Abstract
A total of 128 Streptococcus pneumoniae isolates that were susceptible to penicillin but resistant to non-β-lactam agents were isolated from young carriers in Greece and analyzed by antibiotic susceptibility testing, serotyping, restriction fragment end labeling (RFEL), and antibiotic resistance genotyping. The serotypes 6A/B (49%), 14 (14%), 19A/F (11%), 11A (9%), 23A/F (4%), 15B/C (2%), and 21 (2%) were most prevalent in this collection. Of the isolates, 65% were erythromycin resistant, while the remaining isolates were tetracycline and/or trimethoprim-sulfamethoxazole resistant. Fifty-nine distinct RFEL types were identified. Twenty different RFEL clusters, harboring 2 to 19 strains each, accounted for 76% of all strains. Confirmatory multilocus sequence typing analysis of the genetic clusters showed the presence of three international clones (Tennessee23F-4, England14-9, and Greece6B-22) representing 30% of the isolates. The erm(B) gene was present in 70% of the erythromycin-resistant isolates, whereas 18 and 8% contained the mef(A) and mef(E) genes, respectively. The pneumococci representing erm(B), erm(A), and mef genes belonged to distinct genetic clusters. In total, 45% of all isolates were tetracycline resistant. Ninety-six percent of these isolates contained the tet(M) gene. In conclusion, penicillin-susceptible pneumococci resistant to non-β-lactams are a genetically heterogeneous group displaying a variety of genotypes, resistance markers, and serotypes. This suggests that multiple genetic events lead to non-β-lactam-resistant pneumococci in Greece. Importantly, most of these genotypes are capable of disseminating within the community.
Streptococcus pneumoniae is a common cause of invasive diseases, such as meningitis and bacteremia, and of respiratory tract infections (5). S. pneumoniae isolates that are resistant to penicillin and/or non-β-lactam agents have been frequently reported (6, 12, 35). Resistance of S. pneumoniae to erythromycin and the other macrolides is increasing in many parts of the world (15, 17). Strains resistant to erythromycin are also resistant to azithromycin, clarithromycin, and roxithromycin (37). This global increase in antibiotic-resistant and especially multidrug-resistant pneumococci is the result of the spread of various highly resistant pneumococcal clones (7, 25).
In Greece, the emergence of antibiotic resistance among pneumococcal isolates was recognized in the mid-1990s (30). Recently, pneumococcal isolates susceptible to penicillin and resistant to chloramphenicol, tetracycline, erythromycin, clindamycin, and trimethoprim-sulfamethoxazole (SXT) have been isolated from young Greek carriers. During the period from December 1995 to February 1996, 24% of the pneumococci isolated from healthy carriers attending day care centers were demonstrated to be penicillin-susceptible, non-β-lactam-resistant isolates (29). In a recent study in which 2,448 children younger than 2 years old living in various areas in Greece were screened during a 2-year period (1997 to 1999) for pneumococcal carriage, 15% of the pneumococci appeared to have reduced susceptibility to non-β-lactam agents (31).
The present study was undertaken to investigate the molecular epidemiological characteristics of the Greek pneumococci susceptible to penicillin but resistant to erythromycin and/or other non-β-lactam agents. Furthermore, penicillin, erythromycin, and tetracycline resistance determinants were studied at a molecular level.
MATERIALS AND METHODS
Bacterial isolates.
We studied a collection of 128 S. pneumoniae isolates susceptible to penicillin but resistant to erythromycin and/or other non-β-lactam agents; the isolates were recovered from nasopharyngeal cultures obtained from children during two independent studies in Greece (3, 30, 31). The first study was performed in 338 children attending seven day care centers in the city of Patras in southwestern Greece during the 2-month period from December 1995 to February 1996. In this study, 30 penicillin-susceptible pneumococci resistant to one or more non-β-lactam agents were recovered from 132 carriers. Of these 30 S. pneumoniae isolates, 26 were available for further analysis in the present investigation. The second study, the Hellenic Antibiotic-Resistant Respiratory Pathogens (HARP) study, was conducted from February 1997 to February 1999. Nasopharyngeal cultures for S. pneumoniae were performed in 2,448 children younger than 2 years old living in central and southern Greece. Ninety-five (3.9%) of the 2,448 children attended day care centers. In the HARP study, screening of the children revealed 119 pneumococci which were penicillin susceptible but resistant to non-β-lactam agents. Of these 119 pneumococci, 102 were available for further analysis. Thirteen of these pneumococci were isolated from children attending day care. Only two of these children attended the same day care center.
Bacteriological procedures.
Isolation, identification, and susceptibility testing of the Greek S. pneumoniae isolates were performed by applying standard methods as described previously (30, 31). Penicillin and erythromycin MICs for the Greek isolates were determined by the E-test method (AB Biodisk, Solna, Sweden). Susceptibility to clindamycin, chloramphenicol, tetracycline, and SXT was determined by the disk diffusion method according to the guidelines of the National Committee for Clinical Laboratory Standards (NCCLS) (21). Multidrug resistance was defined as resistance to three or more classes of antimicrobial agents. Pneumococci were serotyped by the capsular swelling method or the latex agglutination technique (30, 33).
Penicillin-binding protein (PBP) genotyping.
Genetic polymorphism of the penicillin resistance genes pbp1a, pbp2b, and pbp2x of the pneumococcal isolates was investigated by restriction fragment length polymorphism (RFLP) analysis of the PCR-amplified genes, as described previously (13).
Detection and analysis of the erm(B) and mef genes.
To detect the presence of erm(B) within the pneumococcal isolates, we used the protocol of Sutcliffe et al. (28). In summary, we amplified the genes by PCR and analyzed the amplified DNA products by agarose gel electrophoresis. The presence of the mef gene was also detected by PCR (28). In order to discriminate between mef(A) and mef(E), PCR-RFLP analysis was performed by the method of Del Grosso et al. (8). The amplicon was digested using BamHI and DraI. The mef(A) gene contains a single BamHI site, which is absent in the mef(E) gene. Digestion of mef(A) and mef(E) with DraI yields two and three fragments, respectively.
Detection of the tet(M) and tet(O) genes.
In order to discriminate between tet(M) and tet(O), a PCR-RFLP analysis was performed as described previously (2). In summary, a PCR mix was made of 25-μl reaction buffer containing 0.5 U of thermostable DNA polymerase, diluted in the buffer supplied by the manufacturer (Integro, Leuvenheim, The Netherlands), 0.2 mM (each) deoxynucleoside triphosphate, 1.5 mM MgCl2, 10 pmol of each primer, and 10 to 50 ng of pneumococcal DNA. Amplification cycling in a programmable thermal controller (PTC-100; MJ Research, Watertown, Mass.) consisted of the following steps: predenaturation for 1 min at 94°C, followed by 30 cycles, with 1 cycle consisting of 1 min at 94°C, 1 min at 57°C, and 1 min at 72°C. Amplification was finished after 3 min at 72°C. A 1.5% agarose gel in 0.045 M Tris-borate buffer with 0.001 M EDTA (0.5× TBE buffer) containing 0.1 μg of ethidium bromide per ml was used to visualize the PCR products.
RFEL typing.
Pneumococcal strain typing by restriction fragment end labeling (RFEL) was performed by the method of van Steenbergen et al. (36) as adapted by Hermans et al. (13). Briefly, purified pneumococcal DNA was digested by the restriction enzyme EcoRI. The DNA restriction fragments were end labeled at 72°C with [α-32P]dATP using DNA polymerase (Goldstar; Eurogentec, Seraing, Belgium). After the radiolabeled fragments were denatured and separated electrophoretically on a 6% polyacrylamide sequencing gel containing 8 M urea, the gel was transferred onto filter paper, vacuum dried (HBI, Saddlebrook, N.Y.), and exposed for variable times at room temperature to ECL hyperfilm (Amersham Laboratories, Amersham, United Kingdom).
Computer-assisted analysis of DNA band patterns.
RFEL autoradiographs were converted to images (Image Master DTS; Pharmacia Biotech, Uppsala, Sweden) and analyzed with a computer (Windows version Gelcompar software version 4; Applied Math, Kortrijk, Belgium). DNA fragments were analyzed as described previously (26). For evaluation of the genetic relatedness of the isolates, we used the following definitions: (i) isolates of a particular RFEL type are 100% identical by RFEL analysis; (ii) an RFEL cluster represents a group of RFEL types that differ in only one band (approximately >95% genetic relatedness); (iii) an RFEL lineage represents a group of RFEL types that differ in less than four bands (approximately >85% genetic relatedness).
International comparison.
The Greek genotypes were compared with an international collection of pneumococcal isolates representing 751 distinct RFEL types originating from 17 different countries in America, Europe, Africa, and Asia (2, 12;M. Sluijter, unpublished observations). The international collection includes the first 16 international pandemic clones described by the Pneumococcal Molecular Epidemiological Network in 2000 (http://www.pneumo.com/physician/pmen_clone_collection.asp) (18).
MLST.
The genotypes of all clusters were verified by multilocus sequence typing (MLST) analysis. For this purpose, a fully automated method for MLST was used as described previously (14), and one or two isolates per cluster were analyzed. The MLST types were compared with the global database at www.mlst.net.
RESULTS
The serotypes 6A/B (49%), 14 (14%), 19A/F (11%), 11A (9%), 23A/F (4%), 15B/C (2%), and 21 (2%) were most prevalent in the collection of 128 Greek pneumococcal isolates. All isolates were invariably susceptible to penicillin, while 84 (65%), 78 (60%), 69 (54%), 63 (49%), and 62 (48%) isolates were resistant to erythromycin, SXT, tetracycline, chloramphenicol, and clindamycin, respectively.
The isolates were classified as penicillin-susceptible erythromycin-resistant (65%) and penicillin-susceptible erythromycin-susceptible (35%) isolates. The latter group was represented by tetracycline- and/or SXT-resistant pneumococci. The 84 penicillin-susceptible erythromycin-resistant pneumococci displayed capsular types 6B (56%), 14 (18%), 19F (13%), 11A (11%), 10A (1.2%), and 15C (1.2%). Of these erythromycin-resistant isolates, 59 (70%), 15 (18%), and 7 (8.3%) isolates carried the erm(B), mef(A), and mef(E) erythromycin resistance determinants, respectively. In four erythromycin-resistant isolates belonging to a serotype 11A clone, an erm(A) gene was previously detected (32). Furthermore, 62 (74%), 57 (68%), 46 (55%), and 42 (50%) of the 84 penicillin-susceptible erythromycin-resistant pneumococci, were also resistant to clindamycin, tetracycline, SXT, and chloramphenicol, respectively. Clindamycin resistance was always identified in combination with erythromycin resistance. All 62 clindamycin- and erythromycin-resistant isolates carried an erm resistance gene. Of the 54 erythromycin-resistant isolates that were resistant to tetracycline, 51 carried the tet(M) resistance gene. The three remaining isolates were negative for both the tet(M) and tet(O) genes.
The 44 penicillin-susceptible erythromycin-susceptible isolates belonged to serotypes 6B (29%), 23F (11%), 14 (6.7%), 21 (6.7%), 1 (4.4%), 6A (4.4%), 15B (4.4%), 18C (4.4%), 19A (4.4%), 8 (2.2%), 10A (2.2%), 11A (2.2%), 16F (2.2%), 19F (2.2%), 20 (2.2%), and 24F (2.2%) and nontypeable serotypes (4.4%).
Sulfamethoxazole resistance was identified in 34 of the 44 penicillin-susceptible erythromycin-susceptible isolates (78%), whereas tetracycline resistance was identified in 12 (27%) isolates. All but one carried the tet(M) resistance gene. All isolates were resistant to a single agent except for one isolate that was resistant to tetracycline and chloramphenicol and two isolates that were resistant to tetracycline and SXT.
Fourteen distinct PBP genotypes were observed. Of the 128 isolates, 76 (59%), 24 (19%), and 4 (3.1%) displayed a known penicillin-susceptible PBP 1A-2B-2X genotype, being 2-2-71, 2-2-3, and 2-2-2, respectively. The remaining isolates displayed alterations in pbp2x (11 distinct types), pbp2b (2 types), and pbp1a (1 type).
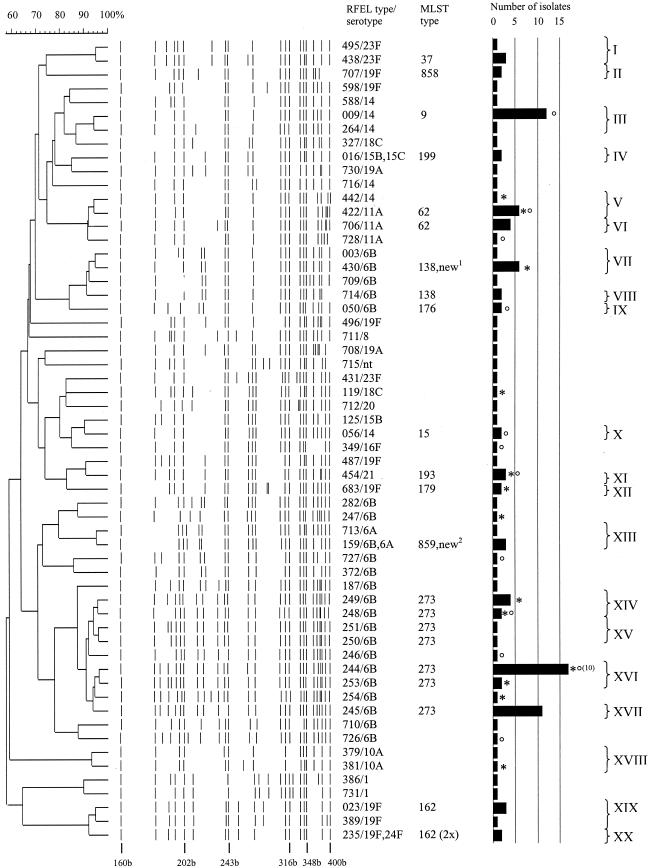
RFEL analysis divided the 128 penicillin-susceptible isolates, which were resistant to one or more non-β-lactam agents into 59 distinct RFEL genotypes (Fig. 1). Ninety-nine isolates belonged to 20 genetic clusters, representing 77% of the isolates and varying in size from 2 to 19 isolates (Table 1). The average cluster size was 5.0 isolates. Five of the 20 clusters contained two serotypes, while the remaining clusters contained only one serotype each. The 20 clusters belonged to 11 lineages. The three largest clusters were cluster III, XVI, and XVII. Cluster III consisted of 13 (10%) isolates and was identical to the pandemic clone England14-9 which was confirmed by MLST analysis (Fig. 1 and Table 1). This cluster belonged to a single predominant lineage of 14 genetically related isolates, representing three RFEL genotypes and harboring the serotypes 11A, 14, and 18C. The serotype 14 pneumococci carried the mef(A) erythromycin resistance determinant and had a low to moderate level of resistance to erythromycin. The remaining pneumococci were resistant to SXT.
FIG.1.
Dendrogram of the 59 RFEL types observed among the 128 Greek S. pneumoniae nasopharyngeal isolates. The molecular sizes of reference bands (in bases [b]), serotypes, PBP types, RFEL types, MLST types, number of isolates per RFEL type, clusters, and cluster codes are depicted. One or two strains per RFEL clusters were analyzed by MLST. Day care center isolates from the 1995 to 1996 study (asterisks) and day care center isolates from the 1997 to 1999 study (degree symbols) are indicated. If more than one day care center isolate belongs to a specific genotype, the total number is displayed in parentheses. Two new strains with new MLST types (new1 and new2) are indicated. These two strains have been submitted to the MLST database. nt, nontypeable.
TABLE 1.
Molecular and phenotypic characteristics of the 129 Greek pneumococcal isolates
| RFEL cluster | No. of isolates/no. of RFEL types | Serotype (no. of strains) | Resistance patterna (no. of strains) | PBP type (no. of strains) | Genotype (no. of strains) |
|---|---|---|---|---|---|
| I | 4/2 | 23F (4) | S | 2-2-3 | |
| II | 2/1 | 19F (2) | TECLS | 2-2-3 | erm(B), tet(M) |
| ECLS | erm(B) | ||||
| III | 13/2 | 14 (13) | E (13) | 2-2-71 | mef(A) |
| IV | 2/1 | 15B | S | 2-2-44 | |
| 15C | ECL | 2-2-3 | erm(B) | ||
| V | 7/2 | 11A (6) | TECL (4) | 2-2-3 (6) | erm(B) (4), tet(M) (4) |
| 14 (1) | S (2) | 2-2-71 (1) | mef(E) (1) | ||
| E (1) | |||||
| VI | 4/1 | 11A (4) | TECL | 2-2-3 | erm(A), tet(M) |
| VII | 7/2 | 6B (7) | S (6) | 2-2-71 | erm(B) (1), tet(M)(1) |
| E (1) | mef(E) | ||||
| VIII | 2/1 | 6B (2) | S (2) | 2-2-71 | |
| IX | 2/1 | 6B (2) | CTECLS | 2-2-71 | Unknown |
| S | |||||
| X | 2/1 | 14 (2) | S (2) | 2-2-71 | |
| XI | 3/1 | 21 | S (3) | 2-2-7 | |
| 2-2-98 | |||||
| XII | 2/1 | 19F (2) | TECL | 2-2-98 | erm(B) (2) |
| tet(M) (2) | |||||
| XIII | 4/2 | 6B (2) | T (3), TS (1) | 2-5-22 (2) | tet(M) (4) |
| 6A (2) | 2-5-99 (1) | ||||
| 2-2-71 (1) | |||||
| XIV | 6/2 | 6B (6) | CTECLS (5) | 2-2-71 | erm(B) (6) |
| CTECL | tet(M) (6) | ||||
| XV | 2/2 | 6B (2) | CTECLS | 2-2-71 | erm(B) (2) |
| CTECL | tet(M) (2) | ||||
| XVI | 19/2 | 6B (19) | CTECLS (17) | 2-2-71 (18) | erm(B) (19) |
| CTECL (1) | 0-0-0 (1) | tet(M) (18) | |||
| TECLS (1) | |||||
| XVII | 11/1 | 6B (11) | CTECLS (11) | 2-2-71 (10) | erm(B) (11) |
| 0-0-0 (1) | tet(M) (11) | ||||
| XVIII | 2/2 | 10A | TECL (1) | 2-2-71 | erm(B) (1), tet(M) (1) |
| S | 2-2-2 | ||||
| XIX | 4/2 | 19F (4) | E | 2-2-2 (4) | met(A) (1) |
| met(E) (3) | |||||
| XX | 2/1 | 19F (1) | S | 2-2-2 (1) | |
| 24F (1) | 2-2-3 (1) |
Abbreviations: C, chloramphenicol; T, tetracycline; E, erythromycin; CL, clindamycin; S, trimethoprim-sulfamethoxazole.
Clusters XVI (19 isolates) and XVII (11 isolates) belonged, together with cluster XIV (6 isolates) and XV (2 isolates), to one predominant lineage, representing 10 RFEL types and all harboring the serotype 6B. Most isolates were resistant to erythromycin, clindamycin, tetracycline, chloramphenicol, and SXT. These pneumococci carried the erm(B) erythromycin resistance determinant and had a high level of resistance to erythromycin. The latter clusters were identical to the pandemic clone Greece6B-22 and closely related to the penicillin-resistant MDR clone Spain6B-2 that has spread from Spain to Iceland in the late 1980s (29).
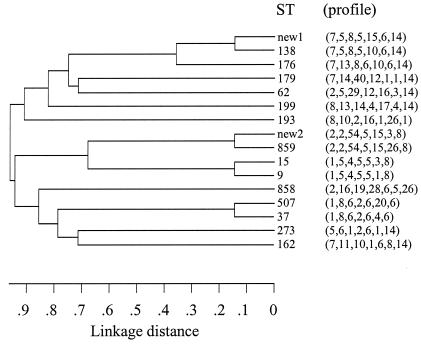
Clusters I (three isolates) and XIX (three isolates) represented genotypes identical to the pandemic clone Tennessee23F-4 and closely related to the pandemic clone Spain9V-3 (six of seven alleles) as described by the Pneumococcal Molecular Epidemiology Network. The genetic relatedness of the observed MLST profiles of the 20 RFEL clusters is depicted in Fig. 2.
FIG. 2.
Genetic relatedness of the 15 MLST sequence types (ST) observed within the 20 RFEL clusters.
The 39 isolates recovered from children attending day care centers displayed 21 different genotypes; 10 isolates displayed unique genotypes, whereas the remaining 29 isolates displayed 13 genotypes belonging to 11 clusters. Cluster XVI contained nine isolates from one day care center, whereas cluster XVII contained three isolates from a second single day care center. The remaining clustering day care center isolates originated from different day care centers (Fig. 1).
DISCUSSION
We evaluated 128 S. pneumoniae isolates that were susceptible to penicillin but resistant to non-β-lactam agents from young carriers in Greece by antibiotic susceptibility testing, serotyping, RFEL, and antibiotic resistance genotyping. In general, the isolates could be divided into two groups: one group consisting of 84 erythromycin-resistant isolates, while the second group of 44 isolates were erythromycin susceptible. Multidrug resistance, i.e., resistance to three or more different classes of antibiotics, was seen predominantly within the first group of isolates, whereas the erythromycin-susceptible isolates predominantly displayed monodrug resistance to tetracycline or sulfamethoxazole. Furthermore, clustering of isolates, which is the result of horizontal spread of the isolates within the community, was higher among multidrug-resistant genotypes. This is in line with previous findings where multidrug-resistant clones have shown to spread from country to country (3, 4, 7, 20, 22, 25, 27, 35). This has led to the classification of pandemic clones by the Pneumococcal Molecular Epidemiological Network (18). Although the majority of these pandemic clones are penicillin resistant, our study as well as previous studies have shown that this is not a prerequisite for clonal spread.
With emerging non-β-lactam resistance among pneumococci, the spread of penicillin-susceptible non-β-lactam-resistant pneumococci has become apparent (1, 24, 29, 34). This is underlined by our observation that, in addition to one large lineage of serotype 6B isolates that were mostly susceptible to penicillin and resistant to erythromycin, tetracycline, chloramphenicol, and sulfamethoxazole, 16 smaller clusters were found. In total, 30% of the isolates displayed a genotype identical to those of the pandemic clones Tennessee23F-4, England14-9, and Greece6B-22 as reported by the Pneumococcal Molecular Epidemiological Network. These findings clearly indicate heterogeneity among the penicillin-susceptible, non-β-lactam-resistant isolates. Furthermore, these clusters represent not only the conjugate vaccine serotypes 6B, 14, 19F, and 23F, but also non-vaccine serotypes, such as 11A, 15B/C, and 21. This observation implicates that non-vaccine serotypes are also able to spread among children; hence, vaccination with pneumococcal conjugate vaccines is not a solution for the emergence of multidrug resistance among pneumococcal isolates.
In this study, 70% of the erythromycin-resistant isolates harbored the erm(B) gene, while 18 and 8% of the isolates contained mef(A) and mef(E) genes, respectively. These data differ from previous findings made by Reinert and coworkers, who observed an almost equal distribution of erm(B) (43%) and mef(E) (56%) genes among erythromycin-resistant isolates in Germany (23). However, our data are in line with a recent study performed in Italy and Vietnam where the majority of the strains also displayed the erm(B) gene (2, 19). In the four low-level erythromycin-resistant isolates displaying an identical RFEL genotype and serotype 11A, the erm(A) gene was previously identified (32).
Genetic analysis of tetracycline resistance genes tet(M) and tet(O) revealed that the tet(M) gene was exclusively observed in 91% of the Greek isolates. This is in line with the Vietnamese study where the tet(M) gene was also exclusively observed (2). In contrast to other studies, the remaining tetracycline-resistant isolates did not harbor the tet(O) gene (16). So far, no other tetracycline resistance determinants have beendescribed; therefore, the underlying mechanism remains unclear. Also in line with the Vietnamese study is the isolation of tet(M)-containing but tetracycline-susceptible strains in Greece, suggesting the presence of a nonfunctional or unexpressed tetracycline resistance gene (2, 9). A similar phenomenon was seen for the PBP genes. Though the majority of the isolates displayed (known) susceptible genotypes, 10% of the isolates displayed alterations in one or two of the three major PBP genes, pbp1a, pbp2b, and pbp2x, which often implicates intermediate penicillin resistance (10). In our study, the observed alterations in pbp1a, pbp2b, and four of the pbp2x alterations were identified previously in non-penicillin-susceptible pneumococci isolated in Thailand, the United States, and The Netherlands (11). Although these alterations have shown to be related to intermediate susceptibility, in all cases an additional alteration in one or two of the PBP genes was present (Sluijter, unpublished). These findings support the hypothesis that not all genetic alterations lead to amino acid substitutions or to substitutions which are relevant for penicillin resistance.
We hypothesize that the ongoing antibiotic pressure will continue the process of alteration and spread of resistance genes among pneumococci. Our study underlines that antibiotic resistance in any form and irrespective of its serotype is of benefit for S. pneumoniae with respect to survival and spread in the community. This implies that despite the introduction of new and effective pneumococcal conjugate vaccines, restrictive use of antibiotics will be of major importance.
REFERENCES
- 1.Bell, J. M., J. D. Turnidge, and R. N. Jones. 2002. Antimicrobial resistance trends in community-acquired respiratory tract pathogens in the Western Pacific Region and South Africa: report from the SENTRY antimicrobial surveillance program (1998-1999), including an in vitro evaluation of BMS284756. Int. J. Antimicrob. Agents 19:125-132. [DOI] [PubMed] [Google Scholar]
- 2.Bogaert, D., N. T. Ha, M. Sluijter, N. Lemmens, R. De Groot, and P. W. Hermans. 2002. Molecular epidemiology of pneumococcal carriage among children with upper respiratory tract infections in Hanoi, Vietnam. J. Clin. Microbiol. 40:3903-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bogaert, D., G. A. Syrogiannopoulos, I. N. Grivea, R. de Groot, N. G. Beratis, and P. W. Hermans. 2000. Molecular epidemiology of penicillin-nonsusceptible Streptococcus pneumoniae among children in Greece. J. Clin. Microbiol. 38:4361-4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castaneda, E., I. Penuela, M. C. Vela, A. Tomasz, et al.. 1998. Penicillin-resistant Streptococcus pneumoniae in Colombia: presence of international epidemic clones. Microb. Drug Resist. 4:233-239. [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 1997. Prevention of pneumococcal disease: recommendations of the Advisory Committee on Immunization Practices (ACIP). Morb. Mortal. Wkly. Rep. 46:1-24. [Google Scholar]
- 6.Corso, A., E. P. Severina, V. F. Petruk, Y. R. Mauriz, and A. Tomasz. 1998. Molecular characterization of penicillin-resistant Streptococcus pneumoniae isolates causing respiratory disease in the United States. Microb. Drug Resist. 4:325-337. [DOI] [PubMed] [Google Scholar]
- 7.Davies, T., R. V. Goering, M. Lovgren, J. A. Talbot, M. R. Jacobs, and P. C. Appelbaum. 1999. Molecular epidemiological survey of penicillin-resistant Streptococcus pneumoniae from Asia, Europe, and North America. Diagn. Microbiol. Infect. Dis. 34:7-12. [DOI] [PubMed] [Google Scholar]
- 8.Del Grosso, M., F. Iannelli, C. Messina, M. Santagati, N. Petrosillo, S. Stefani, G. Pozzi, and A. Pantosti. 2002. Macrolide efflux genes mef(A) and mef(E) are carried by different genetic elements in Streptococcus pneumoniae. J. Clin. Microbiol. 40:774-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doherty, N., K. Trzcinski, P. Pickerill, P. Zawadzki, and C. G. Dowson. 2000. Genetic diversity of the tet(M) gene in tetracycline-resistant clonal lineages of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44:2979-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hakenbeck, R., T. Grebe, D. Zahner, and J. B. Stock. 1999. β-Lactam resistance in Streptococcus pneumoniae: penicillin-binding proteins and non-penicillin-binding proteins. Mol. Microbiol. 33:673-678. [DOI] [PubMed] [Google Scholar]
- 11.Hermans, P. W., K. Overweg, M. Sluijter, and R. de Groot. 2000. Penicillin-resistant Streptococcus pneumoniae: an international molecular epidemiological study, p. 457-566. In A. Tomasz (ed.), Streptococcus pneumoniae: molecular biology and mechanisms of disease. Mary Ann Liebert, Inc., New York, N.Y.
- 12.Hermans, P. W., M. Sluijter, S. Dejsirilert, N. Lemmens, K. Elzenaar, A. van Veen, W. H. Goessens, and R. de Groot. 1997. Molecular epidemiology of drug-resistant pneumococci: toward an international approach. Microb. Drug Resist. 3:243-251. [DOI] [PubMed] [Google Scholar]
- 13.Hermans, P. W., M. Sluijter, T. Hoogenboezem, H. Heersma, A. van Belkum, and R. de Groot. 1995. Comparative study of five different DNA fingerprint techniques for molecular typing of Streptococcus pneumoniae strains. J. Clin. Microbiol. 33:1606-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jefferies, J., S. C. Clarke, M. A. Diggle, A. Smith, C. Dowson, and T. Mitchell. 2003. Automated pneumococcal MLST using liquid-handling robotics and a capillary DNA sequencer. Mol. Biotechnol. 24:303-308. [DOI] [PubMed] [Google Scholar]
- 15.Leclerq, R. 2002. Will resistance to ketolides develop in Streptococcus pneumoniae? J. Infect. 44(Suppl. A):11-16. [PubMed] [Google Scholar]
- 16.Luna, V. A., D. B. Jernigan, A. Tice, J. D. Kellner, and M. C. Roberts. 2000. A novel multiresistant Streptococcus pneumoniae serogroup 19 clone from Washington State identified by pulsed-field gel electrophoresis and restriction fragment length patterns. J. Clin. Microbiol. 38:1575-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lynch, I. J., and F. J. Martinez. 2002. Clinical relevance of macrolide-resistant Streptococcus pneumoniae for community-acquired pneumonia. Clin. Infect. Dis. 34(Suppl. 1):S27-S46. [DOI] [PubMed] [Google Scholar]
- 18.McGee, L., L. McDougal, J. Zhou, B. G. Spratt, F. C. Tenover, R. George, R. Hakenbeck, W. Hryniewicz, J. C. Lefevre, A. Tomasz, and K. P. Klugman. 2001. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the Pneumococcal Molecular Epidemiology Network. J. Clin. Microbiol. 39:2565-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montanari, M. P., M. Mingoia, I. Cochetti, and P. E. Varaldo. 2003. Phenotypes and genotypes of erythromycin-resistant pneumococci in Italy. J. Clin. Microbiol. 41:428-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Munoz, R., T. J. Coffey, M. Daniels, C. G. Dowson, G. Laible, J. Casal, R. Hakenbeck, M. Jacobs, J. M. Musser, B. G. Spratt, et al. 1991. Intercontinental spread of a multiresistant clone of serotype 23F Streptococcus pneumoniae. J. Infect. Dis. 164:302-306. [DOI] [PubMed] [Google Scholar]
- 21.National Committee for Clinical Laboratory Standards. 1999. Performance standards for antimicrobial susceptibility testing, 9th informational supplement. M100-S9. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 22.Overweg, K., P. W. Hermans, K. Trzcinski, M. Sluijter, R. de Groot, and W. Hryniewicz. 1999. Multidrug-resistant Streptococcus pneumoniae in Poland: identification of emerging clones. J. Clin. Microbiol. 37:1739-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reinert, R. R., A. Queck, A. Kaufhold, M. Kresken, and R. Lutticken. 1995. Antimicrobial resistance and type distribution of Streptococcus pneumoniae isolates causing systemic infections in Germany, 1992-1994. Clin. Infect. Dis. 21:1398-1401. [DOI] [PubMed] [Google Scholar]
- 24.Ronchetti, M. P., R. Merolla, S. Bajaksouzian, G. Violo, R. Ronchetti, and M. R. Jacobs. 1998. Antimicrobial susceptibility of Streptococcus pneumoniae from children attending day-care centers in a central Italian city. Clin. Microbiol. Infect. 4:622-626. [DOI] [PubMed] [Google Scholar]
- 25.Sibold, C., J. Wang, J. Henrichsen, and R. Hakenbeck. 1992. Genetic relationships of penicillin-susceptible and -resistant Streptococcus pneumoniae strains isolated on different continents. Infect. Immun. 60:4119-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sluijter, M., H. Faden, R. de Groot, N. Lemmens, W. H. Goessens, A. van Belkum, and P. W. Hermans. 1998. Molecular characterization of pneumococcal nasopharynx isolates collected from children during their first 2 years of life. J. Clin. Microbiol. 36:2248-2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soares, S., K. G. Kristinsson, J. M. Musser, and A. Tomasz. 1993. Evidence for the introduction of a multiresistant clone of serotype 6B Streptococcus pneumoniae from Spain to Iceland in the late 1980s. J. Infect. Dis. 168:158-163. [DOI] [PubMed] [Google Scholar]
- 28.Sutcliffe, J., T. Grebe, A. Tait-Kamradt, and L. Wondrack. 1996. Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 40:2562-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Syrogiannopoulos, G. A., D. Bogaert, I. N. Grivea, N. G. Beratis, R. R. De Groot, and P. W. Hermans. 2001. Molecular epidemiology of penicillin-susceptible, multidrug-resistant serotype 6B pneumococci isolated from children in Greece. J. Clin. Microbiol. 39:581-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Syrogiannopoulos, G. A., I. N. Grivea, N. G. Beratis, A. E. Spiliopoulou, E. L. Fasola, S. Bajaksouzian, P. C. Appelbaum, and M. R. Jacobs. 1997. Resistance patterns of Streptococcus pneumoniae from carriers attending day-care centers in southwestern Greece. Clin. Infect. Dis. 25:188-194. [DOI] [PubMed] [Google Scholar]
- 31.Syrogiannopoulos, G. A., I. N. Grivea, T. A. Davies, G. D. Katopodis, P. C. Appelbaum, and N. G. Beratis. 2000. Antimicrobial use and colonization with erythromycin-resistant Streptococcus pneumoniae in Greece during the first 2 years of life. Clin. Infect. Dis. 31:887-893. [DOI] [PubMed] [Google Scholar]
- 32.Syrogiannopoulos, G. A., I. N. Grivea, A. Tait-Kamradt, G. D. Katopodis, N. G. Beratis, J. Sutcliffe, P. C. Appelbaum, and T. A. Davies. 2001. Identification of an erm(A) erythromycin resistance methylase gene in Streptococcus pneumoniae isolated in Greece. Antimicrob. Agents Chemother. 45:342-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Syrogiannopoulos, G. A., G. D. Katopodis, I. N. Grivea, and N. G. Beratis.2002. Antimicrobial use and serotype distribution of nasopharyngeal Streptococcus pneumoniae isolates recovered from Greek children younger than 2 years old. Clin. Infect. Dis. 35:1174-1182. [DOI] [PubMed] [Google Scholar]
- 34.Syrogiannopoulos, G. A., F. Ronchetti, R. Dagan, I. Grivea, M. P. Ronchetti, N. Porat, T. A. Davies, R. Ronchetti, P. C. Appelbaum, and M. R. Jacobs. 2000. Mediterranean clone of penicillin-susceptible, multidrug-resistant serotype 6B Streptococcus pneumoniae in Greece, Italy and Israel. Int. J. Antimicrob. Agents 16:219-224. [DOI] [PubMed] [Google Scholar]
- 35.Tomasz, A., A. Corso, E. P. Severina, G. Echaniz-Aviles, M. C. Brandileone, T. Camou, E. Castaneda, O. Figueroa, A. Rossi, and J. L. Di Fabio. 1998. Molecular epidemiologic characterization of penicillin-resistant Streptococcus pneumoniae invasive pediatric isolates recovered in six Latin-American countries: an overview. PAHO/Rockefeller University Workshop. Pan American Health Organization. Microb. Drug Resist. 4:195-207. [DOI] [PubMed] [Google Scholar]
- 36.van Steenbergen, T. J., S. D. Colloms, P. W. Hermans, J. de Graaff, and R. H. Plasterk. 1995. Genomic DNA fingerprinting by restriction fragment end labeling. Proc. Natl. Acad. Sci. USA 92:5572-5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Widdowson, C. A., and K. P. Klugman. 1998. Emergence of the M phenotype of erythromycin-resistant pneumococci in South Africa. Emerg. Infect. Dis. 4:277-281. [DOI] [PMC free article] [PubMed] [Google Scholar]