Abstract
Eukaryotic translation initiation factor eIF3, that plays a central role in translation initiation, consists of five core subunits that are present in both the budding yeast and higher eukaryotes. However, higher eukaryotic eIF3 contains additional (non-core) subunits that are absent in the budding yeast. We investigated the role of one such non-core eIF3 subunit eIF3h, encoded by two distinct genes – eif3ha and eif3hb, as a regulator of embryonic development in zebrafish. Both eif3h genes are expressed during early embryogenesis, and display overlapping yet distinct and highly dynamic spatial expression patterns. Loss of function analysis using specific morpholino oligomers indicates that each isoform has specific as well as redundant functions during early development. The morphant phenotypes correlate with their spatial expression patterns, indicating that eif3h regulates development of the brain, heart, vasculature, and lateral line. These results indicate that the non-core subunits of eIF3 regulate specific developmental programs during vertebrate embryogenesis.
Keywords: Eukaryotic Translation Initiation, Translation Initiation Factor 3h subunit, Zebrafish Development, Regulation of early embryogenesis by eIF3h
INTRODUCTION
Initiation of translation in eukaryotic cells requires the essential participation of a dozen protein factors, collectively called eukaryotic translation initiation factors (eIFs) that promote the initial binding of the initiator methionyl-tRNA (Met-tRNAi) to the 40S ribosomal subunit, and subsequent positioning of the resulting 43S preinitiation complex (40S.Met-tRNAi) at the AUG start codon of an mRNA, to form the 48S initiation complex (mRNA.40S.Met-tRNAi). Subsequently, a 60S ribosomal subunit joins the 48S initiation complex to form the 80S ribosomal initiation complex (mRNA.80S.Met-tRNAi) that is competent to undergo peptide bond formation. This overall process involves formation of multiple non-covalent intermediate biochemical complexes in a series of distinct steps that are highly conserved between the unicellular budding yeast Saccharomyces cerevisiae and the higher eukaryotes including plants and mammals. Not only is the function of each initiation factor in the translation initiation pathway conserved but there is also a clear structural homology between the budding yeast and mammalian initiation factors (for review, see Kapp and Lorsch, 2004; Kozak, 1999; Pestova et al., 2007).
Among the translation initiation factors, eukaryotic initiation factor eIF3 is a large heteromeric protein complex that plays a central role in the initiation process (Hinnebusch, 2006; Kapp and Lorsch, 2004; Pestova et al., 2007). The initial binding of eIF3 to free 40S subunits is necessary for the subsequent binding of the initiator Met-tRNAi to generate the stable 43S preinitiation complex. Subsequent recruitment of mRNA to the 43S pre-initiation complex is also mediated by the binding of eIF3 with the initiation factor eIF4G, which in turn interacts with the cap binding initiation factor eIF4E at the 5’ cap structure of the mRNA. Finally, eIF3 has also been shown to play an important role during the scanning of the 43S pre-initiation complex along the mRNA leading to the selection of the AUG start codon. Consistent with this multitude of functions, the subunits of eIF3 are reported to interact with other eIFs that are involved in the initiation process, suggesting that eIF3 functions as a central “hub” (Kapp and Lorsch, 2004) or a “scaffold” (Hinnebusch, 2006) in the assembly of translation initiation complexes.
In view of the well accepted conserved central role of eIF3 in translation initiation, it was somewhat surprising that eIF3 isolated from the budding yeast contains only five distinct subunits, designated eIF3a, eIF3b, eIF3c, eIF3g and eIF3i, whereas the multi-cellular higher eukaryotic eIF3 contains, in addition to the homologs of these five subunits, an additional 6–8 subunits (Hinnebusch, 2006; Kapp and Lorsch, 2004; Pestova et al., 2007). In fact, the genes encoding these additional subunits are absent in the genome of the budding yeast. It has been hypothesized (Hinnebusch, 2006; Kapp and Lorsch, 2004; Pestova et al., 2007) that the five subunits of eIF3 that are common to both the unicellular budding yeast and the higher eukaryotes comprise the “core” subunits that are essential for global translation initiation of all eukaryotic mRNAs. The additional subunits that are only present in eIF3 of higher eukaryotes may either serve as regulators of translation initiation by controlling the translation of specific mRNAs and/or be directly involved in other biological processes. These subunits are designated as the “non-core” subunits and are named eIF3d, eIF3e, eIF3f, eIF3h, eIF3j, eIF3k, eIF3l and eIF3m (Hinnebusch, 2006).
To investigate whether the non-core eIF3 subunits play a regulatory role in translation initiation and/or other biological processes, we chose to study early embryogenesis in a vertebrate. There is now compelling evidence, primarily from studies in Drosophila, C. elegans and Xenopus (Curtis et al., 1995; Groisman et al., 2001; Thompson et al., 2007), that translational control plays an important role in regulating the patterns of protein synthesis and thus the entire process of early embryonic development. In known cases of translational control during development, a regulatory repressor protein binds to a cis-acting sequence at the 3’ UTR of the mRNA. This interacts with the cap-binding initiation factor eIF4E, preventing its interaction with eIF4G. This effectively prevents the eIF4E.eIF4G.eIF3 interaction that is necessary for the binding of the 43S preinitiation complex to the 5’ cap structure of the mRNA (Gebauer and Hentze, 2004; Gray and Wickens, 1998; Hentze et al., 2007; Richter and Sonenberg, 2005; Thompson et al., 2007). The question naturally arises whether this eIF4E.eIF4G.eIF3 interaction, which is critical for the assembly of the translation initiation complex on the mRNA leading to AUG start codon selection, has a regulatory role in translation initiation that is mediated by one or more of the eIF3 non-core subunits during early embryonic development. To test this hypothesis, we used the zebrafish (Danio rerio) as a model, because it provides several distinct experimental advantages (Lele and Krone, 1996.). Here we report our initial studies using zebrafish to evaluate the role of the non-core eIF3 subunit eIF3h during embryonic development. BLAST search analysis in the UCSC genome browser showed that the gene encoding zebrafish eIF3h is duplicated on two distinct chromosomes. We show that two zebrafish eif3h mRNAs are differentially expressed during embryonic development. Loss-of-function experiments demonstrate that these two genes have distinct embryonic requirements, although they also have some common and redundant functions. Overall our studies support the hypothesis that eIF3 non-core subunits have regulatory functions that impact embryonic development.
RESULTS
eIF3h is encoded by two distinct genes in zebrafish
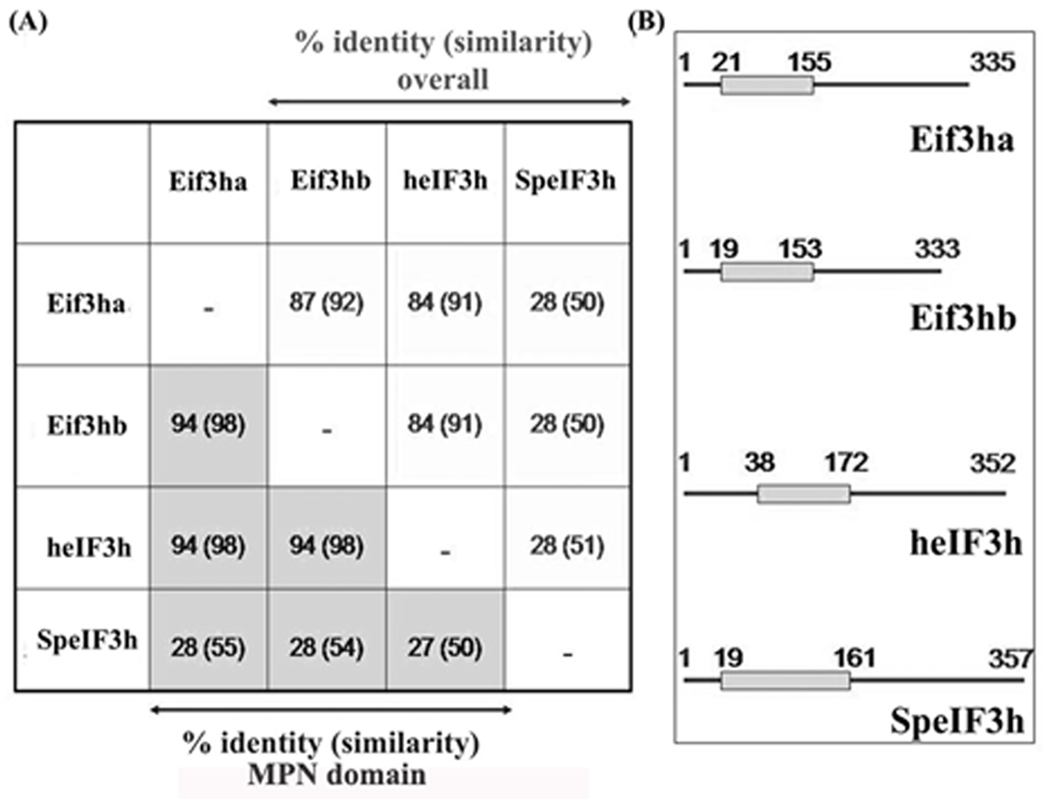
A genome-wide BLAST search revealed that zebrafish eIF3h is encoded by two distinct genes, one located on chromosome 16 and the other on chromosome 19. We have designated these genes as eif3ha and eif3hb, respectively, corresponding to the proteins Eif3ha and Eif3hb, based on the accepted nomenclature system for duplicated genes and proteins in zebrafish. The two predicted proteins (the orthologs of eIF3h) are ~ 87% identical, and each shows ~84% identity with the human protein (Fig. 1A). Each ortholog contains a well-conserved MPN domain (Sanches et al., 2007) near the N-terminus (Fig. 1B) that is characteristic of other multi-subunit protein complexes including the proteasome and the CSN signalosome (Scheel and Hofmann, 2005; von Arnim and Schwechheimer. 2006). The presence of duplicated genes in zebrafish is not unusual (Bahary et al., 2007; Force et al., 1999; Shimeld, 1999). This feature can prove useful for revealing gene functions, since the possibility exists that multiple distinct functions or expression domains for a single human gene may be segregated onto two distinct zebrafish orthologs. Alternatively, the two genes may have redundant functions. Before proceeding to evaluate this further, we first sought to determine if both genes are expressed.
FIG. 1. The zebrafish genome encodes two eif3h genes that are highly conserved with the human homolog.
(A) Percentage identity and similarity in the overall protein structures (white background) and in the single MPN domain (grey background) among the four eIF3h orthologs from zebrafish (represented by Eif3ha and Eif3hb encoded by chromosome 16 and 19, respectively); human (heIF3h), and Schizosaccharomyces pombe (SpeIF3h). For each box, the number represents the identity, while the corresponding similarity is given in the parentheses. (B) Schematic representation of the relative position of the MPN domains of Eif3ha, Eif3hb, heIF3h and SpeIF3h. The lines designate complete protein sequences and the grey boxes represent the MPN domains therein. The numbers indicate the amino acid positions.
Both eif3h genes are expressed at distinct levels and in dynamic patterns during embryogenesis
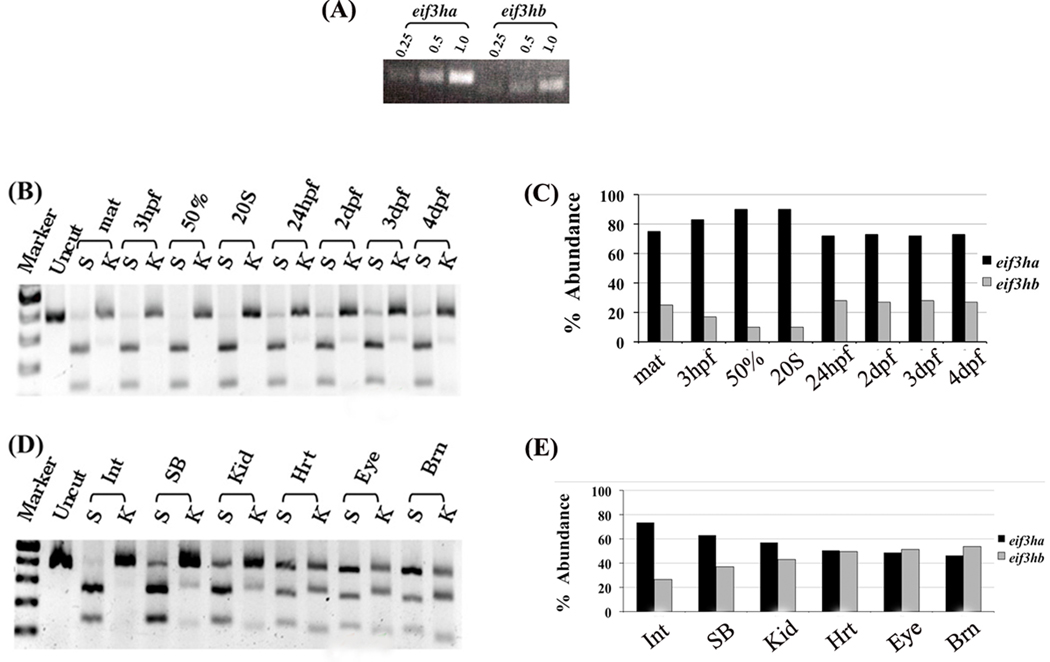
Using two sets of PCR primers, each pair being specific for either the eif3ha or eif3hb transcript, RT-PCR amplification of total zebrafish RNA is expected to yield 432-bp and 365-bp products for eif3ha and eif3hb, respectively. Analysis of total RNA isolated from embryos at 24 hpf showed the presence of both transcripts (Fig. 2A). The conditions used for the RT-PCR reactions were semi-quantitative since the amount of each product generated was directly proportional to the amount of input RNA. The results suggest that the relative level of expression of eif3ha mRNA at 24 hpf is significantly higher than that of eif3hb mRNA. Further confirmation for this observation was obtained using a new single set of forward and reverse primers matching highly conserved sequences of the two genes. In this case, the cDNA derived from the transcript of either gene should be amplified with equivalent efficiency. The PCR product derived from each transcript was distinguished by restriction digestion with either StuI (unique for eif3ha) or KpnI (unique for eif3hb). Using this differential restriction digestion assay, we determined the relative levels of mRNA derived from each gene at various time points during development, as well as in specific adult zebrafish tissues. As shown in Fig. 2B and 2C, eif3ha mRNA is expressed at a significantly higher level than eif3hb throughout embryogenesis. It should be noted that significant levels of eif3hb mRNA, although still much lower compared to eif3ha, start to appear only ~1–2 days post fertilization (dpf). We also evaluated RNA isolated from different organs of adult zebrafish (Fig. 2D and 2E). In contrast to early embryos, both eif3ha and eif3hb transcripts were observed in nearly equivalent amounts in heart, eye, and brain. In other tissues examined, e.g. intestine, swim bladder, and kidney, eif3ha transcripts were observed at a higher level than eif3hb although the relative expression levels vary somewhat depending on the tissues examined. Taken together, these results show that both eif3h genes are expressed, and their relative expression levels vary depending on the embryonic stage or tissue of the adult fish.
FIG. 2. Both zebrafish genes are expressed, with eif3ha transcript levels being predominant during early embryogenesis.
(A) Semi-quantitative RT-PCR analysis of total RNA isolated from 24 hpf embryos using two different pairs of primers that are specific for each ortholog. RT-PCR was performed using 0.25, 0.5 or 1.0 µg of total RNA as substrate. (B–E) Semi-quantitative RT-PCR using a single set of forward and reverse primers that amplify both eif3ha and eif3hb mRNAs followed by diagnostic digestion with either StuI (S) cleaving eif3ha cDNA or KpnI (K) cleaving eif3hb cDNA. (B) Samples were derived from embryos at different developmental stages as indicated; zygote (maternal, mat), 3 hpf, 50% epiboly (50%), 20 somite (20S), 24 hpf, 2, 3 or 4 days post fertilization (dpf). (D) Samples from different adult organs: intestine (Int), Swim-bladder (SB), kidney (Kid), heart (Hrt), eye or brain (Brn). (C) and (E) show the graphical quantification of the gel electrophoresis results shown in (B) and (D), respectively. It should be noted that experiments in panels B and D and the corresponding quantification in panels C and E are representative of several independent experiments.
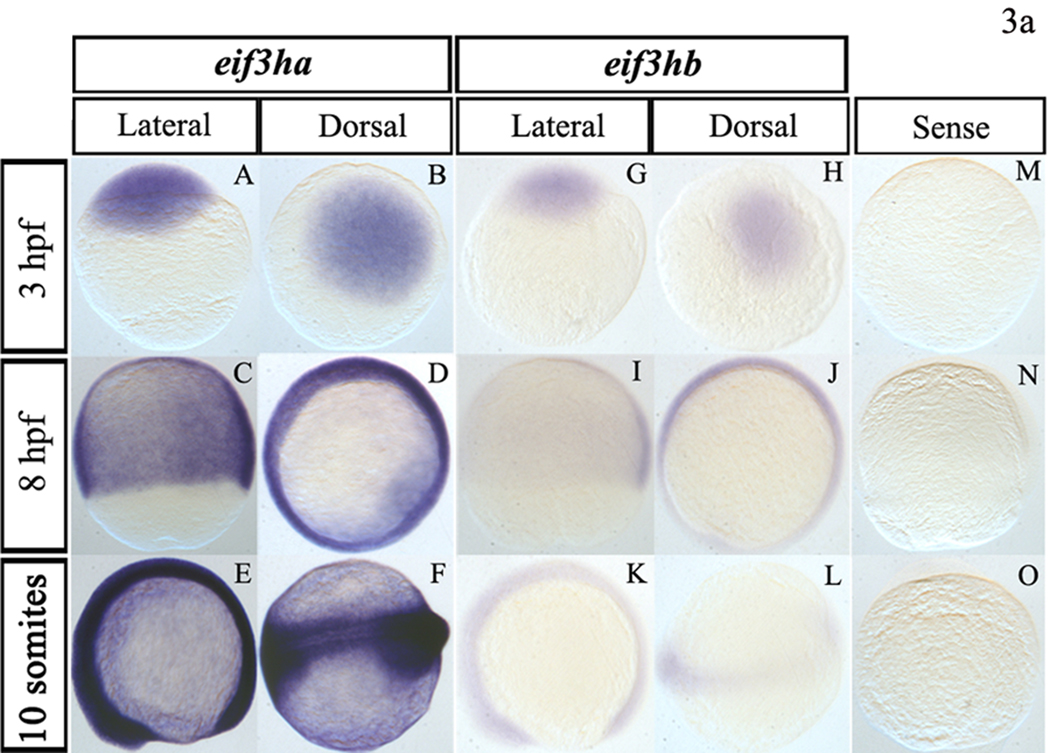
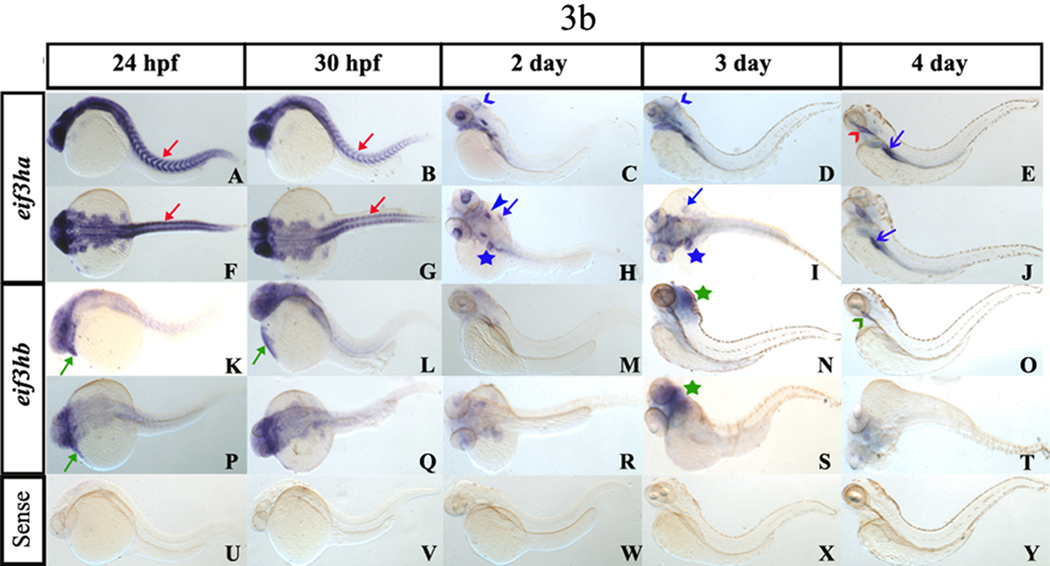
The spatial expression pattern of each eif3h mRNA was investigated by carrying out in situ hybridization experiments using antisense RNA probes labeled with digoxigenin. To minimize potential cross-hybridization between the two transcripts, we designed probes specific for each gene product by subcloning specific regions of each cDNA as described under Materials and Methods. Hybridization of staged embryos with purified probes under stringent conditions showed that with embryos younger than 24 hpf (Fig. 3a), both eif3h transcripts are expressed quite ubiquitously although the eif3ha transcripts appear much more abundant compared to the eif3hb transcript (Fig. 3a, compare panels A–F with panels G–L), as expected based on the RT-PCR data (Fig. 2). However, in embryos staged between 1–4 dpf, spatially restricted and dynamic expression patterns were observed for both eif3h genes (Fig. 3b). At 24 hpf, the eif3ha transcript was found predominantly in brain, eye, and somites (Fig. 3b, panels A and F). In contrast, at this stage, the eif3hb transcripts were restricted much more to the anterior portions of the body as well as to the tissues associated with the developing heart (Fig. 3b, panels K and P). By 30 hpf, the level of eif3ha transcripts decreased and became associated with specific developing organs, e.g. regions of the brain, eye and somites (Fig. 3b, panels B and G). Between 2–3 dpf, eif3ha was expressed specifically in the eye, midbrain-hindbrain boundary, ear, fin bud, and specific regions of the gut tube including the liver (Fig. 3b, panels C, H, D and I). Interestingly, the expression of eif3ha in somites, which was prominent at 24–30 hpf, was not seen by 2 dpf. At 4 dpf, eif3ha transcripts were observed primarily in specific regions of the alimentary canal, including the esophagus and intestinal bulb (Fig. 3b, panels E and J). By 3 dpf, eif3hb transcripts were concentrated more in the brain region (Fig. 3b, panels N and S) showing a marked change in the expression pattern while at 4 dpf, low levels of these transcripts were detected in the esophagus (Fig. 3b, panel O). As a control, we used a sense RNA probe, which failed to generate signal, as expected (Fig. 3a, panels M–O and Fig. 3b, panels U–Y).
FIG. 3. During early embryogenesis, the two eif3h genes are expressed ubiquitously while by 24 hpf the genes are expressed in distinct and restricted patterns.
Panel a, early embryogenesis (<24 hpf). Panels A–F show representative in situ hybridization patterns for eif3ha while Panels G – L are those for eif3hb. For eif3ha, panels A, C, and E represent the lateral view while B, D, and F show the dorsal view. For eif3hb, panels G, K, and I represent the lateral view while H, J, and L show the dorsal view. Panels M – O show the control embryos treated with an eif3ha sense probe. For each of the above panels, the hybridization patterns were determined at 3 hpf, 8 hpf or 10 somites, as indicated. Panel b, in situ hybridization patterns for later stages of embryogenesis (≥24 hpf). Panels A–J for eif3ha: (A – E show lateral views and F – J show dorsal views; panels K – T for eif3hb: (K – O represent lateral views and P – T represent dorsal views). Panels U – Y show control embryos hybridized with an eif3ha sense probe. The specific developmental stages examined are indicated. The red arrows in panels A, B, F, and G highlight the specific expression pattern of eif3ha in the developing somites while the green arrows in panels K, L, and P indicate distinct expression of eif3hb in the cardiac-associated regions. Blue open arrowheads in C and D indicate expression of eif3ha at the midbrain-hindbrain boundary. Blue block arrowheads and blue block arrows show eif3ha expression in otic capsules and fin-buds, respectively, while the blue stars indicate transcripts in the gut tube and liver. Green stars in panels N and S mark the condensed expression of eif3hb at 3 dpf in the brain. At 4 dpf, eif3ha and eif3hb transcripts are present in specific areas of the alimentary canal: eif3ha transcripts in the intestinal bulb (E and J, blue open arrows) and esophagus (E, red open arrowhead) while eif3hb in the esophagus with a relatively weak signal (O, green open arrowhead).
The two eif3h genes have different requirements for embryonic development
We next investigated the effect of blocking the synthesis of each eif3h isoform during early zebrafish embryogenesis. For this purpose, we designed 25-mer antisense morpholino oligomers that would specifically bind to the 5’-UTR region immediately upstream of the AUG start codon of each mature eif3h mRNA, thereby preventing initiation of translation. Distinct morpholino oligomers were also designed to bind to the intron-exon boundary of each of the eif3h pre-mRNAs, thereby blocking the formation of the spliced mature mRNAs. In either case, cellular synthesis of the encoded eIF3h protein isoform should be inhibited, and if both the translational blocker (TB) and the splice blocker (SB) morpholino generate similar phenotypes, this serves to validate the specificity of the eif3h knockdown (Eisen and Smith, 2008). Each of the morpholinos used for this study were also blast-screened (http://genome.ucsc.edu/cgi-bin/hgBlat) genome-wide against other zebrafish mRNAs to exclude mistarget effects. The ability of morpholinos to inhibit the targeted gene activity typically persists for 4 to 6 days, throughout key early stages of zebrafish development.
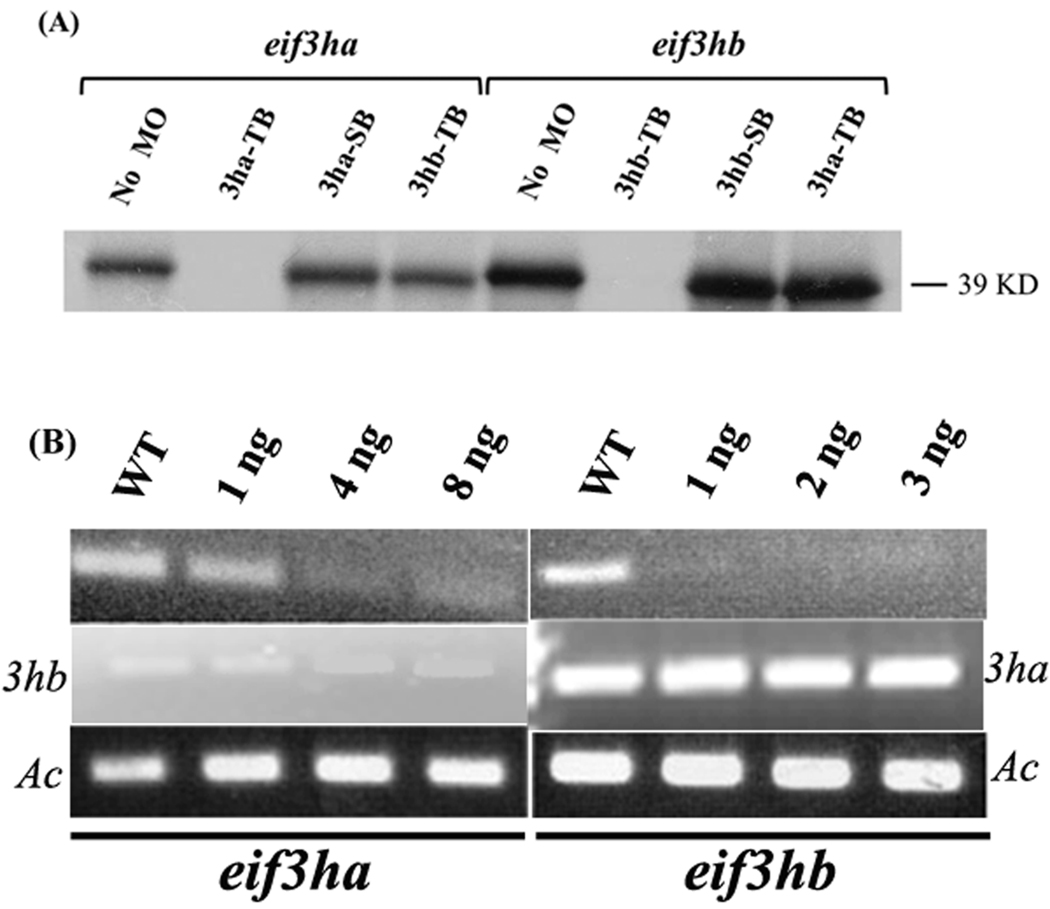
We first confirmed the specificity and activity of each TB morpholino using an in vitro coupled transcription-translation assay system. Addition of the eif3ha-specific morpholino caused a marked inhibition of Eif3ha protein formation in vitro. The specificity of this morpholino was demonstrated by showing that the morpholinos targeted to either the eif3ha pre-mRNA splice site or to the translational start site of eif3hb mRNA failed to inhibit in vitro Eif3ha protein formation (Fig. 4A, the first four lanes). Likewise, in vitro translation of eif3hb mRNA was significantly inhibited by the presence of eif3hb-specific TB morpholino. In this case, neither the eif3ha-specific TB morpholino nor the SB morpholino directed against the eif3hb pre-mRNA inhibits translation of the eih3hb mature mRNA (Fig. 4A, last four lanes). Similarly, the specificity and efficiency of each SB morpholino was verified after injecting fertilized eggs with the corresponding morpholino by isolating total RNA from embryos at 24 hpf (for eif3ha) or 2 dpf (for eif3hb). Subsequent RT-PCR assays showed a clear reduction in the formation of both the mature eif3ha and eif3hb transcripts, using only the corresponding SB morpholino, compared to the control uninjected wild-type embryos, with no cross-targeting (Fig. 4B). Having documented the specificity of morpholino action, we next used these reagents to knockdown gene activity and evaluate developmental phenoptypes. A summary of the morpholinos used in this study and the phenotypes caused by eif3h loss of function, as described below, is provided in Table 1.
FIG. 4. The morpholinos targeting the eif3ha or eif3hb transcripts demonstrate specificity.
(A) In vitro transcription-translation assays were performed in the presence of specific morpholinos as follows: translational blocking morpholino for eif3ha and eif3hb are represented as 3ha-TB and 3hb-TB, respectively while the splice blocking morpholino for each transcript is represented as 3ha-SB or 3hb-SB, respectively. The nonspecific morpholinos (TB morpholino for the other eif3h gene) were also used as indicated. A control reaction without any morpholino (No MO) was also included. The specific DNA constructs used were as indicated. (B) Semi-quantitative RT-PCR assays were carried out using total RNA isolated from splice morphants for eif3ha or eif3hb, as shown. In each case, RT-PCR results for the other eif3h gene was also included to exclude the possibility of cross-targeting by the morpholinos. It should be noted that compared to eif3ha, a very low level of eif3hb transcripts is observed at 24 hpf, in accordance with the in situ hybridization data and RT-PCR analyses shown in Fig. 3 and Fig. 2, respectively. Embryos were injected with the indicated amounts of SB MOs specific for either eif3ha or eif3hb. A wild-type control (WT) was also included. β-actin (Ac) was used as an internal control.
Table 1.
Summary of morpholinos used in the study and associated phenotype(s).
| Morpholino (MO) |
Dose / injection (ng) |
Phenotypes obtained |
Time when the phenotype observed |
No. of Embryos used |
Penetrance of the phenotype |
|---|---|---|---|---|---|
| eif3ha TB MO | 8 – 10 | Brain degeneration | 24 hpf | > 200 | ~ 100 % |
| eif3ha SB MO | 4 – 5 | Brain degeneration | 24 hpf | > 200 | ~ 100 % |
| 3 – 4 | Neuromast defect | 3 dpf | > 100 | ~ 90 – 95 % | |
| Segmental vessel defect |
2.5 – 3 dpf | > 100 | ~ 70 – 75 % | ||
| eif3hb TB MO | 6 – 10 | Brain degeneration | 24 hpf | > 200 | ~ 100 % |
| 4 – 6 | Pericardial edema | 2 dpf | > 200 | ~ 90 – 95 % | |
| eif3hb SB MO | 4 – 5 | Brain degeneration | 24 hpf | > 200 | ~ 100 % |
| 3 – 4 | Pericardial edema | 2 dpf | > 200 | ~ 90 – 95 % | |
| Control MO | Up to 10 | No phenotype | - | > 150 | - |
| eif3c SB MO | 5 | Grossly deformed embryos with overall degeneration |
24 hpf | > 100 | ~ 100 % |
(i) Effects of loss-of-function of eif3ha
Injection of the eif3ha-specific TB morpholino (10 ng/injection) into one-cell staged embryos caused severe defects in the formation of normal brain structures. A representative result of multiple independent sets of morphant embryos (n> 30 – 40 embryos per set) is shown in Fig 5 (compare Fig 5B with Fig 5A). The severity of the phenotype was proportional to the amount of injected morpholino in the 4 – 10 ng range (data not shown). Furthermore, the phenotype obtained is consistent with the expression pattern shown above for eif3ha mRNA (Fig 3b, panels A and F). It should be noted that besides the brain disruption that was consistently seen in these morphant embryos, the morphology of the morphants appeared otherwise grossly normal and the morphants survived for ~ 5 days.
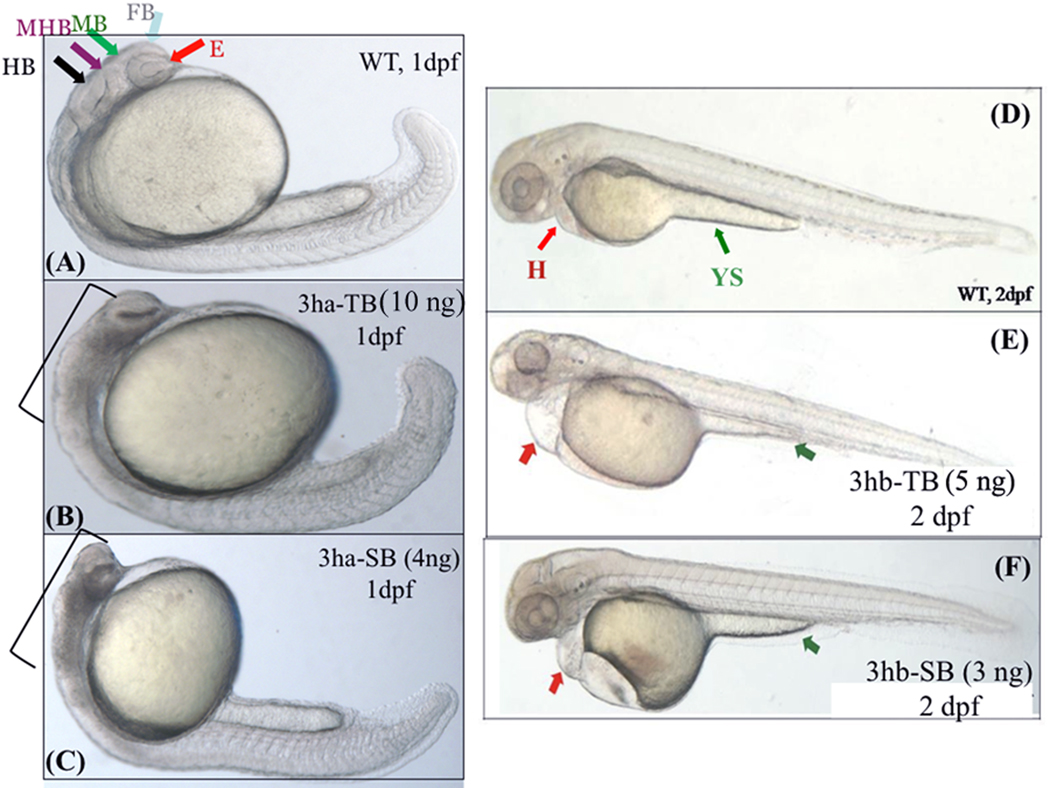
FIG. 5. Each of the eif3h morphants show distinct embryonic phenotypes.
Shown are representative morphant phenotypes obtained by injecting either TB or SB morpholino (MO) for eif3ha (3ha-TB or 3ha-SB) at 1 dpf (B and C) and for eif3hb (3hb-TB or 3hb-SB) at 2 dpf (E and F). In each case, an uninjected wild-type (WT) control (panels A and D) is also included. In panel A, different regions of the normal brain in a WT embryo at 24 hpf are shown: eye (E), forebrain (FB), midbrain (MB), midbrain-hindbrain boundary (MHB), and hindbrain (HB) as indicated by different colored arrows. For the eif3ha morphants (~100%, n ~ 200), following injection of either TB MO (panel B) or SB MO (panel C), gross disruption of all the brain regions are shown using brackets. For the eif3hb morphants (90 – 95%, n ~ 200) at 2 dpf obtained with either TB MO (panel E) or SB MO (panel F), specific defects in the form of edema seen around the heart region (H) and regression of yolk stalk (YS) are indicated with the red and green arrows, respectively.
The specificity of the phenotype observed with the TB morpholino was confirmed by injecting the SB morpholino specifically directed against the exon2-intron2 boundary of the eif3ha pre-mRNA. In multiple independent sets of injections (n > 40 per set; 4 ng/injection), this morpholino severely compromised formation of proper brain and eye structures at the 24 hpf stage, similar to the results observed with the TB morpholino (compare Fig 5C with Fig 5A and also with Fig 5B). We also injected a control morpholino, which should not target any zebrafish mRNA, at the same amount (4ng/injection) and this mrpholino, as expected, did not show any phenotype, similar to the uninjected embryos (Fig S1, Panels A – F). Some morpholinos have been shown to cause brain defects due to off-target effects that stimulate p53-induced apoptosis (Robu et al., 2007). This is not the case for the eif3ha morpholinos, since co-injection of a p53-specific morpholino along with eif3ha morpholino failed to rescue the brain defect (Fig. S2, panels a and b).
(ii) Loss-of-function of eif3hb
Using the eif3hb-specific TB morpholino, we carried out knock-down experiments of eif3hb by injecting sets of 1-cell staged embryos (4 – 10 ng/injection; n > 30 – 40 per set). In contrast to the wild-type embryos, morphant embryos (4 – 6 ng/injection) at 2 dpf showed significant pericardial edema (Fig 5E). Additionally, the morphant embryos appeared to have endoderm or gut defects as indicated by regression of the yolk stalk extension (Fig 5E). It should be noted that the embryos injected with the control morpholino at the same amount (6 ng/injection) did not show any phenotype (Fig S1, Panel G – O). However, a similar phenotype was observed by injecting a specific SB morpholino (~ 3 ng/injection) designed against the intron2-exon2 boundary of the eif3hb pre-mRNA (compare Fig 5F with Fig 5D). Furthermore, while only the pericardial edema phenotype was evident at this dose (3 ng/injection), at a higher dose (4 – 5 ng), the eif3hb-specific SB morpholino caused a consistent brain degeneration phenotype similar to blocking eif3ha (data not shown). This brain degeneration phenotype observed by injecting higher dose of eif3hb-specific morpholino is not due to its targeting of eif3ha mRNA, since there is no reduction of the mature eif3ha mRNA under these conditions (Fig S3).
The two eif3h orthologs have functional redundancy during embryonic development
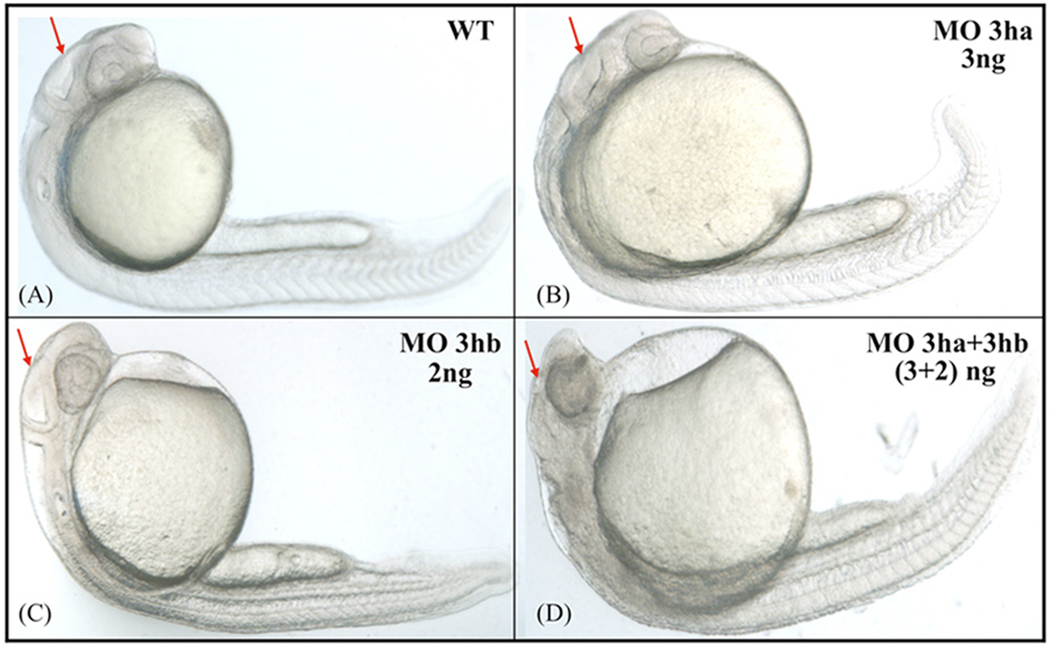
Results presented above show that the morphant phenotypes for the two eif3h orthologs are different, consistent with their distinct expression patterns during zebrafish development. However, the patterns of expression for both genes also appear to overlap, particularly in a common expression domain associated with the developing brain. We therefore tested the effect of blocking the function of both gene products simultaneously. Injection of either 3 ng of eif3ha-SB morpholino or 2 ng of eif3hb-SB morpholino alone failed to generate an obvious phenotype. However, when these two morpholinos were injected together at the same doses, a prominent deterioration of brain structures was observed in morphant embryos at 24 hpf (Fig 6). This result indicates that the reduction of products from one zebrafish eif3h gene can be functionally compensated by the other gene. However, this functional compensation is lost by depletion of both eif3h isoforms below a certain threshold level.
FIG. 6. The two eIF3h orthologs have at least some functional redundancy.
Shown are representative embryos, obtained by injecting at the one cell stage specific SB morpolinos for eif3ha or eif3hb, either separately or in combination, as indicated. A, uninjected wild-type (WT) control; B, 3ng of eif3ha MO (MO 3ha); C, 2ng eif3hb MO (MO 3hb), and D, 3ng MO 3ha + 2ng MO 3hb together (~ 100%, n > 50 – 80). Arrows highlight the relative development of the brain region.
eif3ha displays a striking and dynamic expression pattern in developing somites
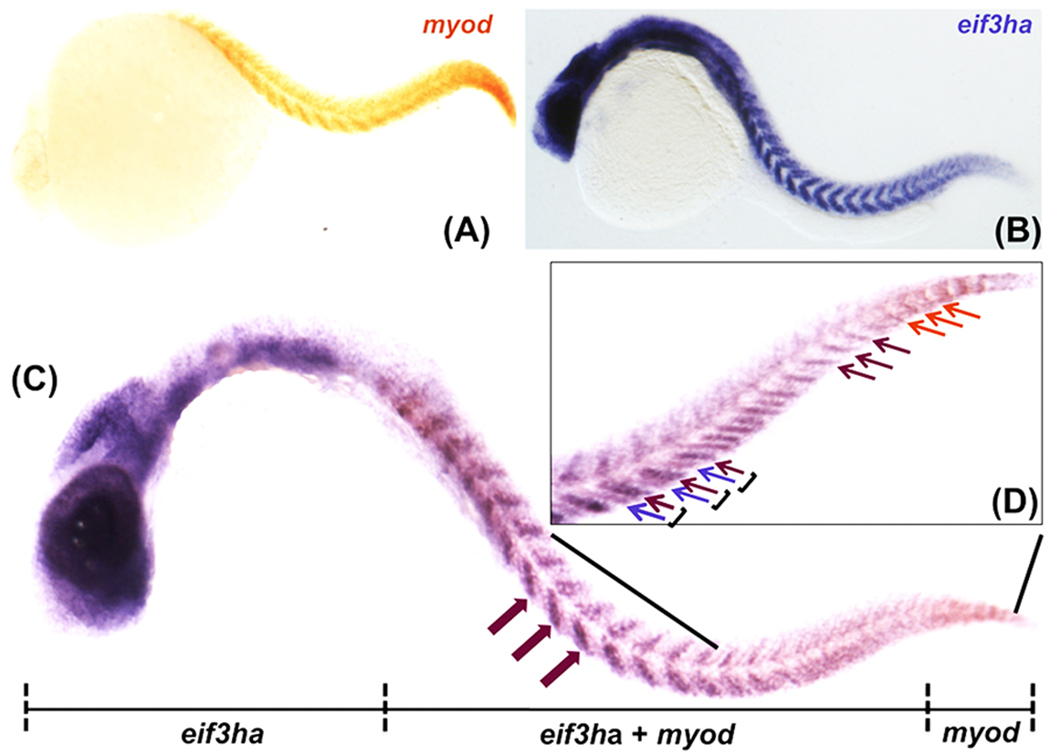
The in situ hybridization experiments presented in Fig 3b showed that at 24–30 hpf the expression pattern of eif3ha transcripts is restricted to a sub-domain of each somite. To evaluate more precisely the specific sub-domain (and therefore progenitor cell types) represented by this pattern, we carried out two-color whole-mount double in situ hybridization experiments using ~26 hpf embryos, and compared the pattern of expression of eif3ha with that of a well-characterized somite marker – myod. Just after somite formation, myod transcripts are known to be restricted to the posterior zone of the somite, generating a clear striped pattern (van Eeden et al., 1996 and see Fig. 7, panel A). This pattern shifts as the somite matures (Weinberg et al., 1996). Older and more developed rostral somites in the trunk region have the highest levels of myod transcripts in the central region along the anterior-posterior axis (Weinberg et al., 1996). We confirmed that the most caudal nascent somites express myod in the expected pattern. However, there was a lack of visible eif3ha transcripts in this domain (Fig. 7, panel D). Rather, expression of eif3ha is initiated in more central developing somites, specifically in the posterior cells of the somites, demonstrated by the complete overlap of the pattern with myod in this region (Fig 7, panel D). In somewhat more rostral somites, the eif3ha expression pattern extended to the anterior cells of each somite where myod transcripts were not observed (Fig 7, panel D). Therefore, in this set of somites eif3ha and myod transcripts are co-expressed in the posterior cells, while eif3ha alone is detected in the anterior half (Fig 7, panel D). Finally, at this stage of development, in the most rostral trunk somites, eif3ha is co-expressed with myod in the somite central domain (Fig 7, panel C). Taken together, these results indicate that eif3ha is expressed in the developing somites in a pattern that depends on the maturation stage of the somite.
FIG. 7. The distinct expression pattern of eif3ha in developing somites overlaps with the well-characterized marker myod.
Panels A and B show the expression for myod and eif3ha transcripts, respectively. Panel C represents the double in situ hybridization using both the probes together. A representative embryo is shown indicating the expression of digoxigenin-labeled eif3ha probe only in the anterior portion, fluorescein-labeled myod probe only in the tail tip and expression of both the transcripts over-lapping in the body trunk region. The magenta block arrows indicate expression of both the transcripts at somite centers. Panel (D) is a closer view of the trunk region including the tail. The regions of expression of only myod or eif3ha are shown using the orange and magenta arrows, respectively. The individual somites are represented using parentheses. Within a somite, the magenta arrow shows the expression of both the mRNAs in the posterior region, while the violet arrow indicates the specific expression of eif3ha mRNA alone in the anterior segment.
eif3ha morphants have defects in lateral line and vascular patterning
While the trunk of the eif3ha morphants appeared grossly normal (see Fig. 5), the specific and dynamic expression pattern of eif3ha mRNA in the somites (Fig. 7) prompted us to investigate more subtle phenotypes that might occur if normal somite development and/or somite-derived signaling pathways were disturbed. For this purpose, we first investigated the zebrafish lateral line, which is a mechano-sensory system present in fish and amphibians related to the human auditory system (Dambly-Chaudiere et al., 2003). The zebrafish lateral line is composed of two sub-domains: the anterior lateral line (ALL) and the posterior lateral line (PLL) that are formed by the head neuromasts and the body/tail neuromasts, respectively. The PLL neuromast progenitors are deposited by an ectodermal placode-derived primordium that migrates toward the tail, starting around 20–22 hpf, from a region just caudal to the otic capsule. Each neuromast forms a hair cell structure surrounded by support cells and is innervated by both the afferent and the efferent neurons from the hindbrain, controlling the sensitivity of the lateral line system to detect touch and water flow. By 2–3 dpf, the deposition of 5–6 neuromasts occurs in a sterotypic fashion in which the neuromasts are arranged in pairs on each side of the body trunk at regular intervals. In addition, the primary PLL also contains 3–4 terminal neuromasts at the tip of the tail (Dambly-Chaudiere et al., 2003). It has been postulated that the regulated deposition of PLL neuromasts is controlled by specific somite-derived signaling cues (David et al., 2002).
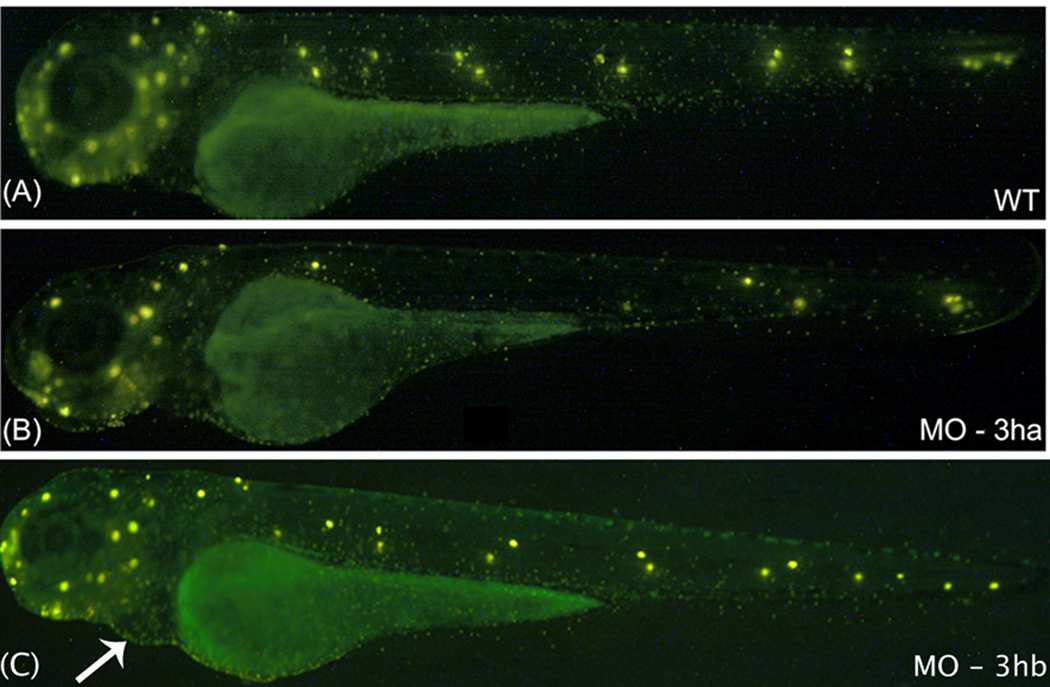
The effect of loss of eif3ha on neuromast formation was investigated in eif3ha morphant embryos (Fig 8). The neuromast pattern is readily observed by staining with the fluorescent vital dye 4-Di-2-ASP (Collazo and Mabee, 1994), which is specifically taken up by the active hair cells (Fig. 8A). We observed that injection of embryos with a low dose (~3 ng/injection) of the eif3ha-specific morpholino (SB MO) generated morphants (approximately 90 – 95%, n ~ 100) at 3 dpf that failed to develop most of the PLL neuromasts in the mid-trunk. Neuromasts at the posterior trunk or the tail tip region appeared to form normally compared to control wild-type embryos (compare Fig 8B with Fig 8A). Neuromast deposition defects could be secondary to brain degeneration, if this disturbed primordium development. However, under these conditions, these deposition-defective eif3ha morphants did not show any brain phenotypes. Similar neuromast defects were also observed in morphant embryos obtained by injecting higher dosage of eif3ha-specific morpholino, although this also led to brain degeneration. In contrast, using low dosage of eif3hb-specific TB MO (2 – 4 ng/injection), 70% of the eif3hb morphants showed apparently normal neuromast deposition, even though pericardial edema started to develop by this stage (Fig. 8C). However, ~ 30% of the eif3hb morphant embryos showed neuromast deposition defects which varied from loss of only PLL (Posterior Laterior Lateral Line) neuromasts to the loss of all PLL and ALL (Anterior Lateral Line) neuromasts. This observation could perhaps be explained by low-level expression of eif3hb in the embryonic somites (See Fig 3b, Panels K, L, P and Q). Alternatively, it could be a secondary effect as a result of the brain degeneration that these morphant embryos tend to acquire as the dose of eif3hb-specific morpholino increases.
FIG. 8. Development of trunk neuromast sensory organs fails in the eif3ha morphants.
Shown are representative embryos following staining with Di-Asp at 3 dpf to identify neuormasts. The representative embryo in panel A demonstrates the normal distribution of neuromasts (bright yellow spots) in an uninjected wild-type (WT) embryo. This is compared with one of the representative embryos injected with either 3 ng of eif3ha SB morpholino (MO-3ha) as shown in panel B or 2 – 4 ng of eif3hb TB morpholino (MO-3hb) as shown in panel C. Note that at this dose, the severe brain defect did not occur using MO-3ha, although most of the trunk neuromasts are absent in the eif3ha morphants (90 – 95 %, n ~ 100). In contrast, for the eif3hb morphants, neuromast formation appears normal even though the cardiac phenotype for this morphant begins to appear (indicated by the white arrow).
Finally, we analyzed the development of the trunk vasculature, which is known to develop in close association with the somite boundaries. In wild-type embryos at 1.5 dpf, the developing intersegmental arteries and veins sprout and elongate dorsally from the dorsal aorta or posterior cardinal vein, respectively, and link to the dorsal longitudinal anastomotic vessel (Isogai et al., 2001; Lawson and Weinstein, 2002). Thus, the intersegmental vessels form a regular metameric structure around the somites in the developing zebrafish trunk. During formation of the intersegmental vessels, mesoderm-derived endothelial progenitors migrate to the dorsal aorta and by subsequently migrating between the somites, reach destination positions in the intersegmental and dorsal longitudinal anastomotic vessels. Individual angioblasts destined to become segmental vessels always leave the dorsal aorta specifically between the ventral regions of the two adjacent somites (Childs et al., 2002). Clearly, establishing this highly regulated pattern requires active participation of somites, both at the level of cellular interactions as well as in the generation of different signaling cues to control angioblast fate.
We investigated the effect of loss of function of eif3ha on vasculature development by injecting eif3ha-specific morpholino in low dosage (SB MO, ~ 3ng / injection) into one-cell stage embryos derived from transgenic fli:gfp reporter fish in which endothelial cells are labeled with GFP. We observed that at 2.5 – 3 dpf, a high proportion of the eif3ha morphant embryos (approximately 70 – 75%, n ~ 100) were highly defective in the normal metameric pattern of segmental vessels in the body trunk. The defect appeared often at the posterior trunk region [Fig 9(a) and (b), Panel C]. In contrast, eif3hb morphants did not show any similar defects [compare panel B with panel A in both Fig. 9(a) and 9(b)]. These results suggest that the well-maintained distance between the intersegmental arteries and vessels seen in the wild-type embryos is defective in the eif3ha morphants, indicating perturbation of the coordinated migration of intersegmental vessel angioblasts. It should be noted that under these conditions the morphant embryos showed no apparent brain phenotypes.
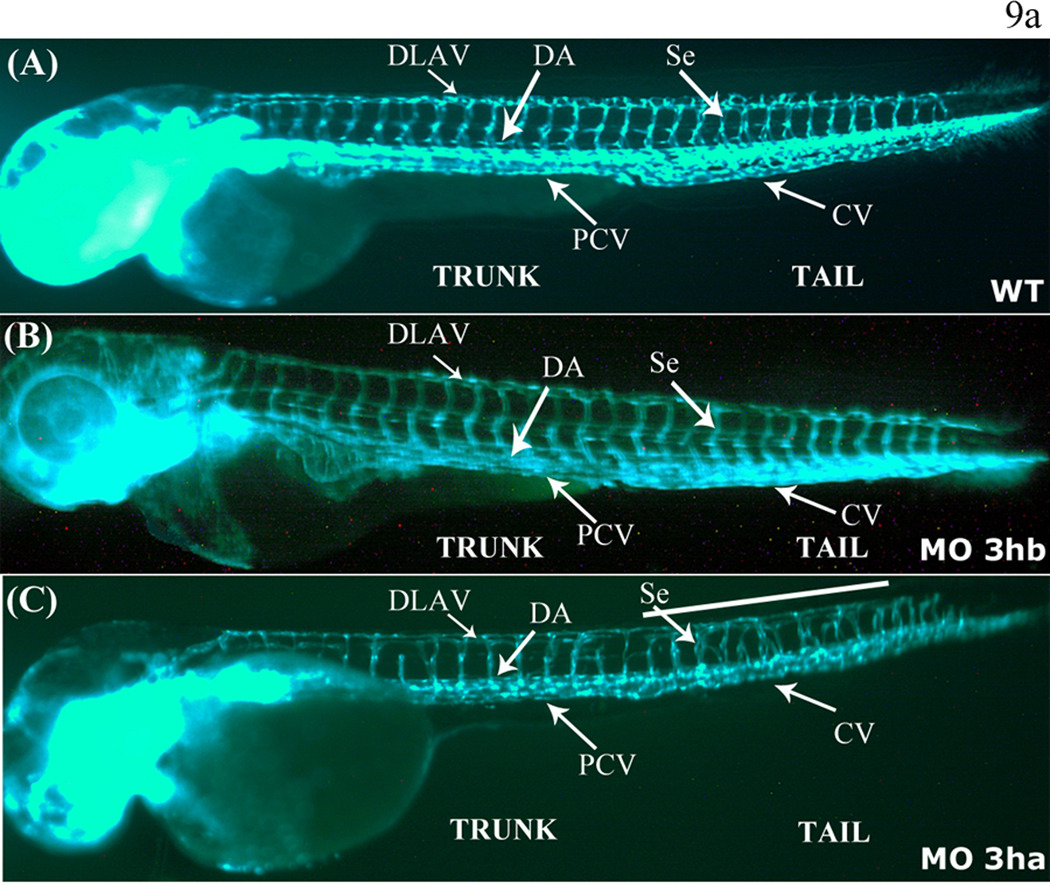
FIG. 9. eif3ha morphants also display defects in segmental vessel formation.
Panel a. Shown are representative embryos at 3 dpf derived from transgenic fli:gfp parents, allowing visualization of the developing vasculature. Panels A and B show the uninjected wild-type (WT) and eif3hb morphant (MO) embryos (6ng/injection), respectively, with normal vasculature pattern, while panel C represents eif3ha morphant embryos with clear defects in segmental vessel formation (70 – 75 %, n ~ 100). Note that the eif3hb morphants received a relatively low dose of morpholino with no apparent brain degeneration phenotype. The caudal vein (CV), posterior cardinal vein (PCV), dorsal aorta (DA), dorsal longitudinal anastomotic vessel (DLAV) are indicated.
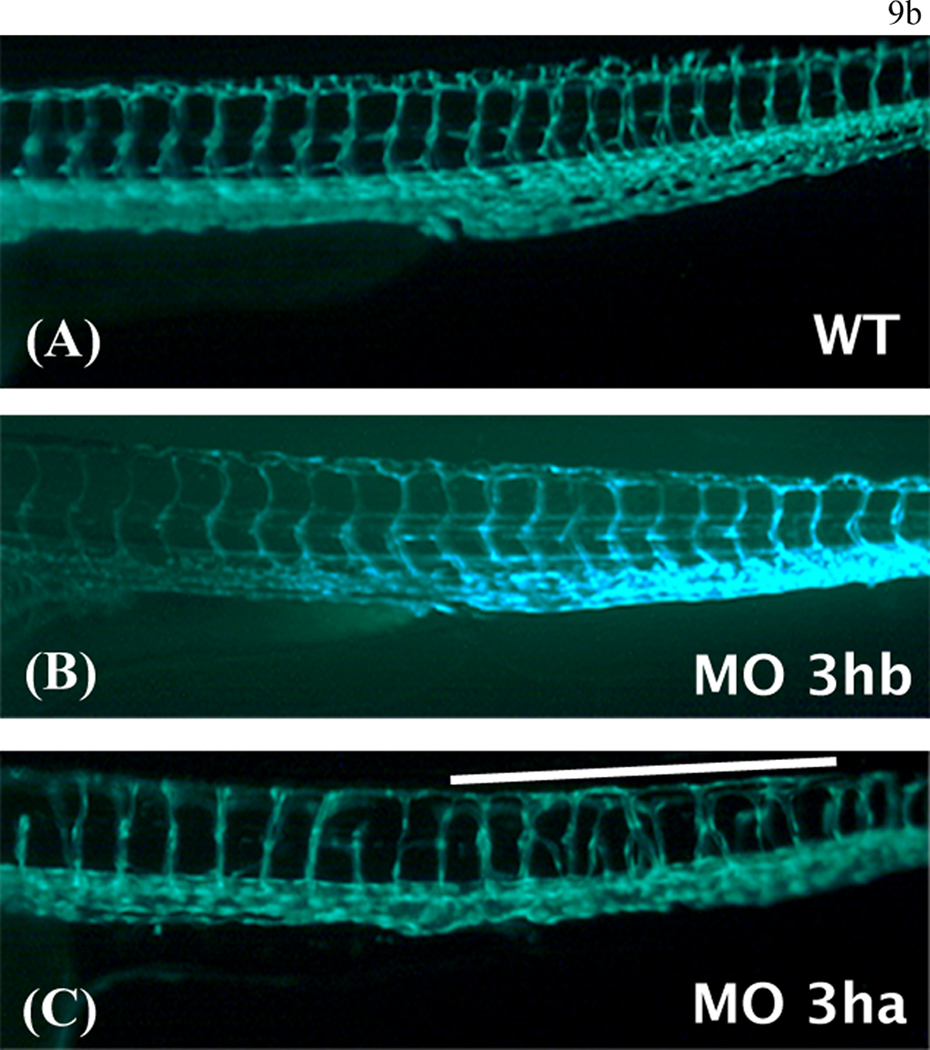
Panel b. Shown are higher magnification views of the mid-trunk region where irregular segmental vessel formation (Se) is most severe in the eif3ha morphants (Panels C) compared to uninjected and eif3hb morpholino injected embryos (Panels A and B, respectively). This region in the eif3ha morphants is indicated with a white line in both panel a and panel b.
DISCUSSION
Analysis of eIF3 in metazoans has been challenging due to its structural complexity. Because the non-core eIF3 subunits that are absent in budding yeast are integral components of the higher eukaryotic eIF3 protein complex, the structure and function of mammalian eIF3 is likely to be considerably different compared to the complex in unicellular budding yeast. A significant step towards understanding the function of the non-core eIF3 subunits came from the observation that in contrast to S. cerevisiae (Hinnebusch, 2006), the genome of the genetically tractable fission yeast Schizzosaccharomyces pombe contains structural homologs of four mammalian non-core eIF3 subunits – eIF3d, eIF3e, eIF3f, and eIF3h (Akiyoshi et l., 2001; Bandyopadhyay et al., 2002; Bandyopadhyay et al., 2000; Crane et al., 2000; Yen et al., 2000; Zhou et al., 2005) and these subunits have been shown to be integral components of the purified eIF3 protein complex (Ray et al., 2008). However, each of the genes encoding eIF3d, eIF3e, and eIF3h was found to be dispensable for growth and viability of the cells (Akiyoshi et l., 2001; Bandyopadhyay et al., 2002; Bandyopadhyay et al., 2000; Crane et al., 2000; Ray et al., 2008 Yen et al., 2000). Additionally, in all three deleted strains the polysome profiles were similar to that observed for the wild-type cells indicating that the global translation process was not affected (Bandyopadhyay et al., 2002; Bandyopadhyay et al., 2000; Ray et al., 2008). However, these deleted strains showed some specific phenotypic properties including severe defects in meiosis/sporulation following conjugation of two haplotype deleted strains (Bandyopadhyay et al., 2002; Bandyopadhyay et al., 2000; Ray et al., 2008). The defect is most severe for deletion of eIF3h (Ray et al., 2008) suggesting the possibility that in fission yeast, these non-essential eIF3 subunits may each be required for efficient translation of a subset of mRNAs, perhaps those encoding proteins required for specific cellular functions. These observations prompted us to examine whether non-core eIF3 subunits function as regulators of gene expression in a vertebrate organism. Since translational control is known to play a key role in regulating gene expression in early embryonic development (for a review see Thompson et al., 2007), and the role of non-core eIF3 subunits is essentially untested in higher animals including vertebrates, we have used zebrafish as a model to study the role of a non-core eIF3 subunit during vertebrate development. The general feasibility of this approach has been investigated in this study for the eIF3h subunit, which in zebrafish is encoded by two distinct genes – eif3ha and eif3hb located on two distinct chromosomes.
Our study shows that these two eif3h genes, both of which are expressed during early embryogenesis, have overlapping yet distinct spatial expression patterns in developing zebrafish embryos and these are highly dynamic. Interestingly, knockdown analysis of each of the two proteins clearly indicated that both eIF3h isoforms have specific as well as overlapping roles during early development of zebrafish. Their knockdown phenotypes correlated with their spatial expression patterns. In addition they share some common expression domains, and above a certain threshold, they can compensate for each other. The eif3ha gene is specifically expressed in embryonic somites in a strikingly dynamic manner, associated with the maturation stage of the developing somite. The partial depletion of eif3ha using a low dosage of morpholino leads to pleiotropic developmental defects related to somite associated signaling cues, such as defective neuromast deposition and irregular angiogenesis in the body trunk region (see Fig. 8 and Fig. 9). During early embryogenesis, tissues associated with heart showed strong expression specifically for eif3hb (Fig. 3b). Furthermore, knockdown of this isoform leads to pericardial edema. Since the cardiac (eif3hb) and vascular/neuromast (eif3ha) phenotypes are not shared by the two sets of morphants, the loss-of-function studies indicate that some specific functions are partitioned between the two orthologs. This could be based on biochemical functions, or simply reflecting different expression pattern or levels of each isoform. However, not all functions are segregated, since the neural degeneration phenotype caused by the double knockdown appears similar to that caused by a high dose of either individual morpholino. The embryo appears to be more sensitive in this case to loss of eif3ha, which probably reflects the higher expression levels of this gene. This illustrates a particularly attractive feature of the zebrafish model, in that some (but not all) specific functions are partitioned between the two orthologs and for the first time we have the possibility to infer about the functional evolution of a bona-fide translation initiation factor during the early development of vertebrates.
Our results support the hypothesis that the non-core eIF3 subunit eIF3h (and perhaps other non-core subunits of eIF3) are involved in specific functions during zebrafish embryonic development. In support of this hypothesis, it should be noted that the gene of a core eIF3 subunit - eIF3c, in contrast to the eif3h genes, is expressed quite ubiquitously (data not shown, but in agreement with the results reported in the ZFIN database – www.zfin.org). Furthermore, when the production of eIF3c was blocked by injecting an eif3c-specific SB morpholino into one cell staged embryos, the morphant embryos, instead of showing any specific defect, appeared to be generally disrupted in overall embryonic development (Fig. S4, see also Table 1) and do not survive beyond approximately 2 dpf. This result is consistent with the requirement of the eIF3 core subunits in global translation of all eukaryotic mRNAs.
The question naturally arises how each of these zebrafish eIF3h orthologs might be involved in regional-specific functions during early embryogenesis. Although none of the zebrafish translation factors including eIF3 has been isolated and characterized, eIF3 has been isolated from a wide variety of eukaryotic sources (mammalian, plant, tumor cells, as well as the unicellular fission yeast S. pombe) and has been shown to be a highly conserved protein complex consisting of 11 – 13 subunits (five core and 6 – 8 non-core subunits) (for a review see refs Hinnebusch, 2006; Kapp and Lorsch, 2004; Ray et. al, 2008). In all cases, the non-core subunit eIF3h was found to be an integral component of the eIF3 protein complex and was never observed to exist in free form in cell free extracts. Translation initiation factor eIF3 is known to be a central component of the 43S ribosomal pre-initiation complex and its presence is essential not only for the pre-initiation complex to bind to the 5’-capped end of the mRNA, but also for its ability to scan through the 5’ UTR of mRNAs leading to AUG selection (Hinnebusch, 2006, Kapp and Lorsch, 2004). It is also well accepted that the structure of the 5’ UTR is a critical component that determines the relative efficiency of scanning (Kozak, 1999.). One attractive hypothesis is that the presence of a particular non-core subunit, e.g. eIF3h, in the eIF3 protein complex may in turn govern the efficiency with which the 43S pre-initiation complex is able to scan through the 5’ UTR of a particular set of mRNAs. Thus the possibility exists that either one or both of these orthologs might be required for tissue-specific translation of a subset of mRNAs. In support of this hypothesis, it was observed (Kim et al., 2007) that in the plant Arabidopsis thaliana, the h subunit of eIF3 increases the efficiency of translation initiation of mRNAs harboring upstream open reading frames (uORFs) in their 5’ UTRs, as well as mRNAs with long 5’ UTRs or coding sequences. These observations were made by microarray comparisons of polysome-associated translating mRNAs isolated from wild-type and eif3h mutant seedlings. Furthermore, it was shown that inactivation of eif3h in this plant, although not lethal, causes pleiotropic developmental defects (Kim et al., 2004). These results justify analogous investigation of zebrafish eIF3h, providing a unique opportunity to define such roles in a vertebrate model.
Notwithstanding the above hypothesis, the possibility remains that eIF3h may also have functions in early development unrelated to direct regulation of translation initiation of specific subsets of mRNAs. For example, it was observed that another non-core subunit of eIF3 - eIF3e in S. pombe, associates with a proteasomal lid subunit (Yen et al., 2003) that regulates the degradation of specific proteins, which is required for mitosis. Future studies will focus on defining the mechanism of action for eIF3h during embryonic development.
In the work presented here, identification of two zebrafish isoforms has also created a new angle to the functional segregation of this non-core subunit. There may exist two distinct populations of zebrafish eIF3 protein complexes depending on the presence of either eif3ha or eif3hb and each eIF3 protein complex might be differentially competent to translate specific subsets of mRNAs. In fact, the presence of distinct forms of eIF3 in fission yeast that differ in their composition of the non-core subunits has been reported (Zhou et al., 2005, Ray et al., 2008). Furthermore, the possibility exists that with a particular eIF3 protein complex containing any one of the zebrafish eIF3h orthologs, the relative efficiency of translation of a distinct set of mRNAs may be dependent on the available concentration of a particular form of eIF3. This may explain our observations (Fig. 8 and Fig. 9) that the appearance of distinct phenotypes is dependent on the extent of knockdown of each eIF3h ortholog. Alternatively, the two isoforms may be biochemically equivalent, and segregated by evolution of cis-regulatory elements that differentially direct expression. These issues may eventually be resolved by rescue experiments that replace one or the other isoform.
Finally, it will be of interest to investigate in the context of zebrafish embryogenesis the function of other eIF3 non-core subunits that are absent in the budding yeast S. cerevisiae. There is evidence that both in fission yeast and in mammalian cells, each non-core subunit performs different biological functions (Guo and Sen, 2000, Holz et al., 2005; Shi et al., 2006; Yen et al,. 2003). In fact, the non-core subunit eIF3e, an integral component of mammalian eIF3, was originally isolated from mouse as an INT6 oncogene (Asano et al., 1997; Marchetti et al., 1995). This eIF3 subunit has also been implicated as a tissue-specific modulator of MEK-ERK signaling in zebrafish embryos (Grzmil et al., 2007). Clearly, each non-core subunit may regulate divergent biological processes in zebrafish, and such characterization may provide insight into their functional evolution.
MATERIALS AND METHODS
Zebrafish strains, fish husbandry and imaging
Zebrafish embryos were maintained at 28.5°C and staged as described (Kimmel et al., 1995). The wild-type (WT) strain was a hybrid of the AB and TU lines. Fish strains AB, TU and transgenic fli:gfp (Lawson and Weinstein, 2002) were obtained from the Zebrafish International Research Center (Eugene, OR). Images were captured under either brightfield or fluorescence light, using a Nikon SMZ1500 fluorescence microscope with an Insight Firewire 2 digital camera and SPOT advanced software.
Plasmids for generating RNA and in situ hybridization probes
The cDNA IMAGE clones for both genes encoding zebrafish eIF3h, designated eif3ha (pDNR-LIB-eif3ha, IMAGE ID: 7154352) and eif3hb (pME18S-FL3-eif3hb, IMAGE ID: 7278934) were obtained from Open Biosystems. The template for whole mount in situ hybridization probe was prepared by PCR amplifying a 410-bp fragment of eif3ha cDNA clone (nt 563 – 973) and ligating into pCRII-TOPO vector (Invitrogen). The forward and reverse PCR primers used were 5’-CTGAGGGGCTGAAGAAGG and 5’-CTGCGATGAGGAGCGTGTC, respectively. The template for eif3hb was a unique 393-bp DNA fragment isolated from pME18S-FL3-eif3hb by PstI/XbaI restriction enzyme digestion (nt 724 – 1117) and subcloned into the pBluescriptII SK(+) vector. For preparing full length eif3ha and eif3hb RNA transcripts, the corresponding cDNAs from pBluescriptII SK(+)-eif3ha and pME18S-FL3-eif3hb, respectively were subcloned into the T7 and T3 RNA polymerase - pCS2+ vector using ClaI/XbaI and XhoI digestion, respectively.
Morpholinos and microinjections
Two morpholino oligomers were designed for each eif3h gene and purchased from Gene Tools (Philomath, OR). For each gene, one morpholino oligomer targeted the 5’ UTR immediately preceding the start codon, in order to block mRNA translation (translation blocker, designated TB), while the second oligomer targeted the exon2-intron2 boundary of each gene to inhibit proper splicing of the corresponding pre-mRNA to form mature mRNAs (splicing blockers, designated SB). The TB morpholino sequences used were: 5’-ATGGATAGGAATCGGAACCAAGCAC for eif3ha and 5’-TTATTATTATCTTGATGAGCGCCGG for eif3hb, while the SB morpholino sequences used were: 5’-GGAGATTGCCGTGCGCCTCACCTTC for eif3ha and 5’-TTAGTCGTTTCAGTCTCCTACCTTC for eif3hb. Blast searches (http://genome.ucsc.edu/cgi-bin/hgBlat) predicted the specificity of each morpholino. Each morpholino was titrated by injection into fertilized eggs to determine a minimal dose that consistently generated the maximal phenotype. All microinjections were performed using a PLI-100 Pico-Injector (Harvard Apparatus).
Reverse Transcription-PCR (RT-PCR)
Staged wild-type and SB morpholino-injected embryos (morphants) were homogenized with Tri-Reagent (Molecular Research Center, OH) and total RNA was isolated. For expression analysis, total RNA (0.25, 0.5 or 1 µg) isolated from 24 hour post fertilization (hpf) embryos was used to generate the corresponding cDNA using Moloney murine leukemia virus (M-MLV) reverse transcriptase and random hexamers (Invitrogen). Primers specific for either eif3ha or eif3hb were used in subsequent PCR reactions for 27 cycles: 5’-TCTCTCCGGAATAACTTTCTAACACG (forward); 5’-CGTGCTGGTAGCTGAACTG (reverse) for eif3ha, and 5’-ATCCCGTAAAGAAACCCCGAA (forward); 5’-CGTGCTGGTAGCTGAACTG (reverse) for eif3hb. The effectiveness of knock down of endogenous mRNAs for either eif3ha or eif3hb morphants was evaluated by RT-PCR. For eif3hb morphants, 0.5 µg of total RNA was amplified for 28 cycles using the above primers, while for eif3ha morphants, 0.25 µg of total RNA was amplified for 25 cycles using 5’-TGCTTCCCT/GTTCCCTCA (forward, a 50% mixture of either T or G represented in bold) and 5’-ATGCCTGTCAGCCAGC-3’ as forward and reverse primers, respectively. In all experiments, β-actin RNA was amplified using 5’-AAGCAGGAGTACGATGAGTCTG (forward) and 5’-GGTAAACGCTTCTTCTGGAATGAC (reverse) as an internal control.
Semi-quantitative RT-PCR
A single set of forward and reverse primers was designed from sequences that are conserved in both eif3ha and eif3hb genes, 5’-TGCTTCCCT/GTTCCCTCA (forward, a 50% mixture of either T or G represented in bold), 5’-ATGAGATGAGAGTTCTTGATGA (reverse). This ensures that the transcripts expressed from either gene were amplified with equivalent efficiencies yielding a single PCR product of 393-bp. Gene-specific transcripts were distinguished by the presence of unique restriction enzyme sites that are specific for each cDNA : StuI for eif3ha, and KpnI for eif3hb. Assays were performed under conditions for semi-quantitative RT-PCR, whereby the amount of PCR product generated was proportional to the input RNA (data not shown). For adult fish, cDNA from 1 µg of total RNA was PCR-amplified for 26 cycles while for staged embryonic samples, 0.5 µg of total RNA was used and cDNA amplified for 27 cycles. The PCR products obtained in each case were digested with StuI or KpnI in separate restriction enzyme reactions, and products quantified after agarose gel electrophoresis.
Whole mount in situ hybridization
Specific probe sequences were designed to avoid cross-hybridization between eif3ha and eif3hb mRNAs, comprised of sequences 724 – 1117 of eif3hb cDNA and 563 – 973 of eif3ha cDNA. Each construct was linearized by digestion with HindIII and anti-sense RNA was transcribed in vitro (Promega) in the presence of digoxigenin-conjugated UTP using RNA polymerase either from bacteriophage T7 (for eif3ha) or T3 (for eif3hb). A control sense RNA probe was also generated for eif3hb after digestion with NotI, followed by transcription with T7 RNA polymerase. The fluorescein-labeled myod probe was generated from a 1.6-kb cDNA present in pBluescript SK vector, linearized by BamHI, using T7 RNA polymerase in the presence of fluorescein-labeled UTP. Whole mount in situ hybridization was carried out essentially as described (Alexander et al., 1998). Essentially, embryos older than 24 hpf were treated with phenylthiourea to prevent pigmentation, permeabilized using proteinase K (10 µg/ml) for 10–30 min. Hybridization was carried out overnight at 70°C in a buffer containing 65% formamide. Signals were detected using alkaline phosphatase-conjugated anti-digoxigenin Fab antibody and NBT/BCIP staining substrate. Embryos were cleared in 2:1 BB:BA solution before being imaged. For two-color in situ hybridization, digoxigenin-labeled eif3ha and fluorescein-labeled myod anti-sense RNA probes were used. The embryos were treated with alkaline phosphatase-conjugated anti-fluorescein Fab antibody fragments and the signal for myod was developed by initially using INT (indonitrotetrazolium chloride)/BCIP phosphatase substrate. Polyvinyl alchohol was used to concentrate the red precipitate. After detecting myod, embryos were washed with a low pH solution (0.1 M glycine-HCl, pH 2.2) for 10 min and subsequently incubated with alkaline phosphatase-conjugated anti-digoxigenin Fab antibody fragments followed by treatment with NBT/BCIP to detect expression of eif3ha.
In vitro transcription and translation
Reactions were carried out using the TNT-Coupled Rabbit Reticulocyte System (Promega). Each reaction contained 0.5 µg of the expression plasmid (pCS2+-eif3ha or pCS2+-eif3hb) with or without 0.5 µg of specific morpholino oligomers. Following incubation for 1.5 hrs at 30°C, 5 µl of each reaction was electrophoresed in a 10% Nu-PAGE-SDS gel (Invitrogen), and the dried gels were subjected to autoradiography to detect 35S -labeled proteins.
Labeling of neuromasts (Di-ASP staining)
Wild-type and morphant embryos were incubated in 200 µM 4-(4-(diethylamino)styryl)-N-methylpyridiniumiodide (4-Di-2-ASP) solution (Dambly-Chaudiere et al., 2003) at room temperature for 5 min and then imaged under fluorescence using a GFP filter.
Supplementary Material
Acknowledgments
This work was supported by grants GM15399 to U.M. and HL064282 and HL056182 to T.E. from the National Institutes of Health and by Cancer Core Support Grant P30CA13330 to the Einstein College of Medicine from the National Cancer Institute.
We thank members of the Evans and Maitra laboratories for their considerable help during the course of this work.
Grant sponsor: For U. Maitra: NIH; Grant number: GM15399
For T. Evans: NIH; Grant numbers: HL064282 and HL056182
References
- Akiyoshi Y, Clayton J, Phan L, Yamamoto M, Hinnebusch AG, Watanabe Y, Asano K. Fission yeast homolog of murine Int-6 protein, encoded by mouse mammary tumor virus integration site, is associated with the conserved core subunits of eukaryotic translation initiation factor 3. J Biol Chem. 2001;276:10056–10062. doi: 10.1074/jbc.M010188200. [DOI] [PubMed] [Google Scholar]
- Alexander J, Stainier DY, Yelon D. Screening mosaic F1 females for mutations affecting zebrafish heart induction and patterning. Dev Genet. 1998;22:288–299. doi: 10.1002/(SICI)1520-6408(1998)22:3<288::AID-DVG10>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Asano K, Merrick WC, Hershey JW. The translation initiation factor eIF3-p48 subunit is encoded by int-6, a site of frequent integration by the mouse mammary tumor virus genome. J Biol Chem. 1997;272:23477–23480. doi: 10.1074/jbc.272.38.23477. [DOI] [PubMed] [Google Scholar]
- Bahary N, Goishi K, Stuckenholz C, Weber G, Leblanc J, Schafer CA, Berman SS, Klagsbrun M, Zon LI. Duplicate VegfA genes and orthologues of the KDR receptor tyrosine kinase family mediate vascular development in the zebrafish. Blood. 2007;110:3627–3636. doi: 10.1182/blood-2006-04-016378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay A, Lakshmanan V, Matsumoto T, Chang EC, Maitra U. Moe1 and spInt6, the fission yeast homologues of mammalian translation initiation factor 3 subunits p66 (eIF3d) and p48 (eIF3e), respectively, are required for stable association of eIF3 subunits. J Biol Chem. 2002;277:2360–2367. doi: 10.1074/jbc.M107790200. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay A, Matsumoto T, Maitra U. Fission yeast Int6 is not essential for global translation initiation, but deletion of int6(+) causes hypersensitivity to caffeine and affects spore formation. Mol Biol Cell. 2000;11:4005–4018. doi: 10.1091/mbc.11.11.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs S, Chen JN, Garrity DM, Fishman MC. Patterning of angiogenesis in the zebrafish embryo. Development. 2002;129:973–982. doi: 10.1242/dev.129.4.973. [DOI] [PubMed] [Google Scholar]
- Collazo A, Fraser SE, Mabee PM. A dual embryonic origin for vertebrate mechanoreceptors. Science. 1994;264:426–430. doi: 10.1126/science.8153631. [DOI] [PubMed] [Google Scholar]
- Crane R, Craig R, Murray R, Dunand-Sauthier I, Humphrey T, Norbury C. A fission yeast homolog of Int-6, the mammalian oncoprotein and eIF3 subunit, induces drug resistance when overexpressed. Mol Biol Cell. 2000;11:3993–4003. doi: 10.1091/mbc.11.11.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis D, Lehmann R, Zamore PD. Translational regulation in development. Cell. 1995;81:171–178. doi: 10.1016/0092-8674(95)90325-9. [DOI] [PubMed] [Google Scholar]
- Dambly-Chaudiere C, Sapede D, Soubiran F, Decorde K, Gompel N, Ghysen A. The lateral line of zebrafish: a model system for the analysis of morphogenesis and neural development in vertebrates. Biol Cell. 2003;95:579–587. doi: 10.1016/j.biolcel.2003.10.005. [DOI] [PubMed] [Google Scholar]
- David NB, Sapede D, Saint-Etienne L, Thisse C, Thisse B, Dambly-Chaudiere C, Rosa FM, Ghysen A. Molecular basis of cell migration in the fish lateral line: role of the chemokine receptor CXCR4 and of its ligand, SDF1. Proc Natl Acad Sci U S A. 2002;99:16297–16302. doi: 10.1073/pnas.252339399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen JS, Smith JC. Controlling morpholino experiments: don't stop making antisense. Development. 2008;135:1735–1743. doi: 10.1242/dev.001115. [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray NK, Wickens M. Control of translation initiation in animals. Annu Rev Cell Dev Biol. 1998;14:399–458. doi: 10.1146/annurev.cellbio.14.1.399. [DOI] [PubMed] [Google Scholar]
- Groisman I, Huang YS, Mendez R, Cao Q, Richter JD. Translational control of embryonic cell division by CPEB and maskin. Cold Spring Harb Symp Quant Biol. 2001;66:345–351. doi: 10.1101/sqb.2001.66.345. [DOI] [PubMed] [Google Scholar]
- Grzmil M, Whiting D, Maule J, Anastasaki C, Amatruda JF, Kelsh RN, Norbury CJ, Patton EE. The INT6 cancer gene and MEK signaling pathways converge during zebrafish development. PLoS One. 2007;2:e959. doi: 10.1371/journal.pone.0000959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Sen GC. Characterization of the interaction between the interferon-induced protein P56 and the Int6 protein encoded by a locus of insertion of the mouse mammary tumor virus. J Virol. 2000;74:1892–1899. doi: 10.1128/jvi.74.4.1892-1899.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze MW, Gebauer F, Preiss T. cis-Regulatory sequences and trans-acting factors in translational control. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2007. pp. 507–544. [Google Scholar]
- Hinnebusch AG. eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem Sci. 2006;31:553–562. doi: 10.1016/j.tibs.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Holtzinger A, Evans T. Gata4 regulates the formation of multiple organs. Development. 2005;132:4005–4014. doi: 10.1242/dev.01978. [DOI] [PubMed] [Google Scholar]
- Holz MK, Ballif BA, Gygi SP, Blenis J. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell. 2005;123:569–580. doi: 10.1016/j.cell.2005.10.024. [DOI] [PubMed] [Google Scholar]
- Isogai S, Horiguchi M, Weinstein BM. The vascular anatomy of the developing zebrafish: an atlas of embryonic and early larval development. Dev Biol. 2001;230:278–301. doi: 10.1006/dbio.2000.9995. [DOI] [PubMed] [Google Scholar]
- Kapp LD, Lorsch JR. The molecular mechanics of eukaryotic translation. Annu Rev Biochem. 2004;73:657–704. doi: 10.1146/annurev.biochem.73.030403.080419. [DOI] [PubMed] [Google Scholar]
- Kim BH, Cai X, Vaughn JN, von Arnim AG. On the functions of the h subunit of eukaryotic initiation factor 3 in late stages of translation initiation. Genome Biol. 2007;8:R60. doi: 10.1186/gb-2007-8-4-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Kim BH, Yahalom A, Chamovitz DA, von Arnim AG. Translational regulation via 5' mRNA leader sequences revealed by mutational analysis of the Arabidopsis translation initiation factor subunit eIF3h. Plant Cell. 2004;16:3341–3356. doi: 10.1105/tpc.104.026880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol. 2002;248:307–318. doi: 10.1006/dbio.2002.0711. [DOI] [PubMed] [Google Scholar]
- Lele Z, Krone PH. The zebrafish as a model system in developmental, toxicological and transgenic research. Biotechnol Adv. 1996;14:57–72. doi: 10.1016/0734-9750(96)00004-3. [DOI] [PubMed] [Google Scholar]
- Marchetti A, Buttitta F, Miyazaki S, Gallahan D, Smith GH, Callahan R. Int-6, a highly conserved, widely expressed gene, is mutated by mouse mammary tumor virus in mammary preneoplasia. J Virol. 1995;69:1932–1938. doi: 10.1128/jvi.69.3.1932-1938.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Lorsch JR, Hellen CUT. The mechanism of translation initiation in eukaryotes. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2007. pp. 87–128. [Google Scholar]
- Ray A, Bandyopadhyay A, Matsumoto T, Deng H, Maitra U. Fission yeast translation initiation factor 3 subunit eIF3h is not essential for global translation initiation, but deletion of eif3h+ affects spore formation. Yeast. 2008;25:809–823. doi: 10.1002/yea.1635. [DOI] [PubMed] [Google Scholar]
- Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, Ekker SC. p53 activation by knockdown technologies. PLoS Genet. 2007;3:e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanches M, Alves BS, Zanchin NI, Guimaraes BG. The crystal structure of the human Mov34 MPN domain reveals a metal-free dimer. J Mol Biol. 2007;370:846–855. doi: 10.1016/j.jmb.2007.04.084. [DOI] [PubMed] [Google Scholar]
- Scheel H, Hofmann K. Prediction of a common structural scaffold for proteasome lid, COP9-signalosome and eIF3 complexes. BMC Bioinformatics. 2005;6:71. doi: 10.1186/1471-2105-6-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Kahle A, Hershey JW, Honchak BM, Warneke JA, Leong SP, Nelson MA. Decreased expression of eukaryotic initiation factor 3f deregulates translation and apoptosis in tumor cells. Oncogene. 2006;25:4923–4936. doi: 10.1038/sj.onc.1209495. [DOI] [PubMed] [Google Scholar]
- Shimeld SM. Gene function, gene networks and the fate of duplicated genes. Semin Cell Dev Biol. 1999;10:549–553. doi: 10.1006/scdb.1999.0336. [DOI] [PubMed] [Google Scholar]
- Thompson B, Wickens M, Kimble J. Translational control in development. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2007. pp. 507–544. [Google Scholar]
- van Eeden FJ, Granato M, Schach U, Brand M, Furutani-Seiki M, Haffter P, Hammerschmidt M, Heisenberg CP, Jiang YJ, Kane DA, Kelsh RN, Mullins MC, Odenthal J, Warga RM, Allende ML, Weinberg ES, Nusslein-Volhard C. Mutations affecting somite formation and patterning in the zebrafish, Danio rerio. Development. 1996;123:153–164. doi: 10.1242/dev.123.1.153. [DOI] [PubMed] [Google Scholar]
- von Arnim AG, Schwechheimer C. Life is degrading--thanks to some zomes. Mol Cell. 2006;23:621–629. doi: 10.1016/j.molcel.2006.08.012. [DOI] [PubMed] [Google Scholar]
- Weinberg ES, Allende ML, Kelly CS, Abdelhamid A, Murakami T, Andermann P, Doerre OG, Grunwald DJ, Riggleman B. Developmental regulation of zebrafish MyoD in wild-type, no tail and spadetail embryos. Development. 1996;122:271–280. doi: 10.1242/dev.122.1.271. [DOI] [PubMed] [Google Scholar]
- Yen HC, Chang EC. Yin6, a fission yeast Int6 homolog, complexes with Moe1 and plays a role in chromosome segregation. Proc Natl Acad Sci U S A. 2000;97:14370–14375. doi: 10.1073/pnas.97.26.14370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen HC, Gordon C, Chang EC. Schizosaccharomyces pombe Int6 and Ras homologs regulate cell division and mitotic fidelity via the proteasome. Cell. 2003;112:207–217. doi: 10.1016/s0092-8674(03)00043-6. [DOI] [PubMed] [Google Scholar]
- Zhou C, Arslan F, Wee S, Krishnan S, Ivanov AR, Oliva A, Leatherwood J, Wolf DA. PCI proteins eIF3e and eIF3m define distinct translation initiation factor 3 complexes. BMC Biol. 2005;3:14. doi: 10.1186/1741-7007-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.