Abstract
In May 2000, the first outbreak of vancomycin-resistant Enterococcus faecium (VREF) was detected in the University Medical Center Utrecht in the nephrology ward. The question arose why some VREF strains spread among hospitalized patients, whereas other strains do not. Thirty patients who were found to be colonized with VREF between May and November 2000 were included in the study. Molecular typing confirmed that 19 of them carried an identical epidemic strain which harbored the esp gene while 11 were colonized by nonepidemic strains that were all esp negative. Acquisition of the outbreak strain was significantly associated with diabetes mellitus, renal transplantation, and extensive use of antibiotics, especially cephalosporins, in the 2-month period before the first isolation of VREF. To establish the duration of colonization, prospective surveillance of VREF carriage for a 6-month period starting from the first isolation of VREF was realized for 20 patients. After 6 months, VREF was still recovered from 60% of carriers of the outbreak strain versus 20% of carriers of nonepidemic strains (P < 0.01). However, antibiotic use during the follow-up period was significantly higher by carriers of the outbreak strain than by carriers of nonepidemic strains. The fact that the outbreak strain was recovered for a longer period of time than nonepidemic strains may facilitate dissemination of the strain. The results support a careful restrictive antibiotic policy for wards at risk for spread of VREF and implementation of isolation precautions for patients who are colonized with esp-positive outbreak strains.
Transmission of vancomycin-resistant Enterococcus faecium (VREF) among patients has been an increasing problem in many American hospitals since the nineties, which mirrors the emergence of methicillin-resistant Staphylococcus aureus. Especially in immunocompromised hosts, VREF strains are important causative agents of nosocomial infections. Such infections are difficult to treat, since VREF strains are no longer susceptible to most antibiotics (7, 23). Therefore, stringent infection control guidelines were recommended by the Centers for Disease Control and Prevention in 1994 to prevent nosocomial spread of VREF (17).
Remarkably, the epidemiology of VREF in the United States is distinct from that in Europe. In The Netherlands, as in other European countries, VREF outbreaks have been reported considerably less frequently than in the United States, but carriage of VREF among healthy individuals in the community is not unusual in Europe (13, 28). This phenomenon has been ascribed to the use of the vancomycin analogue avoparcin as a growth promoter in the livestock industry until it was banned by the European Union in 1997 (27). In the United States, where the use of avoparcin has never been allowed, VREF strains are not recovered from nonhospitalized individuals and rarely recovered from farm animals.
Using molecular typing, it was shown that outbreak strains isolated from hospitalized patients are genetically different from VREF strains present in the fecal flora of nonhospitalized individuals (29). Interestingly, strains recovered from nonhospitalized persons are genetically related to pig isolates (29). In addition, hospital outbreak strains are distinguished by the presence of the variant esp gene (30). This esp gene, which encodes a surface protein, was first identified in Enterococcus faecalis isolates and has been associated with increased virulence (26).
In May 2000, we experienced the first VREF outbreak in our institute (24). It is not known what determines why some VREF strains spread among hospitalized patients, whereas other strains which circulate in the community do not. In this study, we characterized the epidemiology of colonization with the outbreak strain, comparing risk factors for acquisition of the outbreak strain versus nonepidemic VREF strains. In addition, VREF colonization was monitored in carriers of both the outbreak strain and nonepidemic VREF strains for a 6-month period starting from the date of first isolation of VREF, since several studies have demonstrated that many patients colonized by VREF became persistent fecal carriers, especially those who received antibiotic therapy (6, 10, 19, 22).
MATERIALS AND METHODS
Setting and patients.
The University Medical Center Utrecht is a 1,042-bed tertiary care center, where the first VREF outbreak was detected in May 2000. Most patients who were found to be colonized by the outbreak strain were hospitalized in the nephrology ward or were visitors to the hemodialysis ward, but patients with the outbreak strain were also identified in the medical intensive care unit, the neurosurgical ward, and miscellaneous wards (24). The guidelines implemented to prevent nosocomial spread of VREF were based on Hospital Infection Control Practices Advisory Committee guidelines and included separate rooms, the use of gloves and gowns, disinfection of rooms after patients were discharged, and the use of vancomycin was limited as much as possible (17). Weekly surveillance cultures were performed for all patients of the nephrology and hemodialysis wards during the outbreak. Isolation precautions were restricted to patients who were colonized by epidemic VREF strains; no specific measures were installed for carriers of nonepidemic VREF strains (24). Patients colonized by epidemic VREF strains were labeled in the hospital information system so that isolation precautions for these patients could be installed as soon as they entered the hospital again.
VREF-positive patients from whom follow-up cultures had been taken as part of routine surveillance during the outbreak were included in the study. In addition, 38 patients who were found to be colonized by VREF during this outbreak between May and November 2000 and had been discharged from our hospital were sent a letter in which they were asked to participate in this study of VREF carriage.
Microbiology and molecular typing.
Patients were screened for carriage of VREF by culturing rectal swabs in enterococcosel broth (Becton Dickinson, Cockeysville, Md.) supplemented with aztreonam (75 mg/liter) at 37°C. Upon black colorization within 48 h, the broth was subcultured on an enterococcosel agar plate (Becton Dickinson), supplemented with aztreonam and vancomycin (25 mg/liter) at 37°C for 48 h. Black colonies showing gram-positive cocci were identified as VREF by APIstrep (bioMérieux, Marcy l'Étoile, France) and E-test (MIC of vancomycin, ≥32 mg/liter; bioMérieux). VREF isolates were subjected to multiplex PCR to detect vancomycin-resistance genes (van genes) and to esp PCR as described previously (12, 30). To detect clustering, characterization of VREF isolates was performed by pulsed-field gel electrophoresis (PFGE) as described previously (1). In addition, normalized PFGE patterns were imported into BioNumerics software (version 3.0; Applied Maths, Sint-Martens-Latem, Belgium). The outbreak strain was defined as a vanA-positive, esp-positive E. faecium strain, which was distinguished from nonepidemic strains by PFGE, with a cutoff value of 80% genetic similarity for discrimination between distinct clusters of strains. The outbreak strain has been described in reference 30 and was designated Neth-2-1.
Risk factor analysis for VREF acquisition.
Medical records from the included patients were screened for risk factors for acquisition of VREF, including antibiotic use, medical history, invasive procedures, and length of stay. The records of patients who were carriers of the epidemic strain were compared with those of patients who carried nonepidemic strains.
Follow-up of VREF carriage.
Prospective surveillance of VREF carriage was performed to study the duration of colonization. For each patient, the date of first isolation of VREF was designated day 0. For patients who were hospitalized or who were visitors to the hemodialysis ward, surveillance cultures were performed weekly. From then on, VREF carriage in patients was monitored for a period of 6 months. Patients who were discharged from the hospital were asked to take rectal swabs on 3 consecutive days each month for a period of 6 months. They received culture materials and an instruction letter which explained how rectal swabs should be taken. Only patients from whom at least three follow-up cultures had been obtained, including at least one culture after 22 weeks of follow-up, were included in the analysis of the follow-up study. Patients whose last positive cultures were detected before the 22nd week of follow-up and from whom only one or two subsequent negative follow-up cultures were registered were excluded from the analysis. Follow-up of VREF carriage for both patient groups was monitored on a Kaplan-Meier curve. By the end of the follow-up period, patients were classified according to two categories: persisters and converters. Persons whose rectal swabs yielded the original VREF strain after 22 weeks had passed since the first isolation of VREF, whether or not alternating with negative culture results, were designated persisters. Converters were defined as persons from whom the last three (or more) cultures were negative. The average between the week of the last positive culture and the week of the first following negative culture was designated the week of conversion. In addition, carriage of a newly acquired VREF strain was documented. Length of stay and antibiotic use were registered during the whole follow-up period.
Statistics.
Statistical analyses were performed by using t test, Fisher's exact test, and χ2 tests. Multivariate analyses were performed by using logistic regression and a Cox proportional hazard model. P values of <0.05 were considered significant. As a measure of association, relative risks were calculated with their 95% confidence limits.
RESULTS
Patients.
VREF carriers from whom weekly surveillance cultures were available included 11 patients at the hemodialysis ward and 2 patients who were hospitalized in the nephrology department. In addition, a total of 38 patients who were found to be carriers of VREF between May and November 2000 were asked to participate in this study of risk factors for acquisition of the epidemic VREF strain and of the duration of colonization with VREF. Seventeen of them responded positively and were included, 18 patients did not participate, and 3 patients had died before start of the study. Thus, a total of 30 patients were included; none of them developed a clinically evident infection caused by VREF. Demographic and clinical data for these patients are given in Table 1.
TABLE 1.
Variables in patients who contracted the outbreak strain versus patients who contracted nonepidemic strains (univariate analysis)
| Risk factor for acquisitiona | Result for patients with:
|
RRb (95% CI) | |
|---|---|---|---|
| Nonepidemic VREF (n = 11) | Epidemic VREF (n = 19) | ||
| Mean age (yr) (range) | 61.8 (18.0-86.1) | 59.7 (37.7-80.5) | |
| Sex (male/female) | 8/3 | 11/8 | |
| Hospitalization in nephrology ward | 7 (63.6) | 11 (57.9) | |
| Diabetes mellitus | 0 (0) | 6 (31.6)c | 1.7 (1.1-2.6) |
| Hemodialysis | 6 (54.5) | 8 (42.1) | |
| Kidney transplant | 0 (0) | 4 (21.1)d | 1.6 (1.0-2.5) |
| Autologic bone marrow transplant | 2 (18.2) | 0 (0) | |
| Mean length of stay (days) (range) | 20 (0-94) | 40 (0-209) | |
| No antibiotics | 1 (9.1) | 0 (0) | |
| 1 Antibiotic | 6 (54.5) | 4 (21.1) | |
| 2-3 Antibiotics | 2 (18.2) | 13 (68.4)e | 9.7 (1.6-59.7) |
| ≥4 Antibiotics | 2 (18.2) | 2 (10.5) | |
Unless otherwise indicated, all data are numbers of patients, with percentages of the total number of patients indicated in parentheses.
RR, relative risk.
P = 0.037.
P = 0.102.
P = 0.008.
Microbiology and molecular typing.
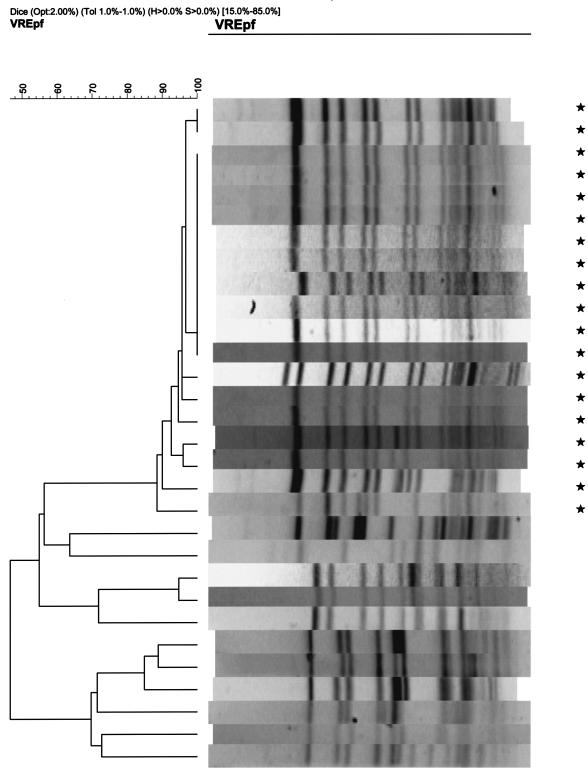
The first VREF isolates from all patients, all from rectal swabs, were available for analysis by PFGE and esp PCR. PFGE analysis revealed at least nine distinct major PFGE types based on a cutoff value of 80% genetic similarity for discrimination between distinct clusters of strains. Nineteen patients were found to be colonized by the nosocomial outbreak strain, which clustered with other epidemic genotypes according to amplified fragment length polymorphism patterns (data not shown) (30). In addition, 11 patients were carriers of nonepidemic strains which were represented by eight other major types (Fig. 1). All isolates which belonged to the outbreak strain harbored the esp gene, whereas this gene was present in none of the nonoutbreak isolates. All patient isolates recovered during the follow-up period were analyzed by PFGE. Each patient from whom VREF was repetitively cultured had a recurrence of colonization with a strain that was indistinguishable from the prior colonizing strain.
FIG. 1.
PFGE patterns of VREF isolates from 30 patients. Lanes marked by asterisks represent isolates belonging to the outbreak strain; the remaining lanes represent isolates belonging to nonepidemic strains.
Risk factors for acquisition of the outbreak strain.
Patients who carried the outbreak strain were compared to patients who were carriers of nonepidemic strains. The receipt of two or more antibiotics and use of cephalosporins and clindamycin during the 2 months prior to the first positive VREF culture were associated with a significantly increased risk for acquisition of the outbreak strain (Table 2). In addition, diabetes mellitus and renal transplantation were also significant risk factors for acquiring the outbreak strain. Two or more antibiotics had been prescribed to 4 of 6 patients with diabetes and to 2 of 4 patients who had undergone renal transplantation. After logistic regression, only the use of cephalosporins during the 2 months prior to the first positive VREF culture was a significantly increased risk for acquisition of the outbreak strain (P = 0.046).
TABLE 2.
Antibiotic use in a 2-month period prior to the first isolation of VREF in patients who contracted the outbreak strain versus patients who contracted nonepidemic strains (univariate analysis)
| Antibiotic(s) | No. (%) of patients with:
|
RRa (95% CI) | P value | |
|---|---|---|---|---|
| Nonepidemic VREF (n = 11) | Epidemic VREF (n = 19) | |||
| Penicillins | 5 (45.5) | 12 (63.2) | 2.1 (0.5-9.3) | 0.346 |
| Cephalosporins | 1 (9.1) | 13 (68.4) | 21.6 (2.2-210.1) | 0.002 |
| Aminoglycosides | 2 (18.2) | 2 (10.5) | 0.5 (0.06-4.4) | 0.552 |
| Clindamycin | 0 (0) | 5 (26.3) | 1.8 (1.3-2.5) | 0.062 |
| Quinolones | 1 (9.1) | 4 (21.1) | 2.7 (0.3-27.5) | 0.397 |
| Glycopeptides | 3 (27.3) | 2 (10.5) | 0.3 (0.04-2.3) | 0.236 |
| Other | 6 (54.5) | 4 (21.1) | 0.2 (0.04-1.1) | 0.060 |
RR, relative risk.
Follow-up study of VREF carriage.
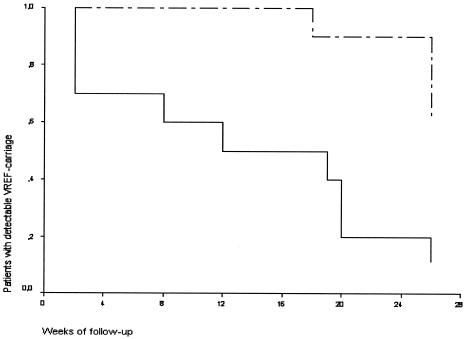
Ten patients (9 carrying the outbreak strain and 1 carrying nonoutbreak strains) were excluded from follow-up during the study. Cultures had not been taken after 22 weeks from five patients; from three other patients, no more than two follow-up cultures were available at all; and two additional patients who showed positive follow-up cultures before the 22nd week showed only one and two subsequently negative cultures, respectively. Thus, 20 of 30 patients fulfilled the inclusion criteria for analysis in the follow-up study. From patients who were colonized by the outbreak strain, an average of 10 follow-up cultures was obtained (range, 3 to 24) while an average of 13 follow-up cultures was available (range, 3 to 23) from patients who were colonized by nonepidemic strains. Forty-seven percent of all follow-up cultures from patients with the outbreak strain were positive versus 15% of all follow-up cultures from patients with nonepidemic strains. One patient who was carrying the outbreak strain at time zero converted; the first negative follow-up culture was followed by a series of four negative cultures only. The remaining nine patients, persisters, had positive cultures in the sixth month of follow-up (Fig. 2). Four of them showed intermittently positive cultures after a series of at least three consecutive negative cultures, with relapse of VREF stool carriage after up to 15 negative cultures over a 3-month period in one patient. In addition, eight carriers of nonepidemic strains converted while the other two were classified as persisters because of positive cultures after a series of negative cultures, as shown in Fig. 2. After 6 months of follow-up, the outbreak strain was detected in significantly more patients than were nonepidemic strains (χ2 = 9.899; P < 0.01). During follow-up, patients with the outbreak strain had used significantly more antibiotics, especially cephalosporins, than patients colonized with nonepidemic strains, whereas no significant differences with regard to hospitalization were noticed between the two groups (Table 3). In a multivariate analysis using a Cox proportional hazard model to control for different lengths of follow-up, however, use of cephalosporins was not significantly associated with prolonged VREF colonization. In this model, carriage of the outbreak strain was the only significant risk factor for prolonged carriage of VREF (hazard ratio, 5.6; 95% confidence interval [CI], 1.2 to 25.7; P = 0.028). In addition to duration of colonization with the initially cultured VREF strain, acquisition of different strain types was documented. No patient who was a carrier of the outbreak strain contracted a new, nonepidemic VREF strain, and none of the carriers of nonepidemic strains was found to be colonized by the outbreak strain over the study period.
FIG. 2.
Kaplan-Meier curve of detectable VREF colonization in patients during a 6-month follow-up period starting from the first positive VREF culture. Patients who were colonized with a particular outbreak strain (dashed line) were compared to patients who carried nonepidemic strains (solid line). The P value was 0.0026 by log rank analysis.
TABLE 3.
Variables for patients who were colonized with the outbreak strain and patients who carried nonepidemic strains during a 6-month follow-up period starting from the first isolation of VREF
| Patient variable during follow-up | Result for patients with:
|
RRa (95% CI) | P value | |
|---|---|---|---|---|
| Nonepidemic VREF (n = 10) | Epidemic VREF (n = 10) | |||
| Mean length of stay (days) (range) | 14.7 (0-57) | 44.4 (5-181) | 0.12 | |
| No. of days on antibiotic | ||||
| Total | 4.8 | 43.3 | 0.009 | |
| Cephalosporins | 1 | 6 | 13.5 (1.2-152.2) | 0.05 |
| Clindamycin | 1 | 3 | 3.8 (0.3-45.6) | 0.58 |
| Vancomycin | 3 | 2 | 0.58 (0.08-4.6) | 1.0 |
RR, relative risk.
DISCUSSION
We studied the epidemiology of colonization with a particular epidemic VREF strain which clustered with other nosocomial outbreak strains and, in accordance with these strains, harbored the esp gene (30). For this purpose, we compared patients who acquired the epidemic VREF strain with patients who carried nonepidemic VREF strains. We showed that acquisition of the outbreak strain was associated with receipt of two or more antibiotics in the 2-month period before the first isolation of VREF. In multivariate analysis, however, only prescription of cephalosporins was significantly associated with acquisition of the outbreak strain. Obviously, clearance of the resident flora by antibiotics was associated with cross-transmission of the outbreak VREF strain between patients. In accordance with this, Fridkin et al., who studied acquisition of a dominant VREF strain in their hospital, reported that acquisition of this dominant strain among other VREF strains was associated with intensity of antibiotic exposure (14). In our study, patients with diabetes mellitus or renal transplantation contracted the epidemic strain significantly more often than nonepidemic strains. Although not supported by statistical significance, prolonged hospitalization tended to be related with acquisition of the outbreak strain as well.
While our study evaluated the risk of contracting a particular outbreak strain among all patients who acquired VREF, most other studies addressed risk factors for the acquisition of any VREF strain. Several of these studies also identified antibiotic therapy, especially treatment with antianaerobic antibiotics, vancomycin, and expanded-spectrum cephalosporins (5, 11, 15-16, 20) as risk factors for acquisition of VREF. It is thought that antibiotics promote the overgrowth of VREF in the intestinal tract, primarily through the inhibition of intestinal anaerobes. Interestingly, Zaas et al. found that diabetes was a risk factor for development of bloodstream infections with VREF in patients with cancer who were colonized with VREF (31). In addition, other factors such as underlying illnesses, compliance with infection control measures, proximity to other VREF-colonized patients, and prolonged or repetitive hospitalization have been reported to affect acquisition of VREF (3, 5, 8, 16, 20, 31).
As patients are identified, it becomes important to know the duration of colonization and when it is safe to remove patients from isolation. The Hospital Infection Control Practices Advisory Committee has recommended using three consecutive negative cultures, at least 1 week apart, for determining clearance of VREF from a previously colonized patient (17). However, it is known that VREF may be detectable for a prolonged period of up to more than 2 years, sometimes despite numerous negative follow-up cultures (22, 25). Unrecognized silent carriage of VREF is among the most important factors leading to person-to-person dissemination in the hospital (9, 21). Therefore, patients who were colonized with the outbreak strain in our hospital were subjected to isolation precautions during the whole study period, even in the case of repetitive negative follow-up cultures.
In course of the study period, VREF strains were detectable in a decreasing number of patients. Prolonged carriage of the outbreak strain was demonstrated in significantly more carriers of the outbreak strain than in carriers of nonepidemic strains. Remarkably, seven patients who showed a series of at least three negative follow-up cultures, which even amounted to 15 over a 3-month period in one patient, were found to be VREF positive again. Obviously, observed relapses of colonization are at least partly due to the limited sensitivity of VREF culture methods (2, 9). In this respect, the rates of isolation of VREF from stool specimens have been reported to be significantly higher than from rectal swabs, and broth enrichment is recommended as well (9, 18). Thus, false-negative cultures may reflect a decrease in the quantity of VREF to an undetectable level rather than true eradication.
We already showed differences between both patient groups, with more diabetics and renal transplant patients carrying the outbreak strain. Moreover, patients with the outbreak strain also received more antibiotics than patients who carried nonepidemic strains during the follow-up period. In addition, the length of stay for patients with the outbreak strain was considerably longer than for patients with nonepidemic strains, but this observation was not supported by statistical significance, probably due to small numbers of patients. Next to strain characteristics, these patient-related factors may affect the duration of colonization with VREF. In accordance, Roghman et al. showed that oncology patients with persistent and intermittent colonization were more likely to have received vancomycin during hospitalization than patients who cleared VREF (25). Byers et al. found that prolonged hospitalization, intensive care, and antibiotic use were each associated with prolonged carriage of VREF (6). Other studies showed that treatment with antianaerobic antibiotics promoted high-density colonization in patients who were colonized by VREF (9, 11).
It has been reported that VREF isolates showed little variation within individual patients (4). In accordance, PFGE patterns of subsequent VREF patient isolates showed that none of the patients who were initially found to be colonized by the outbreak strain acquired another, nonepidemic strain within the follow-up period. In addition, none of the 11 carriers of nonepidemic strains contracted the outbreak strain during the study period, obviously due to successful implementation of isolation precautions for carriers of the outbreak strain.
Some VREF strains, which are recognized in genetic clusters and harbor the esp gene, are more prone to nosocomial dissemination than others (30). This study shows that a particular outbreak strain which belongs to this cluster (strain Neth-2-1 in reference 30) preferentially colonized patients who had been subjected to extensive antibiotic exposure and certain patient categories with underlying illnesses such as diabetes mellitus and renal transplantation. We suggest that a careful restrictive antibiotic policy should be pursued in wards at risk for the spread of VREF to prevent dissemination of epidemic VREF strains. We demonstrated that this outbreak strain was maintained in the fecal flora of the host for a significantly longer period of time than nonepidemic strains, but we cannot exclude the possibility that the extensive use of antibiotics by patients with the outbreak strain during follow-up contributed to the prolonged duration of colonization. Our results confirm that a series of three negative follow-up cultures does not guarantee elimination of VRE and support the importance of prolonged hygienic precautions for patients who are colonized by outbreak strains that harbor the esp gene, especially if these patients are treated (again) with antibiotics. More studies are necessary to elucidate the relative contributions of patient-related factors, antibiotic treatment, and strain characteristics to prolonged duration of VREF colonization.
Acknowledgments
We thank Armand Paauw and Mabel Beitsma for technical assistance.
This study was supported by CVZ/VAZ grant no. 01232.
REFERENCES
- 1.Antonishyn, N. A., R. R. McDonald, E. L. Chan, G. Horsman, C. E. Woodmansee, P. S. Falk, and C. G. Mayhall. 2000. Evaluation of fluorescence-based amplified fragment length polymorphism analysis for molecular typing in hospital epidemiology: comparison with pulsed-field gel electrophoresis for typing strains of vancomycin-resistant Enterococcus faecium. J. Clin. Microbiol. 38:4058-4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baden, L. R., W. Thiemke, A. Skolnik, R. Chambers, J. Strymish, H. S. Gold, R. C. Moellering, and G. M. Eliopoulos. 2001. Prolonged colonization with vancomycin-resistant Enterococcus faecium in long term care patients and the significance of “clearance.” Clin. Infect. Dis. 33:1654-1660. [DOI] [PubMed] [Google Scholar]
- 3.Bonten, M. J. M., S. Slaughter, A. W. Ambergen, M. K. Hayden, J. van Voorhis, C. Nathan, and R. A. Weinstein. 1998. The role of “colonization pressure” in the spread of vancomycin-resistant enterococci. Arch. Intern. Med. 158:1127-1132. [DOI] [PubMed] [Google Scholar]
- 4.Bonten, M. J. M., M. K. Hayden, C. Nathan, T. W. Rice, and R. A. Weinstein. 1998. Stability of vancomycin-resistant enterococcal genotypes isolated from long-term-colonized patients. J. Infect. Dis. 177:378-382. [DOI] [PubMed] [Google Scholar]
- 5.Byers, K. E., A. M. Anglim, C. J. Anneski, T. P. Germanson, H. S. Gold, L. J. Durbin, B. M. Simonton, and B. M. Farr. 2001. A hospital epidemic of vancomycin-resistant Enterococcus: risk factors and control. Infect. Control Hosp. Epidemiol. 22:140-147. [DOI] [PubMed] [Google Scholar]
- 6.Byers, K. E., A. M. Anglim, C. J. Anneski, and B. M. Farr. 2002. Duration of colonization with vancomycin-resistant Enterococcus. Infect. Control Hosp. Epidemiol. 23:207-211. [DOI] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. 1999. National nosocomial infections surveillance (NNIS) system report, data summary from January 1990-May 1999, issued June 1999. Am. J. Infect. Control 27:520-532. [DOI] [PubMed] [Google Scholar]
- 8.D'Agata, E. M. C., W. K. Green, G. Schulman, H. Li, Y.-W. Tang, and W. Schaffner. 2001. Vancomycin-resistant enterococci among chronic hemodialysis patients: a prospective study of acquisition. Clin. Infect. Dis. 32:23-29. [DOI] [PubMed] [Google Scholar]
- 9.D'Agata, E. M. C., S. Gautam, W. K. Green, and Y.-W. Tang. 2002. High rate of false-negative results of the rectal swab culture method in detection of gastrointestinal colonization with vancomycin-resistant enterococci. Clin. Infect. Dis. 34:167-172. [DOI] [PubMed] [Google Scholar]
- 10.Donskey, C. J., C. K. Hoyen, S. M. Das, M. S. Helfand, and M. T. Hecker. 2002. Recurrence of vancomycin-resistant Enterococcus stool colonization during antibiotic therapy. Infect. Control Hosp. Epidemiol. 23:436-440. [DOI] [PubMed] [Google Scholar]
- 11.Donskey, C. J., T. K. Chowdhry, M. T. Hecker, C. K. Hoyen, J. A. Hanrahan, A. M. Hujer, R. A. Hutton-Thomas, C. C. Whalen, R. A. Bonomo, and L. B. Rice. 2000. Effect of antibiotic therapy on the density of vancomycin-resistant enterococci in the stool of colonized patients. N. Engl. J. Med. 343:1925-1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dutka-Malen, S., S. Evers, and P. Courvalin. 1995. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J. Clin. Microbiol. 33:24-27. (Erratum, 33: 1434.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Endtz, H. P., N. van den Braak, A. Van Belkum, J. Kluytmans, J. G. M. Koeleman, L. Spanjaard, A. Voss, A. J. L. Weersink, C. M. J. E Vandenbroucke-Grauls, A. G. M. Buiting, A. Van Duin, and H. A. Verbrugh. 1997. Fecal carriage of vancomycin-resistant enterococci in hospitalized patients and those living in the community in The Netherlands. J. Clin. Microbiol. 35:3026-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fridkin, S. K., D. S. Yokoe, C. G. Whitney, A. Onderdonk, and D. C. Hooper. 1998. Epidemiology of a dominant clonal strain of vancomycin-resistant Enterococcus faecium at separate hospitals in Boston, Massachusetts. J. Clin. Microbiol. 36:965-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fridkin, S. K., J. R. Edwards, J. M. Courval, H. Hill, F. C. Tenover, R. Lawton, R. Gaynes, and J. E. McGowan. 2001. The effect of vancomycin and third-generation cephalosporins on prevalence of vancomycin-resistant enterococci in 126 U. S. adult intensive care units. Ann. Intern. Med. 135:175-183. [DOI] [PubMed] [Google Scholar]
- 16.Henning, K. J., H. Delencastre, J. Eagan, N. Boone, A. Brown, M. Chung, N. Wollner, and D. Armstrong. 1996. Vancomycin-resistant Enterococcus faecium on a pediatric oncology ward: duration of stool shedding and incidence of clinical infection. Pediatr. Infect. Dis. J. 15:848-854. [DOI] [PubMed] [Google Scholar]
- 17.Hospital Infection Control Practices Advisory Committee (HICPAC). 1995. Recommendations for preventing the spread of vancomycin resistance. Infect. Control Hosp. Epidemiol. 16:105-113. [DOI] [PubMed] [Google Scholar]
- 18.Ieven, M., E. Vercauteren, P. Descheemaker, F. Van Laer, and H. Goossens. 1999. Comparison of direct plating and broth enrichment culture for the detection of intestinal colonization by glycopeptide-resistant enterococci among hospitalized patients. J. Clin. Microbiol. 37:1436-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai, K. K., S. A. Fontecchio, A. L. Kelley, Z. S. Melvin, and S. Baker. 1997. The epidemiology of fecal carriage of vancomycin-resistant enterococci. Infect. Control Hosp. Epidemiol. 18:762-765. [PubMed] [Google Scholar]
- 20.MacIntyre, C. R., M. Empson, C. Boardman, D. Sindhusake, J. Lokan, and G. V. Brown. 2001. Risk factors for colonization with vancomycin-resistant enterococci in a Melbourne Hospital. Infect. Control Hosp. Epidemiol. 22:624-629. [DOI] [PubMed] [Google Scholar]
- 21.Martone, W. J. 1998. Spread of vancomycin-resistant enterococci: why did it happen in the United States? Infect. Control Hosp. Epidemiol. 19:539-545. [DOI] [PubMed] [Google Scholar]
- 22.Montecalvo, M. A., H. de Lencastre, M. Carraher, C. Gedris, M. Chung, K. Van Horn, and G. P. Wormser. 1995. Natural history of colonization with vancomycin-resistant Enterococcus faecium. Infect. Control Hosp. Epidemiol. 16:680-685. [DOI] [PubMed] [Google Scholar]
- 23.Murray, B. E. 1998. Diversity among mutidrug-resistant enterococci. Emerg. Infect. Dis. 4:37-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ridwan, B., E. Mascini, N. Van Der Reijden, J. Verhoef, and M. Bonten. 2002. What action should be taken to prevent spread of vancomycin resistant enterococci in European hospitals? Br. Med. J. 324:666-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roghman, M.-C., S. Qaiyumi, R. Schwalbe, and J. G. Morris. 1997. Natural history of colonization with vancomycin-resistant Enterococcus faecium. Infect. Control Hosp. Epidemiol. 18:679-680. [DOI] [PubMed] [Google Scholar]
- 26.Shankar, V., A. S. Baghdayan, M. M. Huycke, G. Lindahl, and M. S. Gilmore. 1999. Infection-derived Enterococcus faecalis strains are enriched in esp, a gene encoding a novel surface protein. Infect. Immun. 67:193-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stobberingh, E., A. Van den Bogaard, N. London, C. Driessen, J. Top, and R. Willems. 1999. Enterococci with glycopeptide resistance in turkeys, turkey farmers, turkey slaughterers, and (sub)urban residents in the south of The Netherlands: evidence for transmission of vancomycin resistance from animals to humans? Antimicrob. Agents Chemother. 43:2215-2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van den Braak, N., A. Ott, A. van Belkum, J. A. J. W. Kluytmans, J. G. M. Koeleman, L. Spanjaard, A. Voss, A. J. L. Weersink, C. M. J. E. Vandenbroucke-Grauls, A. G. M. Buiting, H. A. Verbrugh, and H. P. Endtz. 2000. Prevalence and determinants of fecal colonization with vancomycin-resistant Enterococcus in hospitalized patients in the Netherlands. Infect. Control Hosp. Epidemiol. 21:520-524. [DOI] [PubMed] [Google Scholar]
- 29.Willems, R. J. L., J. Top, N. van den Braak, A. van Belkum, H. Endtz, D. Mevius, E. Stobberingh, A. van den Bogaard, and J. D. A. van Embden. 2000. Host specificity of vancomycin-resistant Enterococcus faecium. J. Infect. Dis. 182:816-823. [DOI] [PubMed] [Google Scholar]
- 30.Willems, R. J. L., W. Homan, J. Top, M. Van Santen-Verheuvel, D. Tribe, X. Manzioros, C. Gaillard, C. M. J. E. Vandenbroucke-Grauls, E. M. Mascini, E. Van Kregten, J. A. Van Embden, and M. J. M. Bonten. 2001. Variant esp gene as a marker of a distinct genetic lineage of vancomycin-resistant Enterococcus faecium spreading in hospitals. Lancet 357:853-855. [DOI] [PubMed] [Google Scholar]
- 31.Zaas, A. K., X. Song, P. Tucker, and T. M. Perl. 2002. Risk factors for development of vancomycin-resistant enterococcal bloodstream infection in patients with cancer who are colonized with vancomycin-resistant enterococci. Clin. Infect. Dis. 35:1139-1146. [DOI] [PubMed] [Google Scholar]