Abstract
Asymptomatic Epstein-Barr virus (EBV) reactivations periodically occur in oral mucosa-associated lymphoid tissues. Until now, EBV reactivation has been diagnosed by serologic profiles that suggest virus replication. Serologic responses, however, are delayed and do not necessarily indicate ongoing replicative activity. The aim of the present study was to establish in healthy carriers parameters for a molecular diagnosis of reactivated EBV infection. Recent studies emphasized the association of an increase in peripheral-B-cell viral load with replicative activity at remote sites. Therefore, real-time PCR was used to quantitate EBV genomes in the peripheral blood mononuclear cells (PBMC) (viral load) and plasma samples (viremia) of 22 healthy EBV-seropositive blood donors over a period of 15 months. Furthermore, transcription of the immediate-early gene encoding BZLF1 was investigated in the PBMC of all volunteers. Serology suggested reactivation in nine donors, of whom all but one showed at least once a significant increase in viral load. Another five individuals also exhibited significant changes in viral load but no serologic response. Of the 13 volunteers with significant increases in viral load, 6 had a period of viremia accompanying the rise in viral load. A stable viral load without viremia and negative serology was seen in eight adults. BZLF1 mRNA was undetectable throughout. We conclude that for healthy subjects serology underestimates the frequency of asymptomatic EBV reactivations. Prospective examination of peripheral viral load and viremia is suitable for the exact diagnosis of EBV reactivation, which might be of advantage for immunocompromised patients in whom EBV reactivations are considerably harmful.
The Epstein-Barr virus (EBV) is a ubiquitous human gammaherpesvirus that shows strong B lymphotropism. It is closely associated with several malignancies, including Burkitt's lymphoma, Hodgkin's disease, and B-cell lymphoma in immunocompromised hosts (for a review, see reference 22). After infection the virus persists latently in the host for life. Like other herpesviruses, EBV reactivates periodically in its host as a means of infecting new B lymphocytes as well as new individuals. The site of long-term persistence is the resting memory B cell (2, 15). In the latently infected host a roughly constant number of infected B cells circulates in the peripheral blood; however, this number varies considerably between individuals (12, 29). In these memory B cells EBV can be detected and the viral load can be quantified (11, 13, 32). Reactivation is thought to occur upon recirculation of EBV-infected resting memory B cells into lymphoid tissue (25). As resting memory B cells respond to signals in the secondary lymphoid organs, EBV-carrying cells are assumed to become reactivated by physiologic signals, e.g., B-cell receptor stimulation (26), in the absence of CD40 activation or viral latent membrane protein-1 expression (1). Response to such stimuli leads to terminal differentiation into plasma cells and initiation of the viral replicative cycle, characterized by expression of ZEBRA (BamHI-Z fragment, EBV replication activator) as well as of viral capsid antigen or glycoprotein gp350/220, leading to release of infective virions. When virions are released into peripheral circulation, the result may be viremia. In parallel, new naive B lymphocytes become infected in the oropharyngeal lymphoid tissue, and after a phase of EBV-driven transformation and differentiation into a germinal-center-like phenotype, these cells are released into the periphery as resting memory B cells (25). An increase in peripheral viral load (typically expressed as the number of EBV genome copies per microgram of DNA) is thought to register this event.
In the immunocompetent individual the occurrence of EBV reactivation leading to immortalization of B lymphocytes is strongly regulated by cytotoxic T lymphocytes specific for lytic as well as latent antigens (22). Under healthy conditions, this system is well balanced and almost no specific symptoms indicate these events, but in patients under immunosuppression, e.g., after solid organ or stem cell transplantation or with an acquired T-cell immunodeficiency like human immunodeficiency virus infection, viral reactivation is suspected to cause severe complications. Several studies have connected lytic activity and reactivation with rejection episodes in transplant recipients (3, 8), posttransplant lymphoproliferative disorder (PTLD) (20, 23, 33), and clinical disease activity in patients with multiple sclerosis (34). Immunosuppression seems to trigger not only reactivation but also physically or psychologically challenging situations that result in diminished cell-mediated immunity (6, 14, 18).
Until now, EBV reactivation has been diagnosed only by means of serology (8, 16, 24), by determination of transformational activity in throat washings (20, 36), or by EBV DNA analysis of saliva samples (10, 16, 18). Serology, however, reflects reactivation only retrospectively and does not represent the actual event. Especially under immunosuppression, serologic responses may be delayed, but as immediate diagnosis of EBV reactivation is of importance to prevent or avert severe complications in certain patient groups, we wanted to investigate whether direct molecular markers can be used to immediately and reliably diagnose EBV reactivation. We assessed peripheral-B-cell viral load, plasma viremia, and expression of ZEBRA mRNA in a 15-month follow-up of 22 healthy EBV carriers and correlated our findings to serologic responses indicative of EBV reactivation.
MATERIALS AND METHODS
Study population.
EDTA-anticoagulated whole blood of 22 healthy voluntary blood donors (18 men and 4 women; median age, 55 years) was examined over a period of 15 months. The health of the blood donors was screened by physicians each time they donated blood. Donors with symptoms indicative of upper respiratory tract infection or general malaise were excluded from the study for the respective time points. Donors were monitored irregularly dependent on their voluntary appearance for blood donation, but with a minimal time period between two donations of 10 (men) or 12 (women) weeks to prevent iron deficiency according to the German guidelines for blood donation. For all donors, whole blood cell counts were determined at each time point by use of a Beckman Coulter STKS hematology analyzer.
EBV serology.
EBV serology was determined using two different sets of enzyme-linked immunosorbent assays (ELISA). The first ELISA employed purified recombinant EBV antigens (Euroimmun, Gross Grönau, Germany) and assessed immunoglobulin G (IgG) antibodies against Epstein-Barr nuclear antigen 1 (EBNA1) as well as IgM antibodies against EBV early antigen (EA). Seropositivity was defined for EBNA1 IgG as values of >20 relative units/ml and was defined for EA IgM as a ratio (extinction sample/extinction calibrator) of >1. Individuals positive for EBNA1 IgG were considered to have latent EBV infection. Serologic EBV reactivation was defined in latently infected donors with seropositivity for EA IgM. Results were compared to those of a set of ELISA employing a mixture of latent and lytic EBV antigens and determining levels of whole anti-EBV IgG, IgM, and IgA (Enzygnost Dade-Behring) (24, 31). Plasma samples were preabsorbed with anti-IgG antibodies prior to analysis of IgM and IgA. IgG and IgM seropositivity was defined with a final optical density (OD) of >0.1, and IgA positivity was defined with an OD of >0.6. IgG anti-EBV antibody values were quantified with the α-method as recommended by the manufacturer, and the results were expressed as international units per milliliter. Donors with positive EBV IgM values or with IgG values of >650 IU/ml and positive IgA values were considered to have reactivated EBV infection as described elsewhere (24).
DNA isolation.
Blood samples were collected in 9-ml EDTA-containing tubes (Sarstedt, Nürnbrecht, Germany) and forwarded to the laboratory, where they were centrifuged at 600 × g for 10 min within 6 h after collection. Serum was aliquoted in portions of 1 ml and immediately stored at −80°C until further use. Peripheral blood mononuclear cells (PBMC) were prepared by standard density centrifugation (Ficoll separation solution; Biochrom, Berlin, Germany) and divided into equal shares. One part was directly stored at −80°C, while the other was mixed with guanidine isothiocyanate-containing lysis buffer (QIAGEN, Hilden, Germany) containing 1% 2-mercaptoethanol and then also stored at −80°C. DNA was isolated from 0.5 × 107 to 1 × 107 PBMC and from 1 ml of plasma of each sample by using the QIAamp DNA blood mini kit (PBMC) and midi kit (plasma) (QIAGEN) according to the manufacturer's protocol. The extracted DNA was eluted in 200 μl of elution buffer. DNA concentrations were measured by spectrophotometer (Ultrospec 2000; Pharmacia Biotech, Freiburg, Germany) at a wavelength of 260 nm. PBMC DNA was adjusted to a concentration of 50 μg/ml.
Real-time PCR for EBV genome quantification.
For detection and quantification of EBV DNA in PBMC and plasma samples, an EBV-specific multiplex real-time PCR assay was employed using the ABI PRISM 7700 sequence detection system (Perkin-Elmer [PE] Applied Biosystems, Foster City, Calif.) as described recently (11). This assay amplifies part of the EBNA1 gene together with a sequence of the human C-reactive protein (CRP) gene that serves as a control for amplifiability as well as for quantification of genomic DNA. EBNA1 and genomic CRP DNA were distinguished with probes labeled with two different reporter dyes. The master mix consisted of 2× universal mastermix (PE Applied Biosystems), a 300 nM concentration of each EBNA1, primer, a 40 nM concentration of each CRP primer, a 250 nM concentration of the EBNA1, and a 100 nM concentration of the CRP probe as well as DNase-free water for a final reaction volume of 50 μl, including 10 μl of PBMC DNA and 20 μl of plasma DNA, respectively. The thermal cycler conditions have already been described previously (11). With each analysis, calibration curves were run in parallel using DNA extracted from the Namalwa cell line. Namalwa DNA dilutions as well as samples were always run in duplicate and with two negative water blanks (no template controls) in every analysis. Correct amplification of the EBNA1 amplicon was proven by blank end vector cloning (Invitrogen) after amplification with Pfu DNA polymerase (GIBCO). Gel electrophoresis of the amplification product showed a single band at approximately 108 bp. Sequencing of the vector insert revealed a sequence identical to part of the EBNA1 open reading frame (GenBank accession no. V01555).
Calculation of the number of EBV copies per microgram of PBMC DNA was performed as reported earlier (11). The number of EBV copies per milliliter of plasma (N) was calculated as follows: N = AEBV × 10/3.3, with AEBV being the amount of EBV-specific DNA (picograms of Namalwa DNA), 10 being the factor for adjusting elution volume and sample volume to 1 ml of plasma (1/ml), and 3.3 being the conversion factor for single-copy genes (picograms of DNA/copy) (11). The amount of genomic DNA in plasma preparations was calculated from the coamplified CRP sequence as follows: C = ACRP × 10, with C being the amount of genomic DNA per milliliter of plasma, ACRP being the amount of CRP-specific DNA (picograms of Namalwa DNA), and 10 being the factor for adjusting elution volume and sample volume to 1 ml of plasma. The detection limits of the two assays were determined by using different plasmid concentrations (pCMVEBNA [Invitrogen], containing the whole EBNA1 gene) either amplified in the presence of 0.5 μg of EBV-negative human DNA or EBV-negative human plasma, respectively. Detection probabilities—as given in Table 1—were calculated by Probit analysis by employing eight replicates of each plasmid dilution and SPSS software.
TABLE 1.
Detection probabilities of multiplex real-time PCR for simultaneous detection of EBV-specific and genomic DNA
| Detection probabilitya (%) | EBV genomes detected in:
|
|
|---|---|---|
| PBMC (no. of copies/μg of DNA) | Plasma (no. of copies/ml of plasma) | |
| 50 | 3.2 | 12.2 |
| 95 | 5.5 | 17.5 |
| 99 | 8.6 | 19.8 |
Probabilities were calculated by means of Probit analysis using SPSS software. Eight replicates of known plasmid copy numbers containing the whole BKRF1 open reading frame (EBNA1) were amplified in the presence of 0.5 μg of EBV-negative DNA for assessment of detection limits in PBMC. For detection limits in plasma, eight replicates of known plasmid copy numbers were isolated from the background of 1 ml of EBV-negative plasma and amplified.
RNA isolation and RT.
Total RNA from up to 107 PBMC was extracted by the QIAGEN spin column method (RNeasy mini kit) exactly as described in the manufacturer's protocol manual. The extracted RNA was eluted in 30 μl of RNase-free water. Ten microliters of eluted RNA was then directly transcribed into cDNA by means of the TaqMan Gold reverse transcription (RT)-PCR kit (PE Applied Biosystems) according to the manufacturer's protocol. The total volume employed in the RT reaction was 30 μl. Cycling parameters were as follows: 10 min at 25°C for annealing of random hexamers, 30 min at 48°C for RT, and 5 min at 95°C for RT inactivation.
Real-time RT-PCR for ZEBRA mRNA detection.
A novel multiplex real-time PCR assay was developed using the ABI PRISM 7700 sequence detection system for detection of ZEBRA mRNA encoded by the viral open reading frame BZLF1. DNA sequence was obtained from the GenBank sequence database, accession number V01555. Primer-probe sequences were designed by means of Primer Express Software (PE Applied Biosystems) and purchased from TIB MOLBIOL, Berlin, Germany. Considering two known polymorphisms of the BZLF1 open reading frame (GenBank accession no. M20820 and M20821), primers and probe were situated within the aligned consensus sequence with OMIGA software (version 1.1.3; Oxford Molecular, Oxford, United Kingdom). BZLF1 sequences read as follows: for the forward primer, 5′ TCG CAT TCC TCC AGC GAT; for the reverse primer, 5′ AAC CTG GAG ACA ATT CTA CTG TTC AA; and for the probe, 5′ VIC-CAC CAA TGT CTG CTA GCT GTT GTC CTT GGT. Checkerboard titration of the MgCl2 versus the probe concentrations as well as of the forward primer versus the reverse primer concentrations were performed for BZLF1. Furthermore, the optimal annealing and extension temperature (from 58 to 64°C) was assessed. BZLF1 was coamplified together with a DNA sequence of the human β-actin gene (β-actin control reagents; PE Applied Biosystems). By using limited β-actin primers, coamplification of β-actin was shown not to affect the amplification efficiency of BZLF1 (Fig. 1). To determine the threshold concentration of BZLF1 detection, a semilogarithmic dilution series ranging from 100 to 10−4.5 cDNAs from the EBV-positive Burkitt's lymphoma cell line P3HR-1/13 (kindly provided by H. Wolf, Regensburg, Germany) was used. BZLF1 cDNA was reliably detected up to a dilution of 10−3.5. The following amplification conditions were set: an initial 10 min at 95°C, followed by 45 cycles of 15 s at 95°C and 1 min at 58°C. PCR runs were carried out with a total volume of 50 μl containing 5 μl of cDNA. The final concentrations of primers and probes were as follows: 400 nM for BZLF1 primers, 300 nM for the BZLF1 probe, 36 nM for β-actin primers, and 30 nM for the β-actin probe, plus a 1.5 mM concentration of additional MgCl2. Again, every sample was run in duplicate together with two water blanks (no template controls) and two samples containing cDNA from the P3HR1/13 cell line serving as a positive control.
FIG. 1.
Effect of β-actin cDNA coamplification on ZEBRA cDNA amplification efficiency and detection limit by using multiplex real-time PCR. Ct, cycle of threshold; error bars, SD.
RESULTS
Stability assessment of EDTA-anticoagulated blood samples.
To distinguish viremia from EBV DNA released from ruptured B lymphocytes, we first determined PBMC stability dependent on storage time. EDTA-anticoagulated blood samples of eight immunosuppressed patients after kidney transplantation were kept without any further processing at 4°C for 0, 3, 6, 12, 24, 48, and 72 h. Afterwards, plasma was separated from cells by gentle centrifugation (as described above). PBMC were isolated, and the amounts of viremia, viral load, and levels of genomic DNA in plasma were measured. At 0 h no EBV-specific DNA was detected in any plasma sample. In six of eight donors, PBMC harbored EBV DNA ranging in number from 65 to 18,110 copies/μg of DNA. Genomic DNA was detectable immediately after blood drawing in four of eight plasma samples in amounts ranging from 0.3 to 2.4 ng/ml (mean value, 1.2 ng/ml). Thereafter, genomic DNA values increased to a mean value of 3.1 ng/ml at 72 h of storage. On the other hand, EBV DNA was amplified in the plasma of only two patients after 12 or 24 to 72 h of storage, respectively. While these samples had PBMC viral loads of 65 and 472 copies/μg of DNA, even samples with viral loads as high as 10,012 and 18,110 copies/μg of DNA, respectively, did not show EBV-specific DNA amplification in any plasma sample. We concluded that a time span of up to 6 h between blood drawing and processing was safe to distinguish between viral particle DNA and EBV DNA from destroyed B cells.
In addition, we quantified amounts of genomic DNA found in plasma samples of our study population of healthy blood donors. Genomic DNA was detected in the majority of samples (90 of 134; 67%). However, no correlation was observed between viremia and levels of genomic DNA. The mean value of genomic DNA was 2.6 ng/ml for all plasma samples containing genomic DNA, except those with detectable viremia; the mean value for samples displaying viremia was 1.8 ng/ml and thus considerably lower. Regarding the low numbers of EBV virions found in plasma samples (range, 10 to 35 copies/ml), no attempt was undertaken to prove infectivity of the viral DNA by means of cord blood transformation assay.
Serology.
Each of the 22 blood donors was determined to be seropositive for EBNA1 IgG. Three individuals had persistent EA IgM antibodies, whereas in one donor a single conversion to EA IgM between the fifth and sixth donation was registered. Thus, 4 out of 22 subjects (18.2%) showed evidence of reactivation by means of the Euroimmun ELISA. Using the Enzygnost ELISA, all 22 healthy subjects were positive for EBV IgG, but for only 3 of them were titers >650 IU/ml. Of these, two were also positive for EBV IgA; thus, these subjects have reactivated or persistently active EBV infection. A detectable EBV IgM was seen in six donors, with five individuals showing persistence of EBV IgM throughout. Of these, three were also positive for EBV IgA. Another five donors positive for EBV IgA but with normal IgG titers and negative for IgM were considered not to have had EBV reactivation (donors D, F′, S, V, and Z). In summary, 9 out of 22 individuals (40.9%) had reactivated EBV infection as shown by means of serology (Table 2).
TABLE 2.
Serologic profiles indicative of EBV reactivation found in 10 out of 22 healthy subjects during 15 months of follow-up
| ELISA | Anti-EBV antigen | Follow-up samplea | Resultb for donor
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| B | E | G | H | J | M | O | U | Y | |||
| Euroimmun | EA IgM | 1 | + | − | − | − | + | − | + | − | − |
| 2 | + | − | − | − | + | − | + | − | − | ||
| 3 | + | − | − | − | + | − | + | − | − | ||
| 4 | + | − | − | − | + | − | + | − | − | ||
| 5 | + | − | − | − | + | − | + | − | − | ||
| 6 | − | + | + | − | + | − | − | ||||
| 7 | − | + | − | ||||||||
| Enzygnost Dade-Behring | EBV IgM | 1 | + | + | − | − | + | − | + | − | + |
| 2 | + | + | − | − | + | − | − | − | + | ||
| 3 | + | + | − | − | + | − | + | − | + | ||
| 4 | + | + | − | − | + | − | − | + | + | ||
| 5 | + | + | − | − | + | − | − | − | + | ||
| 6 | + | − | + | − | − | − | + | ||||
| 7 | − | − | + | ||||||||
| EBV IgA | 1 | + | + | + | + | − | + | − | − | − | |
| 2 | + | + | + | + | − | + | − | − | − | ||
| 3 | + | + | + | − | − | + | − | + | − | ||
| 4 | + | + | + | + | − | + | − | + | − | ||
| 5 | + | + | + | + | − | + | − | − | − | ||
| 6 | + | + | − | + | − | − | − | ||||
| 7 | + | − | − | ||||||||
| EBV IgG | 1 | 412 | 189 | 707 | 108 | 188 | 501 | 364 | 196 | 212 | |
| 2 | 75 | 193 | 800 | 87 | 149 | 738 | 339 | 194 | 333 | ||
| 3 | 385 | 178 | 896 | 124 | 162 | 783 | 409 | 189 | 397 | ||
| 4 | 347 | 292 | 648 | 165 | 213 | 923 | 447 | 223 | 416 | ||
| 5 | 197 | 139 | 681 | 188 | 203 | 1,007 | 353 | 125 | 328 | ||
| 6 | 182 | 188 | 201 | 666 | 318 | 274 | 378 | ||||
| 7 | 161 | 289 | 442 | ||||||||
Follow-up samples were obtained within a period of 15 months with at least 10 (men) or 12 (women) weeks between two appointments.
Results are reported as positive or negative, except for those obtained with EBV IgG, which are given in international units per milliliter. Boldface type is used for titer that are >650 IU/ml, which in the presence of EBV IgA were regarded as indicative of reactivation.
EB viral load and plasma viremia in blood donors with serologic EBV reactivation.
The nine healthy adults with positive serologic reactivation markers exhibited individual patterns of viremia and viral load during 15 months of follow-up. In three donors viremia that preceded or accompanied significant viral load increases was observed. The individual courses of PBMC viral load could be combined into two major categories: group I, donors with a single or repeated conversion from undetectable to detectable viral load (n = 5), and group II, donors with significant increases in viral load when viral load was detectable continuously (n = 3). An increase in viral load was considered to be significant if any value exceeded the upper threefold standard deviation (SD) of the individual mean viral load. The mean individual load was calculated from all values, not considering obvious peak values. Blood donor U could not be assigned to either group.
Examples of representative individuals with conversion to detectable viral loads (group I) are given in Fig. 2. Donors B, E, and O had persistent anti-EBV/EA IgM titers throughout but only showed short episodes of positive viral load, which were accompanied by viremia in donors B (not shown) and E. Donor H also showed a single phase of detectable viral load, although EA IgM was negative first. However, EA IgM turned positive after viral load had already fallen below the detection threshold again. Donor M showed a fluctuating viral load with two episodes of detectable viral load on his first and second as well as fourth and fifth appointments. In response to the first viral load increase he developed remarkable high EBV IgG titers, while anti-EBV IgA stayed positive throughout.
FIG. 2.
Group I contains donors who again exhibit a detectable viral load and have serologic evidence of EBV reactivation during 15 months of follow-up.
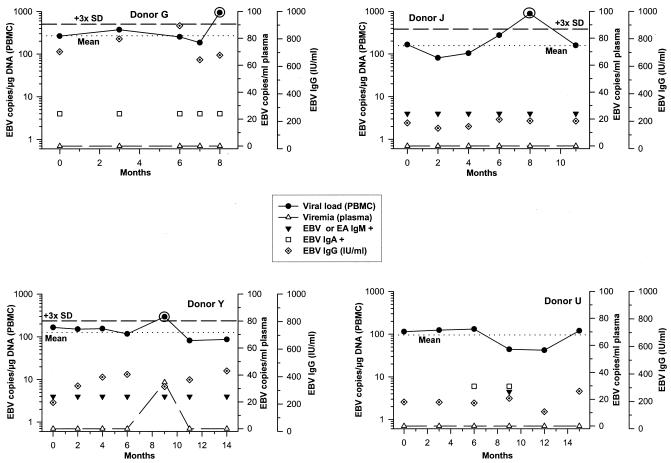
In group II, all PBMC samples were constantly positive for EBV DNA (Fig. 3). Viral loads of donors G, J, and Y, however, were not stable and increased significantly after 8 to 9 months. At these appointments, viral load levels were each beyond the upper threefold SD of the individual mean viral load (Fig. 3), while viremia was detectable only in donor Y. Despite persisting anti-EBV/EA IgM titers in donors J and Y, only a single viral load increase was observed in both donors. Similarly, donor G consistently displayed IgG titers of >650 IU/ml accompanied by positive anti-EBV IgA but showed a significant rise in viral load only at his last appointment. Donor U neither had viremia nor showed an obvious viral load increase despite conversion to detectable EBV IgA and IgM levels after 6 and 9 months, respectively.
FIG. 3.
Group II contains donors with serologic evidence of EBV reactivation and significant increases in viral load during 15 months of follow-up (except donor U, who did not show obvious changes in viral load). Black circled values of viral load represent obvious peak values not considered for calculation of the mean individual viral load (indicated by dotted lines; the upper threefold SD of the mean is indicated by short dashes).
Molecular parameters of EBV reactivation in donors without serologic evidence.
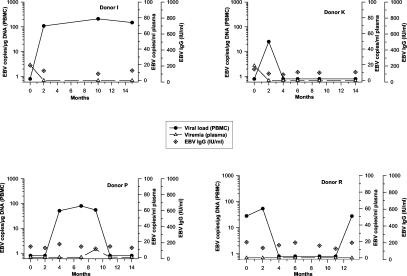
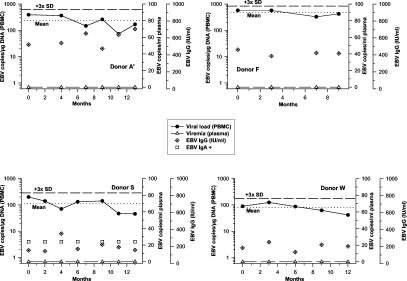
For the 13 blood donors without serologic reactivation markers, the following results were determined. Three more donors (donors I, K, and P) showed viremia accompanying or preceding conversion to detectable viral loads. Donors L (not shown) and R each revealed two episodes of detectable viral load during follow-up, which afterwards turned negative again. Virus reactivation has to be considered in these five donors, although EBV or EA IgM remained undetectable and viremia was not observed (Fig. 4). The remaining eight study subjects (donors A′, D, F, F′, S, V, W, and Z) exhibited stable viral loads, with conspicuously large amounts in donors F (mean, 472 EBV copies/μg of DNA) and A′ (mean, 233 EBV copies/μg of DNA) (Fig. 5). At no time point was viremia assessed for any of these eight individuals. None of the viral load values rose above the upper threefold SD of the mean individual load. Despite considerably high levels of viral load, they were thus not regarded as having undergone viral reactivation. At no time point was ZEBRA mRNA detected in PBMC of any sample. Leukocyte numbers were stable in all blood donors and within the normal range (4.3 to 10/nl).
FIG. 4.
Virologic parameters in donors without serologic evidence of EBV reactivation but significant changes in viral load and viremia.
FIG. 5.
Donors without serologic or virologic evidence of reactivated EBV infection. Mean individual viral loads are indicated by dotted lines; the upper threefold SD of the mean is indicated by short dashes.
In summary, 15 of 22 individuals (68.2%) of our collective survey had evidence of reactivated EBV infection by means of virologic and/or serologic investigations.
DISCUSSION
The aim of our study was to define molecular EBV reactivation markers in healthy EBV carriers, under the supposition that this group shows a close association between serologic and virologic reactivation parameters. We concluded from earlier studies that viremic episodes together with distinct increases in peripheral-B-cell viral loads might represent virus replication at remote mucosa-associated lymphoid tissues (12, 20, 36). The present study indicates that in healthy subjects serologic responses suggestive of virus replication are indeed accompanied by short episodes of increased viral load, although the different kinetics of serology and virologic markers suggest different control mechanisms: serologic investigations did not determine exact reactivation time points; on the contrary, serology of most donors suggested a continuous replicative activity. Moreover, changes of EBV viral load together with viremia occurred more frequently as opposed to serologic reactivation profiles. The reason for this remains uncertain but might be due to a lower sensitivity of the serologic assays used. In the only donor with serologic evidence of EBV reactivation despite stable viral loads and lack of viremia (donor U), a phase of viremia or viral load increase might have been missed owing to the long examination intervals.
A detectable viral load was observed in all individuals at least once; 98 of 138 PBMC samples contained EBV DNA (71%). By using our multiplex real-time PCR assay, we have already shown earlier that the cross-sectional detection rate of EBV DNA was 84.6% in PBMC of healthy blood donors (11). The detection rate for EBV DNA in a recent study was 28% for bone marrow recipients by means of nested semiquantitative PCR (28). Another study assessing Light Cycler-based PCR found a detectable viral load in 37.5% of healthy individuals (4). A study investigating the value of semiquantitative nested PCR in the detection of the BamHI W repeat region of the EBV genome came up with detection rates of 55.6% for healthy subjects (7). Our quantitative multiplex real-time PCR method thus appears to be more sensitive but equally suitable for routine diagnosis than other conventional or real-time PCR assays, although we have to keep in mind the different samples' backgrounds.
A previous study of healthy individuals reported a viremia detection rate of 10% by means of conventional PCR and centrifugation of concentrated serum (5). In our study group, plasma samples of six blood donors (27%) contained EBV DNA during 15 months of follow-up. Viremic episodes usually preceded significant changes in viral load. Viral load increases were seen without viremia, while the latter did not occur alone. We conclude that for high-risk patients, e.g., those at high risk for PTLD, quantification of viral load together with determination of plasma EBV DNA levels is advisable for an immediate diagnosis of EBV reactivation as viremia appears first.
In contrast to recent studies (13, 17) but corresponding to the opinion of Hopwood et al. (7), we refrain from stating single predictive values of viral load or viremia for reactivation or even PTLD. Our data clearly show that a single high viral load does not necessarily have to be seen as reactivation parameter. This is in accordance with our observations of transplant patients (30) as well as with results from Gärtner et al. (B. C. Gärtner, K. Kortmann, M. Schäfer, N. Mueller-Lantzsch, U. Sester, H. Kaul, and H. Pees, Letter, J. Clin. Microbiol. 38:2458, 2000), who reported no correlation between single viral load determinations and serologic parameters for different patients groups or healthy controls.
Contrary to data from previous studies (9, 19, 27), we did not manage to amplify BZLF1-specific mRNA in samples from our healthy cohort. This negative result might be due to the fact that those previous studies analyzed BZLF1 mRNA expression partly in solid-organ transplant recipients (9, 27) and patients with infectious mononucleosis (19). However, it is noteworthy that two other studies investigating transplant recipients also did not find ZEBRA mRNA even by nested RT-PCR (21, 35). We therefore conclude that ZEBRA mRNA detection in peripheral B cells is not appropriate for diagnosis of asymptomatic EBV reactivation—at least with healthy subjects.
In conclusion, we suggest that viremic episodes and conversion to detectable viral load levels or, in individuals with a consistently detectable viral load, an increase beyond the upper threefold SD of the mean individual load has to be regarded as viral reactivation. Although prospective monitoring was necessary to detect significant changes in viral load, molecular analysis instead of serologic examinations might help to better define actual EBV replication. Appropriate diagnosis of EBV reactivation might be favorable for immunocompromised patients in whom EBV reactivations are considerably harmful.
Acknowledgments
This work was supported by grant 800/401 (B3) from the University of Lübeck School of Medicine.
We are indebted to the work of Nina Klapproth, Karen Wittich, and Stefanie Wichmann for expert technical assistance. We thank all volunteers of the Center for blood donation, University of Lübeck, Lübeck, Germany, for their participation in this study.
REFERENCES
- 1.Adler, B., E. Schaadt, B. Kempkes, U. Zimber-Strobl, B. Baier, and G. W. Bornkamm. 2002. Control of Epstein-Barr virus reactivation by activated CD40 and viral latent membrane protein1. Proc. Natl. Acad. Sci. USA 99:437-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babcock, G. J., and D. A. Thorley-Lawson. 1998. EBV persistence in memory B cells in vivo. Immunity 9:395-404. [DOI] [PubMed] [Google Scholar]
- 3.Babel, N., F. Schwarzmann, N. Prang, M. Jaeger, H. Wolf, F. Kern, H. D. Volk, and P. Reinke. 2001. Association between Epstein-Barr virus infection and late acute transplant rejection in long-term transplant patients. Transplantation 72:736-738. [DOI] [PubMed] [Google Scholar]
- 4.Brengel-Pesce, K., P. Morand, A. Schmuck, M. J. Bourgeat, M. Buisson, G. Bargues, M. Bouzid, and J. M. Seigneurin. 2002. Routine use of real-time quantitative PCR for laboratory diagnosis of Epstein-Barr virus infections. J. Med. Virol. 66:360-369. [DOI] [PubMed] [Google Scholar]
- 5.Gan, Y., L. Sullivan, and J. W. Sixbey. 1994. Detection of cell-free Epstein-Barr virus DNA in serum during acute infectious mononucleosis. J. Infect. Dis. 170:436-439. [DOI] [PubMed] [Google Scholar]
- 6.Gleeson, M., D. B. Payne, J. P. Austin, J. Lynn Francis, R. L. Clancy, W. A. McDonald, and P. A. Fricker. 2002. Epstein-Barr virus reactivation and upper respiratory illness in elite swimmers. Med. Sci. Sports Exerc. 34:411-417. [DOI] [PubMed] [Google Scholar]
- 7.Hopwood, P., L. Brooks, R. Parratt, B. J. Hunt, B. Maria, T. J. Alero, Y. Magdi, and D. H. Crawford. 2002. Persistent Epstein-Barr virus infection: unrestricted latent and lytic viral gene expression in healthy immunosuppressed transplant recipients. Transplantation 74:194-202. [DOI] [PubMed] [Google Scholar]
- 8.Hornef, M. W., G. Bein, L. Fricke, J. Steinhoff, H. J. Wagner, W. Hinderer, H. H. Sonneborn, and H. Kirchner. 1995. Coincidence of Epstein-Barr virus reactivation, cytomegalovirus infection and rejection episodes in renal transplant recipients. Transplantation 60:474-480. [DOI] [PubMed] [Google Scholar]
- 9.Hornef, M. W., H. J. Wagner, L. Fricke, G. Bein, and H. Kirchner. 1995. Immunocytochemical detection of Epstein-Barr virus antigens in peripheral B lymphocytes after renal transplantation. Transplantation 59:138-140. [DOI] [PubMed] [Google Scholar]
- 10.Ikuta, K., Y. Satoh, Y. Hoshikawa, and T. Sairenji. 2000. Detection of Epstein-Barr virus in salivas and throat washings in healthy adults and children. Microbes Infect. 2:115-120. [DOI] [PubMed] [Google Scholar]
- 11.Jabs, W. J., H. Hennig, M. Kittel, K. Pethig, F. Smets, P. Bucsky, H. Kirchner, and H. J. Wagner. 2001. Normalized quantification by real-time PCR of Epstein-Barr virus load in patients at risk for posttransplant lymphoproliferative disorders. J. Clin. Microbiol. 39:564-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khan, G., E. M. Miyashita, B. Yang, G. J. Babcock, and D. A. Thorley-Lawson. 1996. Is EBV persistence in vivo a model for B cell homeostasis? Immunity 5:173-179. [DOI] [PubMed] [Google Scholar]
- 13.Kimura, H., M. Morita, Y. Yabuta, K. Kuzushima, K. Kato, S. Kojima, T. Matsuyama, and T. Morishima. 1999. Quantitative analysis of Epstein-Barr virus load by using a real-time PCR assay. J. Clin. Microbiol. 37:132-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mehta, S. K., D. L. Pierson, H. Cooley, R. Dubow, and D. Lugg. 2000. Epstein-Barr virus reactivation with diminished cell-mediated immunity in Antarctic expeditioners. J. Med. Virol. 61:235-240. [DOI] [PubMed] [Google Scholar]
- 15.Miyashita, E., B. Yang, G. J. Babcock, and D. A. Thorley-Lawson. 1997. Identification of the site of EBV persistence in vivo as a resting B cell. J. Virol. 71:4882-4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Obel, N., M. Hoier-Madsen, and H. Kangro. 1996. Serological and clinical findings in patients with serological evidence of reactivated Epstein-Barr virus infection. APMIS 104:424-428. [DOI] [PubMed] [Google Scholar]
- 17.Ohga, S., E. Kubo, A. Nomura, H. Takada, N. Suga, E. Ishii, A. Suminoe, T. Inamitsu, A. Matsuzaki, N. Kasuga, and T. Hara. 2001. Quantitative monitoring of circulating Epstein-Barr virus DNA for predicting development of posttransplantation lymphoproliferative disease. Int. J. Hematol. 73:323-326. [DOI] [PubMed] [Google Scholar]
- 18.Payne, D. A., S. K. Mehta, S. K. Tyring, R. P. Stowe, and D. L. Pierson. 1999. Incidence of Epstein-Barr virus in astronaut saliva during space flight. Aviat. Space Environ. Med. 70:1211-1213. [PubMed] [Google Scholar]
- 19.Prang, N. S., M. W. Hornef, M. J.äger, H. J. Wagner, H. Wolf, and F. M. Schwarzmann. 1997. Lytic replication of Epstein-Barr virus in peripheral blood: analysis of viral gene expression in B lymphocytes during infectious mononucleosis and in the normal carrier state. Blood 89:1665-1677. [PubMed] [Google Scholar]
- 20.Preiksaitis, J. K., F. Diaz-Mitoma, F. Mirzayans, S. Roberts, and D. L. J. Tyrrell. 1992. Quantitative oropharyngeal Epstein-Barr virus shedding in renal and cardiac transplant recipients: relationship to immunosuppressive therapy, serologic responses, and risk of posttransplant lymphoproliferative disease. J. Infect. Dis. 166:986-994. [DOI] [PubMed] [Google Scholar]
- 21.Qu, L., M. Green, S. Webber, J. Reyes, E. Demetrius, and D. Rowe. 2000. Epstein-Barr virus gene expression in the peripheral blood of transplant recipients with persistent virus loads. J. Infect. Dis. 182:1013-1021. [DOI] [PubMed] [Google Scholar]
- 22.Rickinson, A. B., and E. Kieff. 2001. Epstein-Barr virus, p. 2575-2627. In D. M. Knipe and P. M. Howley (ed.), Fields virology, vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 23.Rooney, C. M., S. K. Loftin, M. S. Holloday, M. K. Brenner, R. A. Krance, and H. Heslop. 1995. Early identification of Epstein-Barr virus-associated post-transplantation lymphoproliferative disease. Br. J. Haematol. 89:98-103. [DOI] [PubMed] [Google Scholar]
- 24.Schaade, L., M. Kleines, and M. Häusler. 2001. Application of virus-specific immunoglobulin M (IgM), IgG, and IgA antibody detection with a polyantigenic enzyme-linked immunosorbent assay for diagnosis of Epstein-Barr virus infections in childhood. J. Clin. Microbiol. 39:3902-3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thorley-Lawson, D. A. 2001. Epstein-Barr virus: exploiting the immune system. Nat. Immunol. 1:75-82. [DOI] [PubMed] [Google Scholar]
- 26.Tovey, M. G., G. Lenoir, and J. Begon-Lours. 1978. Activation of latent Epstein-Barr virus by antibody to human IgM. Nature 276:270-272. [DOI] [PubMed] [Google Scholar]
- 27.Vajro, P., S. Lucariello, F. Migliaro, E. Sokal, B. Gridelli, A. Vegnente, R. Iorio, F. Smets, I. Quinto, and G. Scala. 2000. Predictive value of Epstein-Barr virus genome copy number and BZLF1 expression in blood lymphocytes of transplant recipients at risk for lymphoproliferative disease. J. Infect. Dis. 181:2050-2054. [DOI] [PubMed] [Google Scholar]
- 28.Venard, V., A. S. Carret, N. Pascal, B. Rihn, P. Bordigoni, and A. Le Faou. 2000. A convenient semi-quantitative method for the diagnosis of Epstein-Barr virus reactivation. Arch. Virol. 145:2211-2216. [DOI] [PubMed] [Google Scholar]
- 29.Wagner, H. J., G. Bein, A. Bitsch, and H. Kirchner. 1992. Detection and quantification of latently infected B lymphocytes in Epstein-Barr virus-seropositive, healthy individuals by polymerase chain reaction. J. Clin. Microbiol. 30:2826-2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wagner, H. J., L. Fischer, W. J. Jabs, M. Holbe, K. Pethig, and P. Bucsky. 2002. Longitudinal analysis of Epstein-Barr viral load in plasma and peripheral blood mononuclear cells of transplanted patients by real-time polymerase chain reaction. Transplantation 74:656-664. [DOI] [PubMed] [Google Scholar]
- 31.Wagner, H. J., M. Hornef, J. Feldner, and H. Kirchner. 1994. Determination of IgG- and IgM-antibodies to Epstein-Barr virus associated antigens in blood donors by a novel enzyme-linked immunosorbent assay. Lab. Med. 18:165-167. [Google Scholar]
- 32.Wagner, H. J., W. Jabs, F. Smets, M. Wessel, L. Fischer, G. Offner, H. Kirchner, and P. Bucsky. 2000. Real-time polymerase chain reaction (RQ-PCR) for the monitoring of Epstein-Barr virus (EBV) load in peripheral blood mononuclear cells. Klin. Padiatr. 212:206-210. [DOI] [PubMed] [Google Scholar]
- 33.Wagner, H. J., M. Wessel, W. Jabs, F. Smets, L. Fischer, G. Offner, and P. Bucsky. 2001. Patients at risk for development of posttransplant lymphoproliferative disorder: plasma versus peripheral blood mononuclear cells as material for quantification of Epstein-Barr viral load by using real-time quantitative polymerase chain reaction. Transplantation 72:1-8. [DOI] [PubMed] [Google Scholar]
- 34.Wandinger, K., W. Jabs, A. Siekhaus, S. Bubel, P. Trillenberg, H. Wagner, K. Wessel, H. Kirchner, and H. Hennig. 2000. Association between clinical disease activity and Epstein-Barr virus reactivation in MS. Neurology 55:178-184. [DOI] [PubMed] [Google Scholar]
- 35.Yang, J., Q. Tao, I. W. Flinn, P. G. Murray, L. E. Post, H. Ma, S. Piantadosi, M. A. Caligiuri, and R. F. Ambinder. 2000. Characterization of Epstein-Barr virus-infected B cells in patients with posttransplantation lymphoproliferative disease: disappearance after rituximab therapy does not predict clinical response. Blood 96:4055-4063. [PubMed] [Google Scholar]
- 36.Yao, Q. Y., A. B. Rickinson, and M. A. Epstein. 1985. A re-examination of the Epstein-Barr virus carrier state in healthy seropositive individuals. Int. J. Cancer 35:35-42. [DOI] [PubMed] [Google Scholar]