on experiments as shown in b.
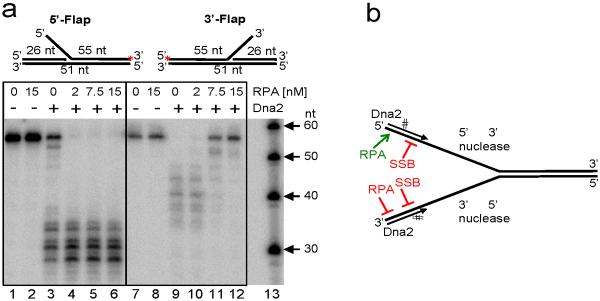
Although consistent with in vivo results1, our findings are surprising because characterization of Dna2 nuclease suggests that both strands should be fully degraded14. To determine the basis for the observed strand bias in end resection, we used DNA containing either 5'- or 3'-ssDNA tails that were used previously to study Dna218. In the absence of RPA, Dna2 does indeed degrade both 5'- and 3'-terminated ssDNA (Fig. 3a, lanes 3 and 9). Unexpectedly, however, RPA blocks degradation of only the 3'-terminated strand; inhibition is concentration dependent and is maximal when RPA exceeds the amount required to saturate the ssDNA (lanes 10–12). In contrast, RPA stimulates the 5' → 3' nucleolytic capacity of Dna2 (lanes 4–6). Thus, RPA enforces discrimination of the 3'- and 5'- terminated strands. These results are in agreement with the interpretation of earlier observations using G4-containing DNA15. To confirm the observations using a substrate that resembles a DNA end unwound by a helicase, we also used a synthetic DNA duplex containing a Y-structure (Supplementary Fig. 5a,b). In agreement, the results showed that RPA blocked nucleolytic degradation of the 3'-ssDNA arm, whereas it stimulated degradation of the 5'-ssDNA arm (Supplementary Fig. 5a,b). The nuclease activity of Dna2 was limited to the ssDNA region, as reported14 and Dna2 did not unwind the 31 bp dsDNA (Supplementary Fig. 5a). The effect of RPA is species-specific, as SSB completely inhibited Dna2 nuclease activity (Supplementary Fig. 6), consistent with the reported physical interaction between RPA and Dna219,20. These results collectively show that RPA selectively enhances the 5'→3' degradative capacity of Dna2 while repressing the 3'→5' degradative activity (Fig. 3b). This specific interaction with RPA thus alters the functionality of Dna2 and explains the strand bias of DNA end resection.
Figure 3. RPA promotes 5'→3' degradation by Dna2 and inhibits 3'→5' degradation.
a, Duplex DNA substrates containing either a 5'- or 3'-ssDNA flap (red asterisk indicates the 32P-label) incubated with Dna2 (15 nM) and indicated RPA. b, Illustration summarizing results from panel a and Supplementary Figs. 4 and 5 showing modulation of ssDNA nuclease activities of Dna2.
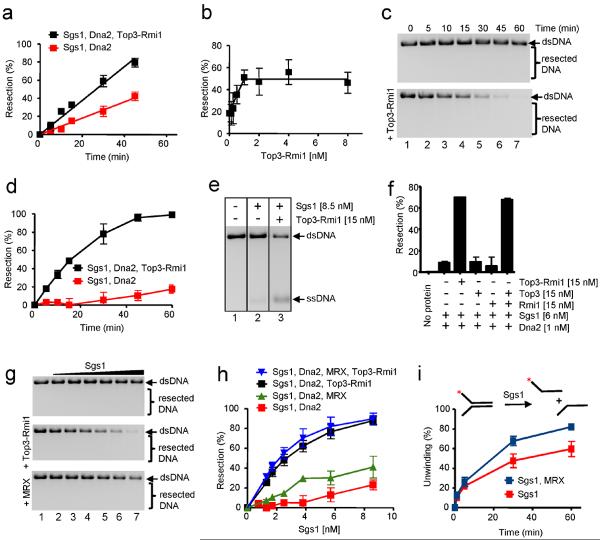
Sgs1 also physically interacts with Top35 and Rmi121,22 to dissolve double Holliday junctions to complete recombination (P. C., J. Plank, C. Bachrati, I. Hickson, S. C. K., submitted). Surprisingly, in vivo, both top3Δ and rmi1Δ mutants showed similar defects to an sgs1Δ mutant in resection, suggesting that the functional unit in resection is an Sgs1-Top3-Rmi1 complex4, although it remained possible that the proteins are required for Sgs1 protein stability in vivo22. When examined in vitro (Fig. 4a and Supplementary Fig. 7a), Top3-Rmi1 stimulated resection by Dna2-Sgs1-RPA by ~2-fold. A titration with Top3-Rmi1 at a fixed Sgs1 and Dna2 concentration (each 0.5 nM) shows concentration-dependent stimulation with saturation occurring at ~1 nM Top3-Rmi1 (Fig. 4b), suggesting that the proteins function in a nearly equimolar protein complex.
Figure 4. Top3-Rmi1 and MRX complexes stimulate DNA resection by Dna2-Sgs1-RPA.
a, Resection kinetics: Sgs1 (0.3 nM), Dna2 (1 nM), RPA (3 μM), and Top3-Rmi1 heterodimer (10 nM). b, Top3-Rmi1-dependent stimulation of DNA resection: Sgs1 (0.5 nM), Dna2 (0.5 nM) and RPA (3 μM). c, Resection kinetics in high salt buffer (5 mM Mg2+ and 100 mM Na+): Sgs1 (7.5 nM), Dna2 (1 nM), RPA (3 μM ), and Top3-Rmi1 heterodimer (15 nM). d, Quantification of experiments as in c. e, DNA unwinding in high salt buffer; RPA (3 μM). f, Resection in high salt buffer; RPA (3 μM). g, Stimulation of resection in high salt buffer by Top3-Rmi1 (15 nM) and MRX (20 nM) using Dna2 (1 nM), RPA (3 μM), Sgs1 (0, 1.3, 1.7, 2.5, 3.8, 5.7 and 8.6 nM). h, Quantification of experiments as in g. i, Unwinding of Y-structure DNA in high salt buffer: Sgs1 (200 pM); RPA (2.25 nM); and MRX (5 nM).
To determine the basis for this stimulation, we examined the effect of Top3-Rmi1 on the activities of Sgs1 and Dna2. The Y-structure oligonucleotide substrate (Supplementary Fig. 5) was used because it permits a quantitative evaluation of either the helicase activity of Sgs112 or the nuclease activity of Dna2. Sgs1 is the most active RecQ-helicase reported12, yet unexpectedly, we discovered that Top3, Rmi1, and Top3-Rmi1 stimulated the initial rate of DNA unwinding by Sgs1 (Supplementary Fig. 7b). The Sgs1 concentration required for half-maximal unwinding is 68 pM in the absence of Top3-Rmi1, and only 30 pM in the presence of Top3-Rmi1, corresponding to a 2.3-fold increase in apparent affinity (Supplementary Fig. 7c). We found that 10-fold more Top3 than Top3-Rmi1 was required to stimulate the helicase activity of Sgs1 (Supplementary Fig. 7e,f). The stimulatory effect of Top3-Rmi1 was completely RPA-dependent, as no increase in unwinding by Sgs1 was observed in the absence of RPA (Supplementary Fig. 7g,h). Furthermore, Top3-Rmi1 increased the DNA-dependent ATPase activity of Sgs1 in the presence of RPA ~2.5-fold (Supplementary Fig. 7d). Thus, Top3-Rmi1 promotes the helicase activity of Sgs1 by increasing its affinity to DNA. However, Top3-Rmi1 did not stimulate the nuclease activity of Dna2 (Supplementary Fig. 8, lane 2 vs. 4). We therefore conclude that Top3-Rmi1 stimulates DNA end resection by recruiting Sgs1 to DNA, rather than by potentiating the nuclease activity of Dna2.
To further examine the stimulation of Sgs1-Dna2 by Top3-Rmi1, we used elevated concentrations of Mg2+ (5 mM magnesium acetate) and Na+ (100 mM sodium acetate), which are suboptimal for Sgs1 helicase activity, thereby better revealing substrate specificity12, and which are more representative of in vivo conditions. As expected, higher salt concentrations significantly inhibited DNA end resection by Dna2-Sgs1-RPA (Fig. 4c,d). However, addition of Top3-Rmi1 resulted in a striking restoration of resection (~11-fold stimulation). Top3-Rmi1 also stimulated unwinding of the 2.7 kb DNA substrate by Sgs1 alone (Fig. 4e). Finally, under these conditions, both Top3 and Rmi1 are required for the stimulation of resection, because neither alone is effective (Fig. 4f; Supplementary Fig. 9), in agreement with the in vivo resection data4. Therefore, the stimulatory role of Top3-Rmi1 in DNA end resection is evident when conditions are more physiological. Collectively, these results show that Top3 and Rmi1 stimulate DNA end resection by promoting the helicase activity of Sgs1 by enhancing its affinity for DNA.
In vivo, DSB resection is delayed in the physical absence of MRX2,4, whereas both the yield and resection rate in cells expressing nuclease-deficient MRX (mre11-D56N) are indistinguishable from wild type10. These findings led Llorente and Symington to suggest that MRX is needed as a structural complex to recruit a resection nuclease which is not Exo110. In agreement, in vivo observations by Budd and Campbell revealed that Dna2 nuclease can function in X-ray repair in the absence of Mre11 nuclease, but not in the physical absence of Mre1111. To determine whether MRX has a role in our reconstituted biochemical system, we used the in vitro conditions that revealed a nearly essential role for Top3-Rmi1. We found that MRX modestly promoted DNA end resection by Dna2-Sgs1-RPA, but the magnitude of the stimulation (~2-fold) was far less than for Top3-Rmi1 (Fig. 4g,h and Supplementary Fig. 10a,b). When Top3-Rmi1 was present, MRX did not further stimulate resection by Dna2-Sgs1-RPA (Fig. 4h). MRX modestly promoted DNA unwinding by Sgs1 (Fig. 4i and Supplementary Fig. 10c,d), whereas it did not affect Dna2 nuclease activity (Supplementary Fig. 8 (lane 2 vs. 6). These findings are consistent with the previously reported physical interaction between Sgs1 and Mre116, and we propose that MRX stimulates DNA end resection by recruiting Sgs1 to the DSBs.
In this work, we reconstituted elements of the machinery that processes DSBs in preparation for DNA repair. Our biochemical findings show that Sgs1, Dna2, and RPA are the minimal essential components. By promoting the binding of Sgs1 to DNA, the Top3-Rmi1 complex is stimulatory, and is effectively essential at approximately physiological conditions. The MRX complex can stimulate resection modestly, consistent with a recruitment function in resection in vivo that can be bypassed. MRX has a high affinity for DNA ends and is a sensor of DSBs in vivo23. Mre11 interacts directly with Sgs16, suggesting that stimulation of resection by MRX might be due to recruitment of Sgs1 to DNA ends, consistent with our biochemical results and in vivo interpretations10,11. In addition to signaling and recruitment functions, MRXS nuclease is essential during meiosis to remove Spo11 from the 5'-terminated strand of DSBs24, and in mitotic cells to remove adducts of topoisomerase II covalently linked to DSB ends that arise from drug treatment3,25. Thus, the MRXS proteins are especially important for initial processing of DNA ends that contain adducts which could impede processing. In the absence of such adducts, Sgs1-Dna2-RPA in a complex with Top3-Rmi1 comprise the core of one of the two major pathways for extensive DNA resection. We show that Top3-Rmi1 and MRX potentiate resection by recruiting Sgs1 to the site of the break, and propose that this recruitment is consistent with both the requirement for Top3-Rmi1 in vivo and the delay when MRX is physically absent. Sgs1 interacts with Dna2 to resect DNA in an RPA-regulated manner. Based on the in vitro data presented here, we propose a model for DNA end resection by Dna2-Sgs1-RPA (Supplementary Fig. 1). First, the Sgs1-Top3-Rmi1 complex is recruited to a DSB in a step that can be augmented by the physical presence of MRX. Sgs1 unwinds dsDNA, and the single-strands of DNA are coated with RPA. Concomitantly, and mediated through direct interaction with Sgs1, Dna2 preferentially degrades the 5'-terminated DNA strand. RPA promotes the degradation of this strand and inhibits the degradation of the 3'-terminated strand. Thus, this reconstituted reaction recapitulates steps required to resect a DSB to produce a 3'-ssDNA overhang. Results that parallel the major findings reported here have been independently obtained by the Sung laboratory (Niu et al., (2010), this issue of Nature). Finally, this eukaryotic resection complex shows intriguing functional parallels to the resection machine of bacteria, RecBCD (or AddAB), where the recombination-promoting complex (post Chi-recognition) comprises a helicase subunit (RecB) unwinding 3'→5' (equivalent to Sgs1); a slower translocation subunit (RecD) traveling 5'→3' on the complementary strand (equivalent to Dna2); and a nuclease domain that is threaded onto the end of a DNA strand but that acts endonucleolytically to process the 5'-terminated DNA strand to produce 3'-tailed duplex DNA26. Furthermore, RecBCD delivers RecA to the 3-terminated processed ssDNA via an essential interaction with a RecA-loading domain26,27; because the C-terminus of Sgs1 physically interacts with Rad5128, it is also possible that Sgs1 and its homologues may coordinate processing with DNA pairing in a related manner in eukaryotes29.
Methods Summary
Unless indicated otherwise, the DNA substrate used for end resection experiments was linear pUC19 dsDNA (2.7 kb). The reaction products were separated by electrophoresis, and visualized by ethidium bromide staining. For clarity, we present all DNA gels as inverted images. The interaction of Sgs1 with Dna2 was determined using Ni2+-nitrilotriacetic acid (NTA) pull-down assays using Dna2 tagged with His6 and MBP-tagged Sgs1. The directionality of resection was determined by hybridization using radiolabeled strand-specific oligonucleotide probes. All oligonucleotide-based DNA substrates were 32P-labeled, and visualized by autoradiography. The resection assays with the dsDNA substrates containing either 5'- or 3'-ssDNA flaps were carried out as described previously15. Helicase assays were carried out as described previously12.
Methods
DNA substrates
The dsDNA substrates containing 5'- or 3'-ssDNA flaps were prepared as described previously18. The oligonucleotide-based DNA substrates were described previously12. Unless otherwise indicated, the DNA substrate used for resection assays was unlabelled pUC19 dsDNA that had been linearized with Hind III and purified by phenol-chloroform extraction and ethanol precipitation.
Proteins
Sgs1, RPA, SSB, and RecQ proteins were expressed and purified as described12,31–33. Top3, Rmi1, and Top3-Rmi1 heterodimer were prepared as described (P.C., J.L. Plank, and S.C.K., submitted). All of the above proteins are free of nuclease contamination. Srs2, Pif1, and MRX proteins were generous gifts from Xavier Veaute (Institute of Cellular and Molecular Radiation Biology, France); Jean-Baptiste Boulé and Alain Nicolas (Institut Curie, Paris); and Patrick Sung (Yale University), respectively.
Purification of Dna2
Dna2 was expressed from a modified pGAL:DNA29 vector that contains N-terminal Flag and HA tags and a C-terminal His6-tag, in protease-deficient S. cerevisiae strain WDH66834. This protocol describes purification from 4 liters of cell culture. Yeast cells were grown to an OD600 ~ 0.6 in a standard synthetic SD medium, lacking uracil, and supplemented with both glycerol (3%) and lactic acid (2%). Expression of Dna2 was induced by adding galactose (2%) for 6 hours. All subsequent steps were carried out on ice or at 4°C. Pelleted cells were resuspended in 40 ml TBSG-PI buffer (25 mM Tris HCl (pH 7.5), 100 mM NaCl, 10% glycerol, 5 mM β-mercaptoethanol, 10 μg/ml leupeptin, 1 mM phenylmethylsulphonyl fluoride (PMSF) and protease-inhibitor cocktail (Sigma, P8340, diluted 1:1000)), and lysed in French press. The lysed cells were collected by centrifugation at 58,000 g for 30 minutes; the supernatant was transferred to a new centrifuge tube and spun again as above. The cleared extract was then supplemented with imidazole (10 mM) and incubated batch-wise with 5 ml Ni2+-NTA agarose (Qiagen) for 1 hour. The resin was washed extensively with TBSG-PI buffer containing imidazole (10 mM) batch-wise and the bound proteins were eluted from the column with 400 mM imidazole in TBSG-PI. The fractions containing proteins were pooled, and loaded on a HiTrap Heparin HP column (5 ml; GE Healthcare) at 2.5 ml/minute in HEP buffer A (25 mM Tris HCl (pH 7.5), 100 mM NaCl, 10% glycerol, 1 mM β-mercaptoethanol, 10 μg/ml leupeptin, 1 mM PMSF and protease-inhibitor cocktail (Sigma, P8340, diluted 1:1000)), and eluted with HEP buffer B (the same as HEP buffer A, but with 600 mM NaCl) at 2.5 ml/minute. Fractions containing proteins were pooled, diluted 1:1 with TBS buffer (50 mM Tris HCl (pH 7.5), 150 mM NaCl), and incubated batch-wise with M2 anti-FLAG affinity resin (0.5 ml; Sigma) for 30 minutes. The resin was then washed with TBS buffer, and Dna2 was eluted with TBS supplemented with 3XFLAG peptide (150 μg/ml; Sigma). Fractions containing protein were pooled, diluted with ½ volume water and 1 volume of Q buffer A (25 mM Tris HCl (pH 7.5), 100 mM NaCl, 10% glycerol, and 5 mM β-mercaptoethanol), and then loaded on HiTrapQ column (1 ml; GE Healthcare) at 0.8 ml/minute. The column was washed with Q buffer A, and Dna2 was eluted with Q buffer B (the same as Q buffer A, but with 600 mM NaCl). Fractions containing protein were pooled, small aliquots were frozen in liquid nitrogen, and stored at −80 °C. The final protein concentration was quite low, and thus estimated by densitometry by comparison with dilution series of broad range protein marker (BioRad) on 10% polyacrylamide gel stained with Coomassie Brilliant Blue. The protein yield was ~3.7 μg and concentration ~27 nM. An identical procedure without the FLAG step was employed to prepare the enriched Dna2 protein used for the affinity pull-down experiments. This Dna2 preparation was more concentrated but less pure (~20%), as shown previously15. The Dna2 protein used in Fig. 3 was prepared as described previously9.
DNA resection assays
The resection assays with the dsDNA substrates containing either 5'- or 3'-ssDNA flaps were carried out as described previously15. All other resection assays contained, unless indicated otherwise, 25 mM Tris acetate (pH 7.5), 1 mM dithiothreitol, 2 mM magnesium acetate, 250 μg/ml BSA, 1 mM ATP, 1 mM phosphoenolpyruvate (Sigma), 80 U/ml pyruvate kinase (Sigma), 200 ng linear pUC19 DNA substrate (7.6 nM molecules; 41 μM nucleotides) and the indicated proteins. Reactions that were conducted at the “high salt” condition, where indicated, were in standard buffer containing 100 mM sodium acetate and 5 mM magnesium acetate. Unless otherwise indicated, the reactions were assembled on ice, initiated by adding ATP, and carried out for 30 minutes at 30°C, in a volume of 15 μl. The reactions were terminated with 5 μl of stop buffer (150 mM EDTA, 2% SDS, 30% glycerol, 0.1% bromphenol blue) and 1 μl of proteinase K (14–22 mg/ml, Roche) for 30 minutes at 30 °C, unless otherwise indicated, and analyzed by electrophoresis in 1% agarose in the presence of 0.05 μg/ml ethidium bromide. Gels were analyzed using an AlphaImager HP (Alpha Innotech) imaging station, and are presented as the inverted image. Resection was quantified by measuring disappearance of the substrate dsDNA band. All error bars show the standard error from 2–5 independent experiments as determined by GraphPad Prism 5.0.
To analyze the directionality of resection by hybridization with radiolabeled oligonucleotide probes (as in Fig. 2), a standard reaction was first carried out as described above. Upon termination, the reaction was diluted so that the DNA concentration was 1 nM (molecules). The oligonucleotide probe, which was 32P-labeled at the 5'-terminus (2 nM, molecules), was added to the diluted mixture, as well as PNK buffer (New England Biolabs) to final concentrations of 7 mM Tris-HCl (pH 7.6), 1 mM MgCl2, and 0.5 mM dithiothreitol. The sequences of the oligonucleotide probes are: for 5'-resection at 0 nt (GCATGCCTGCAGGTCGACTC), 100 nt (GGCGTTACCCAACTTAATCG), 300 nt (AGCCAGCCCCGACACCCGCC), and for 3'-resection at 0 nt (GAGTCGACCTGCAGGCATGC), 100 nt (CGATTAAGTTGGGTAACGCC), and 300 nt (GGCGGGTGTCGGGGCTGGCT). The mixture was then heated to 70 °C for 5 minutes, and cooled to room temperature over ~ 2 hours. The products were then separated by electrophoresis in 1% agarose and analyzed by Storm 860 PhosphorImager (GE Healthcare). The percentage of DNA hybridization was calculated from the proportion of annealed vs. free oligonucleotides, assuming 100% efficiency of annealing.
Helicase assays
The helicase assays were carried out as described previously12. The products were separated by native 10% polyacrylamide gel electrophoresis and detected by autoradiography.
Ni2+-nitrilotriacetic acid (NTA) pull-down assays
The Sgs1 protein used for Ni2+-NTA pull-down experiments contained the MBP tag but lacked the His10-tag12. The Dna2 protein used for the pull-down experiments contained a His6-tag, and was prepared by a procedure similar to that published previously15, which provides a higher yield but lower purity of the recombinant polypeptide. The identity of proteins from both purifications was verified by western blotting (data not shown). Purified proteins (1 – 2 μg in a final volume of 150 μl) were incubated together at room temperature in 20 mM Tris HCl (pH 7.5), 10% glycerol, 0.1% NP40, 100 mM NaCl and 10 mM imidazole, with or without 12.5 Units of Benzonase nuclease (Novagen) for 20 minutes at room temperature. Ni2+-NTA agarose (50 μl, Qiagen) was then added, incubated for 30 minutes, and the resin was washed with buffer. The bound proteins were eluted in the same buffer containing 600 mM imidazole, and 20% of eluate was analyzed by electrophoresis in 8% polyacrylamide, stained with Sypro Orange (Invitrogen), and detected by Storm 860 PhosphorImager (GE Healthcare).
Supplementary Material
Acknowledgements
We thank J.-B. Boulé, A. Nicolas, X. Veaute, B. Rad, J. L. Plank and P. Sung for purified proteins and DNA substrates, L. Symington for discussions, and to W. D. Heyer and the members of the Kowalczykowski and Campbell laboratories for their comments on the manuscript. We are particularly grateful to Patrick Sung for communicating his results to us prior to their publication. This work was supported by a Swiss National Science Foundation Fellowship (P.C.), and National Institutes of Health Grants GM-78666 (J.L.C), GM-41347 (S.C.K.), and GM-62653 (S.C.K.).
References
- 1.Krogh BO, Symington LS. Recombination proteins in yeast. Annu. Rev. Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- 2.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gangloff S, McDonald JP, Bendixen C, Arthur L, Rothstein R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol. Cell. Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiolo I, et al. Srs2 and Sgs1 DNA helicases associate with Mre11 in different subcomplexes following checkpoint activation and CDK1-mediated Srs2 phosphorylation. Mol. Cell. Biol. 2005;25:5738–5751. doi: 10.1128/MCB.25.13.5738-5751.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Szankasi P, Smith GR. A role for exonuclease I from S. pombe in mutation avoidance and mismatch correction. Science. 1995;267:1166–1169. doi: 10.1126/science.7855597. [DOI] [PubMed] [Google Scholar]
- 8.Budd ME, Choe WC, Campbell JL. DNA2 encodes a DNA helicase essential for replication of eukaryotic chromosomes. J. Biol. Chem. 1995;270:26766–26769. doi: 10.1074/jbc.270.45.26766. [DOI] [PubMed] [Google Scholar]
- 9.Budd ME, Choe W, Campbell JL. The nuclease activity of the yeast DNA2 protein, which is related to the RecB-like nucleases, is essential in vivo. J. Biol. Chem. 2000;275:16518–16529. doi: 10.1074/jbc.M909511199. [DOI] [PubMed] [Google Scholar]
- 10.Llorente B, Symington LS. The Mre11 nuclease is not required for 5' to 3' resection at multiple HO-induced double-strand breaks. Mol. Cell. Biol. 2004;24:9682–9694. doi: 10.1128/MCB.24.21.9682-9694.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Budd ME, Campbell JL. Interplay of Mre11 nuclease with Dna2 plus Sgs1 in Rad51-dependent recombinational repair. PLoS ONE. 2009;4:e4267. doi: 10.1371/journal.pone.0004267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cejka P, Kowalczykowski SC. The full-length Saccharomyces cerevisiae Sgs1 protein is a vigorous DNA helicase that preferentially unwinds holliday junctions. J. Biol. Chem. 2010;285:8290–8301. doi: 10.1074/jbc.M109.083196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cobb JA, Bjergbaek L, Shimada K, Frei C, Gasser SM. DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J. 2003;22:4325–4336. doi: 10.1093/emboj/cdg391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bae SH, Seo YS. Characterization of the enzymatic properties of the yeast Dna2 helicase/endonuclease suggests a new model for Okazaki fragment processing. J. Biol. Chem. 2000;275:38022–38031. doi: 10.1074/jbc.M006513200. [DOI] [PubMed] [Google Scholar]
- 15.Masuda-Sasa T, Polaczek P, Peng XP, Chen L, Campbell JL. Processing of G4 DNA by Dna2 helicase/nuclease and replication protein A (RPA) provides insights into the mechanism of Dna2/RPA substrate recognition. J. Biol. Chem. 2008;283:24359–24373. doi: 10.1074/jbc.M802244200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zierhut C, Diffley JF. Break dosage, cell cycle stage and DNA replication influence DNA double strand break response. EMBO J. 2008;27:1875–1885. doi: 10.1038/emboj.2008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X, Stith CM, Burgers PM, Heyer WD. PCNA is required for initiation of recombination-associated DNA synthesis by DNA polymerase delta. Mol. Cell. 2009;36:704–713. doi: 10.1016/j.molcel.2009.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kao HI, Veeraraghavan J, Polaczek P, Campbell JL, Bambara RA. On the roles of Saccharomyces cerevisiae Dna2p and Flap endonuclease 1 in Okazaki fragment processing. J. Biol. Chem. 2004;279:15014–15024. doi: 10.1074/jbc.M313216200. [DOI] [PubMed] [Google Scholar]
- 19.Bae SH, Bae KH, Kim JA, Seo YS. RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature. 2001;412:456–461. doi: 10.1038/35086609. [DOI] [PubMed] [Google Scholar]
- 20.Bae KH, et al. Bimodal interaction between replication-protein A and Dna2 is critical for Dna2 function both in vivo and in vitro. Nucleic Acids Res. 2003;31:3006–3015. doi: 10.1093/nar/gkg422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mullen JR, Nallaseth FS, Lan YQ, Slagle CE, Brill SJ. Yeast Rmi1/Nce4 controls genome stability as a subunit of the Sgs1-Top3 complex. Mol. Cell. Biol. 2005;25:4476–4487. doi: 10.1128/MCB.25.11.4476-4487.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang M, et al. RMI1/NCE4, a suppressor of genome instability, encodes a member of the RecQ helicase/Topo III complex. EMBO J. 2005;24:2024–2033. doi: 10.1038/sj.emboj.7600684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stracker TH, Theunissen JW, Morales M, Petrini JH. The Mre11 complex and the metabolism of chromosome breaks: the importance of communicating and holding things together. DNA Repair (Amst) 2004;3:845–854. doi: 10.1016/j.dnarep.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 24.Neale MJ, Pan J, Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436:1053–1057. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hartsuiker E, Neale MJ, Carr AM. Distinct requirements for the Rad32(Mre11) nuclease and Ctp1(CtIP) in the removal of covalently bound topoisomerase I and II from DNA. Mol. Cell. 2009;33:117–123. doi: 10.1016/j.molcel.2008.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dillingham MS, Kowalczykowski SC. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol. Mol. Biol. Rev. 2008;72:642–671. doi: 10.1128/MMBR.00020-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anderson DG, Kowalczykowski SC. The translocating RecBCD enzyme stimulates recombination by directing RecA protein onto ssDNA in a χ-regulated manner. Cell. 1997;90:77–86. doi: 10.1016/s0092-8674(00)80315-3. [DOI] [PubMed] [Google Scholar]
- 28.Wu L, Davies SL, Levitt NC, Hickson ID. Potential role for the BLM helicase in recombinational repair via a conserved interaction with RAD51. J. Biol. Chem. 2001;276:19375–19381. doi: 10.1074/jbc.M009471200. [DOI] [PubMed] [Google Scholar]
- 29.Nimonkar AV, Ozsoy AZ, Genschel J, Modrich P, Kowalczykowski SC. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc. Natl. Acad. Sci. U. S. A. 2008;105:16906–16911. doi: 10.1073/pnas.0809380105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Handa N, Morimatsu K, Lovett ST, Kowalczykowski SC. Reconstitution of initial steps of dsDNA break repair by the RecF pathway of E. coli. Genes Dev. 2009;23:1234–1245. doi: 10.1101/gad.1780709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.LeBowitz J. Biochemical mechanism of strand initiation in bacteriophage lambda DNA replication. Johns Hopkins University; Baltimore, MD: 1985. [Google Scholar]
- 32.Harmon FG, Kowalczykowski SC. RecQ helicase, in concert with RecA and SSB proteins, initiates and disrupts DNA recombination. Genes Dev. 1998;12:1134–1144. doi: 10.1101/gad.12.8.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alani E, Thresher R, Griffith JD, Kolodner RD. Characterization of DNA-binding and strand-exchange stimulation properties of y-RPA, a yeast single-strand-DNA-binding protein. J. Mol. Biol. 1992;227:54–71. doi: 10.1016/0022-2836(92)90681-9. [DOI] [PubMed] [Google Scholar]
- 34.Solinger JA, Lutz G, Sugiyama T, Kowalczykowski SC, Heyer WD. Rad54 protein stimulates heteroduplex DNA formation in the synaptic phase of DNA strand exchange via specific interactions with the presynaptic Rad51 nucleoprotein filament. J. Mol. Biol. 2001;307:1207–1221. doi: 10.1006/jmbi.2001.4555. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.