Abstract
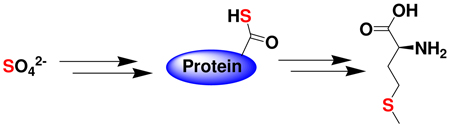
Thiocarboxylated proteins are important intermediates in a variety of biochemical sulfide transfer reactions. Here we identify a protein thiocarboxylate dependent methionine biosynthetic pathway in Wolinella succinogenes. In this pathway, the carboxy terminal alanine of a novel sulfur transfer protein, HcyS-Ala is removed in a reaction catalyzed by a metalloprotease, HcyD. HcyF, an ATP-utilizing enzyme, catalyzes the adenylation of HcyS. HcyS acyl-adenylate then undergoes nucleophilic substitution by bisulfide produced by Sir to give the HcyS thiocarboxylate. This adds to O-acetylhomoserine to give HcyS-homocysteine in a PLP-dependent reaction catalyzed by MetY. HcyD mediated hydrolysis liberates homocysteine. A final methylation completes the biosynthesis. The biosynthetic gene cluster also encodes the enzymes involved in the conversion of sulfate to sulfide suggesting that sulfate is the sulfur source for protein thiocarboxylate formation in this system.
Keywords: Methionine biosynthesis, protein thiocarboxylate, sulfite reductase, fluorescence assay
INTRODUCTION
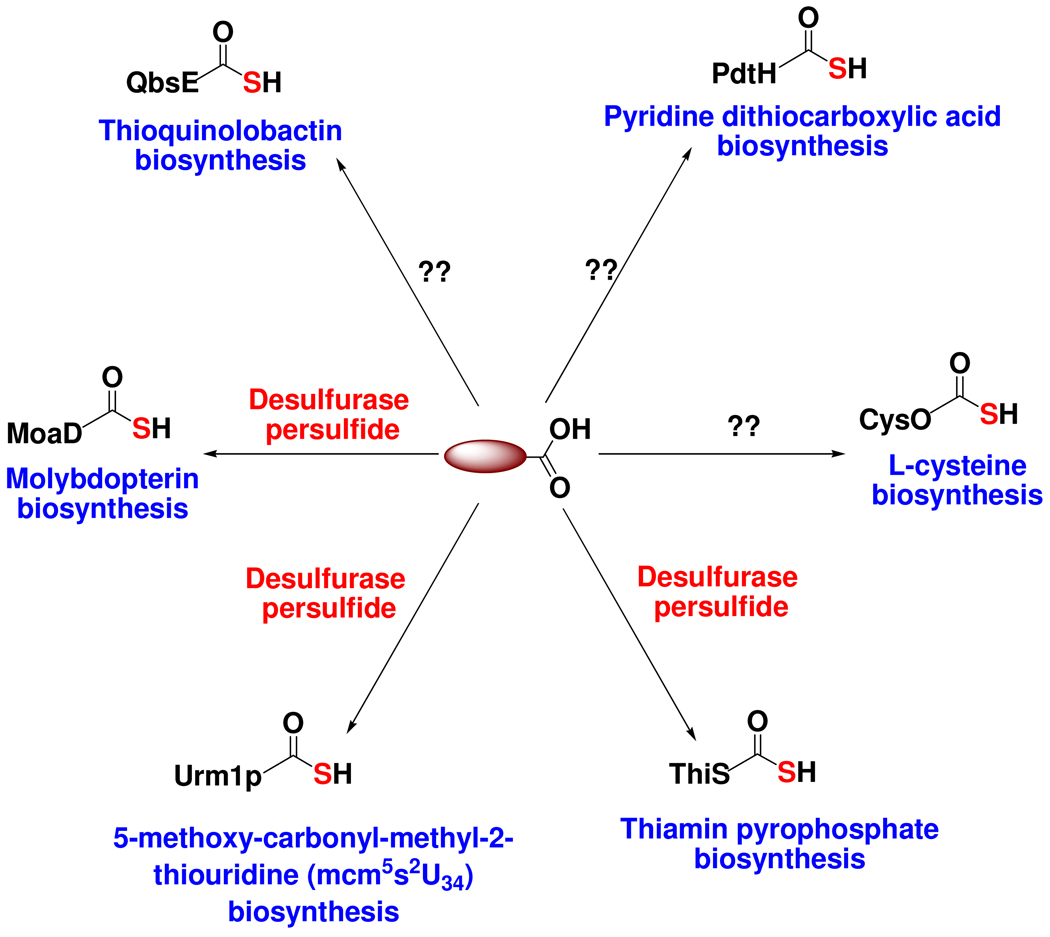
Sulfur-containing natural products are widely distributed in nature e.g. amino acids, carbohydrates, proteins, nucleic acids, antibiotics, cofactors, siderophores, alkaloids etc.1,2 Three major sulfur sources have been identified in bacterial metabolism: free sulfide used for example in cysteine biosynthesis, protein persulfides (R-S-SH) used for example in iron sulfur cluster biosynthesis and protein thiocarboxylates (R-COSH). Protein thiocarboxylates are members of a growing family of biosynthetic sulfide donors and are involved in a variety of biosynthetic pathways, including vitamin B1 (ThiS-COSH)3, molybdopterin (MoaD-COSH)4, cysteine (CysO-COSH)5, thioquinolobactin (QbsE-COSH)6, 2-thioribothymidine (TtuB-COSH)7 and 5-methoxy-carbonyl-methyl-2-thiouridine (Urm1p-COSH)8. A significant gap in our understanding of the biosynthetic role of protein thiocarboxylates is the identification of the sulfur source for thiocarboxylate formation. For ThiS-COSH9, MoaD-COSH4,10 and Urm1p8,11, a desulfurase persulfide is the sulfur donor; for the others shown in Figure 1, the sulfur source is unknown.
Figure 1.
Protein thiocarboxylates function as sulfide donors in the biosynthesis of a variety of natural products. In many cases, the sulfur source for thiocarboxylate formation is unknown.
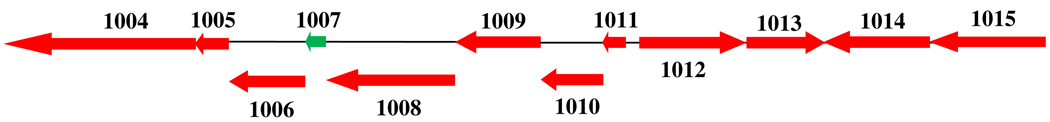
A search for ThiS-COOH orthologs in the SEED database (http://theseed.uchicago.edu/FIG/index.cgi), revealed in Wolinella succinogenes the gene for a putative thiocarboxylate-forming protein clustered with the sulfate assimilation proteins and putative methionine biosynthetic genes (Figure 2). This protein thiocarboxylate/sulfate assimilation clustering pattern, and variations of it, was seen in many other micro-organisms (e.g. Clostridium kluyveri, Clostridium thermocellum, Desulfitobacterium hafniense, Carboxydothermus hydrogenoformans Z-2901, Caldicellulosiruptor saccharolyticus DSM 8903, Alkaliphilus metalliredigens QYMF, Geobacter metallireducens GS-15, Geobacter uraniireducens Rf4, Acidovorax sp. JS42 and Pelodictyon luteolum DSM 273) suggesting that the genes are functionally related and that sulfate may be used as the sulfur source for protein thiocarboxylate formation in these systems.
Figure 2.
The putative thiocarboxylate forming protein, WS1007 (shown in green), clustered with sulfate assimilation proteins – WS1004, WS1008, WS1009 and WS1010, and homocysteine biosynthetic proteins, WS1012 and WS1015 in Wolinella succinogenes. Hypothetical functions for these genes are given in Table 1.
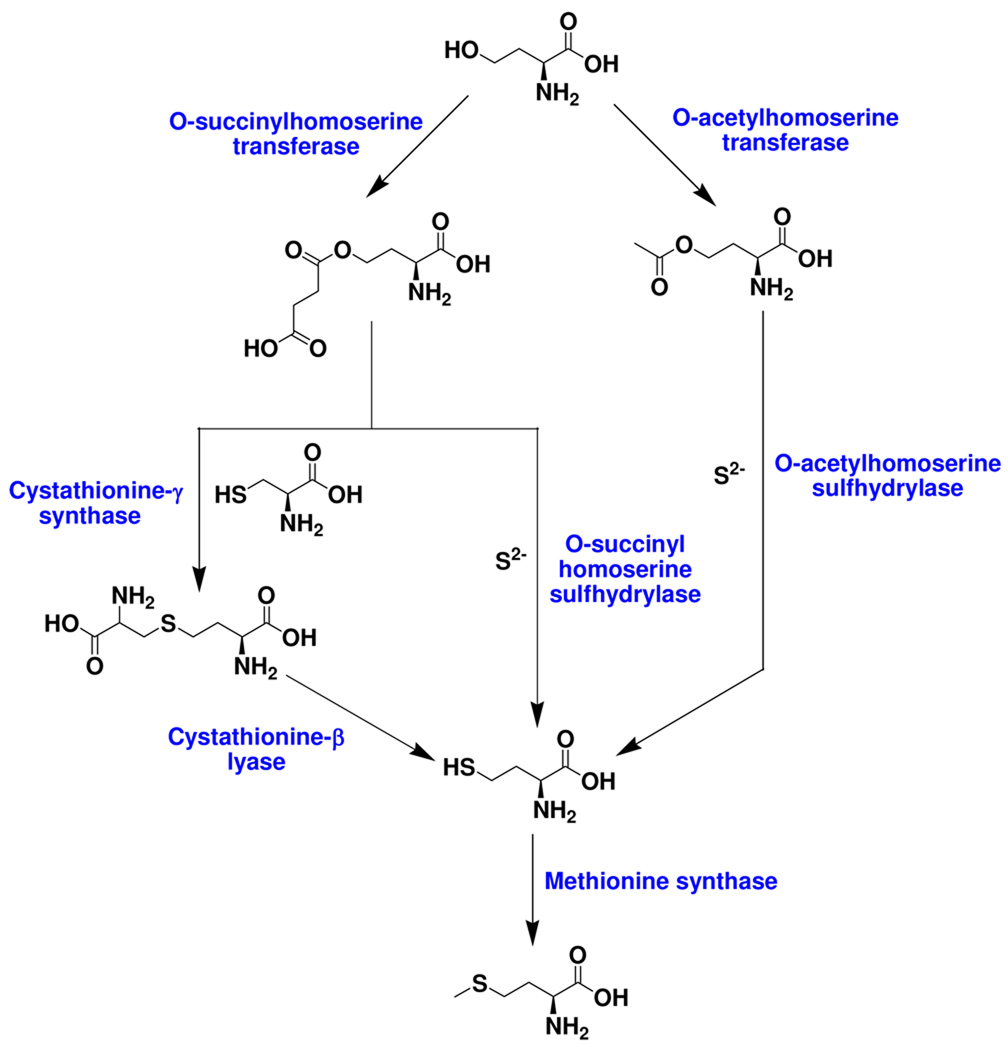
The well-characterized biosynthesis of methionine is outlined in Figure 3. In this pathway homocysteine formation occurs either by direct addition of sulfide to O-succinylhomoserine or O-acetylhomoserine, or by breakdown of cystathionine12 (Figure 3). In this paper, we describe the identification of a new protein thiocarboxylate-dependent methionine biosynthetic pathway in Wolinella succinogenes and identify sulfate as the probable sulfur donor.
Figure 3.
The three well-characterized pathways for L-methionine biosynthesis. The transsulfuration pathway that makes homocysteine from cysteine through cystathionine is absent in W.succinogenes (http://seed-viewer.theseed.org/seedviewer.cgi).
2 Materials and Methods
2.1 Materials
W.succinogenes genomic DNA (ATCC 29543D-5) was purchased from ATCC (Manassas, VA). pTYB1, SapI, NdeI, XhoI and chitin beads were bought from New England Biolabs (Ipswich, MA). EMD biosciences (Gibbstown, NJ) supplied Luria-Bertani, glycerol, chloroform, methanol, acetonitrile, potassium phosphate and ammonium acetate. IPTG, amplicillin and kanamycin were procured from Lab Scientific (Livingston, NJ). L-cysteine, arabinose, DL-methionine, D-glucose, M9 minimal salts, formic acid, EDTA, urea, D2O, triton X-100, methyl viologen, TCEP, O-acetyl-L-serine, O-succinyl-L-homoserine, DL-homocysteine, 10% Pt on activated carbon, hydroxycobalamin hydrochloride, S-adenosylmethionine chloride, 3-mercaptopropionic acid and o-phthalaldehyde were purchased from Sigma-Aldrich (St.Louis, MO). Ferrous ammonium sulfate, nickel sulfate, chloramphenicol, boric acid, Na2SO3 and Tris.HCl were acquired from Fisher Scientific (Fairlawn, NJ). CaCl2, ZnSO4, MgCl2, MgSO4 and NaCl were from Mallinckrodt. Microcon YM-10 (MWCO 10 kDa), YM-3 (MWCO 3 kDa) and amicon (MWCO 5 kDa) cellulose filters were obtained from Millipore Corporation (Billerica, MA). Aminolevulinic acid, ATP and sodium sulfide were from Acros. O-acetyl-L-homoserine and 5-DL-methyltetrahydrofolic acid calcium salt trihydrate was from TRC Canada (North York, Ontario). Typhoon trio and chelating sepharose fast flow (used for Ni-NTA affinity purification) were products of GE healthcare biosciences (Piscataway, NJ). All bacterial cultures were grown in a New Brunswick Scientific (Edison, NJ) Excella E25 shaker incubator and lysed by sonication using Misonix sonicator 3000 (Misonix Inc., Farmingdale, NY). Absorbance data was obtained on a Cary 300 Bio UV-visible spectrophotometer (Varian, Palo Alto, CA). The glove box was made by Coy Laboratory products (Grass lakes, MI). ESI-MS analysis was performed using an Esquire-LC_00146 instrument (Bruker, Billerica, MA) in the positive ion mode. MALDI-MS data were recorded in positive mode on an Applied Biosystems Voyager STR (matrix: sinapinic acid). LC-MS data were obtained on an Agilent 1200 capillary HPLC system interfaced to an API QSTAR Pulsar Hybrid QTOF mass spectrometer (Applied Biosystems/MDS Sciex, Framingham, MA) equipped with an electrospray ionization (ESI) source. Liquid chromatography (LC) separation was achieved using a Phenomenex Jupiter C4 microbore column (150 × 0.50 mm, 300 Å) at a flow rate of 10 µL/min. MALDI-MS and LC-MS data were provided by the Laboratory of Biological Mass spectrometry at Texas A&M University, College Station, Texas. All the protein concentrations were measured by the Bradford assay13. The protein stock samples are in 100 mM Tris, 150 mM NaCl, 2 mM TCEP, 30% glycerol, pH 8.0 unless otherwise mentioned. Methionine-auxotroph E.coli B834(DE3), harboring the iron sulfur cluster biosynthetic genes in the vector pDB1282, was a gift from Dr. Squire Booker.14
2.2 Methods
Cloning and over-expression
The sir, hcyD, hcyF, hcyS-ala, metY, metZ and metE genes, from Wolinella succinogenes FDC 602W, were inserted between NdeI/XhoI restriction sites of the THT vector (Table 1s). This vector is a pET-28 derived vector which allows attachment of a modified 6xHisTag followed by a TEV protease site onto the N-terminus of the expressed protein. Salmonella typhimurium cysG (siroheme synthase) was cloned into the pACYCDuet vector. All the genes, except sir, were over-expressed in E.coli BL21(DE3). Luria-Bertani cultures containing 40 mg kanamycin per liter were grown at 37°C till an OD600 of 0.6, cooled to 15°C and then induced with a final concentration of 500 µM IPTG before continuing growth at 15°C for another 12 –16 h. The sir gene was co-expressed with the S. typhimurium cysG and the A. vinelandii IscS cluster in E.coli B834(DE3). M9 minimal medium (1.5 L) was supplemented with 30 mL 20% glucose, 3 mL of 1 M MgSO4, 150 µL of 1 M CaCl2, 120 mg DL-methionine, 150 mg ampicillin and 60 mg each of kanamycin and chloramphenicol, inoculated with a starter culture and grown at 37°C to an OD600 of 0.1. At this point, 3.75 g of L-arabinose, 88 mg of ferrous ammonium sulfate and 90 mg of L-cysteine were added and the culture was shaken at 100 rpm till the OD600 reached 0.6. The culture was then cooled for 4 h at 4°C, 45 mg of aminolevulinic acid and IPTG (final concentration = 0.5 mM) were added and the culture was incubated with shaking at 15°C for 12–16 h.
All the cultures were harvested by centrifugation and the cell-pellets were lysed by sonication on ice. The proteins were purified by Ni-NTA affinity chromatography at 4°C. All buffers contained 1 mM TCEP. After purification, all proteins were buffer exchanged into 100 mM Tris, 150 mM NaCl, 2 mM TCEP, 30% glycerol, pH 8.0 and stored in frozen aliquots at −80°C.
To make HcyS-COSH and HcyS-(DL)-homocysteine, hcyS (with the C-terminal alanine removed, see below) was inserted between the NdeI/SapI restriction sites in pTYB1, an intein encoding plasmid15. The HcyS-Intein fusion was over-expressed in E.coli BL21(DE3) as follows: Luria-Bertani cultures containing 100 mg of ampicillin per liter were grown at 37°C till an OD600 of 0.6–0.8 when the temperature was reduced to 15°C and the cultures were induced with IPTG (final concentration = 0.5 mM). Further growth was carried out at 15°C for 12–16 h with constant shaking. The cells were harvested by centrifugation and lysed by sonication on ice in 20 mM Tris, 500 mM NaCl, 1 mM EDTA, 0.1% Triton X-100, pH 7.8. The samples were then loaded onto a chitin column (20 mL) at a flow rate of 0.5 mL/min and washed with 300 mL of 20 mM Tris, 500 mM NaCl, 1 mM EDTA, pH 7.8 at a flow rate of 2 mL/min. Cleavage of the HcyS-Intein fusion was carried out at 4°C for 12–16 h with 30 mL of 50 mM Na2S to give HcyS-COSH or with 30 mL of 50 mM DL-homocysteine to yield HcyS-(DL)-homocysteine. Proteins were buffer exchanged into 100 mM Tris, 150 mM NaCl, 2 mM TCEP, 30% glycerol, pH 8.0 by dialysis using a Novagen D-tube dialyzer Maxi (MWCO 3.5 kDa) and stored as frozen aliquots at −80°C.
Activity assay for HcyD (putative metalloprotease)
200 µL of 611 µM HcyS-Ala were treated with 6 µL of 2.4 mM HcyD and 3 µL of 10 mM ZnSO4 at room-temperature for 2 h. The samples were desalted into 200 µL of 50 mM NH4OAc. 200 µL of acetonitrile and 2 µL of HCOOH were then added and the sample was then analyzed for HcyS formation by positive-mode ESI-MS.
For alanine detection, HcyS-Ala and HcyD were buffer exchanged twice into 50 mM potassium phosphate, pH 8.0 using Bio-rad bio-spin 6 columns. 85 µL of 28 mM HcyS-Ala was treated with 100 µL of 1.62 mM HcyD at room-temperature for 2 h. The sample was freeze-dried and re-dissolved in D2O. The proteins were removed using YM-3 microcon (washed extensively with D2O to remove glycerol before loading the protein) and the filtrate was analyzed by 1H-NMR on a Varian 500 MHz spectrometer.
Activity assay for HcyF (putative HcyS adenylating enzyme)
30 µL of 3.98 mM HcyS-Ala, 23 µL of 1.76 mM HcyF, 30 µL of 0.88 mM HcyD, 3 µL of 10 mM ATP and 6 µL of 10 mM MgCl2 were mixed and incubated at room-temperature for 15 min. The reaction was quenched with an equal volume of 12 M urea, the proteins were removed using a YM-10 microcon and the samples analyzed for AMP formation by HPLC (Agilent 1200, Supelco supelcosil 15 cm × 4.6 mm, 3 µm LC-18-T column) using the following gradient at a flow rate of 1 mL/min: solvent A is water, solvent B is 100 mM potassium phosphate, pH 6.6, solvent C is methanol. 0 min: 100% B; 7 min: 10% A, 90% B; 12 min: 25% A, 60% B, 15% C; 17 min: 25% A, 10% B, 65% C; 19 min: 100% B, 25 min: 100% B. Controls lacking HcyF, HcyD and HcyS were similarly run and analyzed.
Preparation of reduced methyl viologen
Methyl viologen (149.5 mg) was dissolved in 12.5 mL of 50 mM potassium phosphate, pH 8.0 in a 15 mL centrifuge tube. 10% Pt on activated carbon (15 mg) was added. Argon was bubbled through the solution for 5 min followed by hydrogen for 30 min. The sample was quickly sealed with parafilm and centrifuged for 5 min to remove the catalyst. The supernatant containing the reduced methyl viologen was then transferred to a new 15 mL centrifuge tube in an oxygen free glove-box. The concentration of the reduced methyl viologen was measured at 600 nm (extinction coefficient 1.3 × 104 M−1cm−1)16.
Sir-mediated HcyS-COSH formation detected using lissamine rhodamine sulfonyl azide and LC-MS
30 µL of 3.98 mM HcyS-Ala, 30 µL of 1.33 mM HcyF, 10 µL of 1.61 mM HcyD and 9 µL of 379 µM Sir were mixed. The sample was then transferred to a glove-box and allowed to stand for 2 h to allow for the sample to become anerobic. 30 µL of 10 mM ATP, 4 µL of 1 M MgCl2, 2.5 µL of 100 mM Na2SO3 and 100 µL of 3.5 mM reduced methyl viologen were then added. After 15 min, the sample was quenched by exposing to air with shaking.
For gel analysis, the sample was buffer exchanged into 100 µL of 50 mM NH4OAc, 6 M urea, pH 6.0 using a Bio-rad bio-spin 6 column and then treated with 7.4 µL of 15 mM lissamine rhodamine sulfonyl azide for 15 min in the dark at 26°C.17 The sample was then desalted by chloroform:methanol precipitation and analyzed by SDS-PAGE (15% tris-glycine gel). The fluorescence image of the labeled protein in the gel was obtained on a Typhoon Trio imager (excitation: 532 nm green laser; emission: 580-nm band-pass filter (580 BP 30)). A similar sample, lacking HcyD, was prepared as a control.
For MS analysis, the sample was heated at 100°C for 5 min and left to cool to room-temperature for an hour. The precipitated protein was removed by filtration. The filtrate, containing small proteins, was then concentrated and buffer exchanged into 50 mM NH4OAc, pH 6.0 using a Bio-rad bio-spin 6 column and analyzed by LC-MS.
HcyS-COSH is the sulfur donor for homocysteine biosynthesis
95 µL of 159 µM HcyS-COSH and 11 µL of 1.4 mM MetY were incubated with 1.3 µL of 10 mM O-acetyl-L-serine or O-acetyl-L-homoserine. The samples were incubated at room-temperature for 1 h. They were then buffer exchanged into 50 mM NH4OAc, pH 6.0 and analyzed by MALDI-MS. The time-period of incubation was later reduced to 2 min to avoid formation of the 7,912 Da adduct (Supplementary material, Figure 3s).
Sulfide at high concentrations is also a sulfur source for homocysteine biosynthesis
80 µL of 1.4 mM MetY (in 50 mM potassium phosphate, pH 8.0) was mixed with 320 µL of 50 mM potassium phosphate, 80% D2O, pH 8.0 along with 50 µL each of 100 mM sodium sulfide and 100 mM O-acetyl-L-homoserine. The sample was incubated for 1 h at 26°C. An identical sample lacking MetY was prepared as a control. Samples were freeze-dried, re-dissolved in 100% D2O and analyzed by 1H-NMR on Varian 300 MHz instrument.
Reaction of HcyS-COSH with MetZ
2.9 µL of 1.4 mM MetZ and 95 µL of 42 µM HcyS-COSH were incubated with the appropriate substrate – 0.4 µL of 10 mM O-acetyl-L-serine, 0.4 µL of 10 mM O-acetyl-L-homoserine and 0.4 µL of 10 mM O-succinyl-L-homoserine for 3 min at room-temperature. The samples were buffer exchanged into 50 mM NH4OAc, pH 6.0 and analyzed by MALDI-MS.
HcyD catalyzed homocysteine release from HcyS-homocysteine
95 µL of 337 µM HcyS-(DL)-homocysteine were mixed with 3.6 µL of 880 µM HcyD and incubated at room-temperature for 5 min. The sample was then treated with 98.6 µL of 9 M urea, buffer exchanged into 50 mM NH4OAc, pH 6.0 using a Bio-rad bio-spin 6 chromatography column and analyzed by MALDI-MS. An identical reaction mixture lacking HcyD was prepared as a control.
For small molecule analysis, 190 µL of 42 µM HcyS-COSH and 8 µL of 1 mM MetY were mixed, buffer-exchanged into 50 mM potassium phosphate, pH 8.0 and concentrated to 100 µL using a millipore ultrafree centrifugal filter device (NMWL = 5000 Da). 0.4 µL of 250 mM TCEP and 0.8 µL of 10 mM O-acetyl-L-homoserine were then added. After incubation at room-temperature for 5 min, 10 µL of 76 µM HcyD in 50 mM potassium phosphate, pH 8.0 was added. After an additional 5 min at room-temperature, protein was removed using a YM-3 microcon and the sample was treated with methanolic o-phthalaldehyde18–20 (1 mM final concentration) and analyzed immediately by HPLC (Agilent 1200 using Supelco supelcosil LC-18-T (15 cm × 4.6 mm, 3 µm)) using the following gradient at a flow rate of 1 mL/min: solvent A is water, solvent B is 100 mM potassium phosphate, pH 6.6, solvent C is methanol. 0 min: 100% B; 7 min: 10% A, 90% B; 12 min: 25% A, 60% B, 15% C; 17 min: 25% A, 10% B, 65% C; 19 min: 100% B, 25 min: 100% B.
HcyD catalyzed hydrolysis of HcyS-COSH
95 µL of 42 µM HcyS-COSH were mixed with 3.6 µL of 88 µM HcyD and incubated at room-temperature for 5 min. The sample was then treated with 99.5 µL of 9 M urea, buffer exchanged into 50 mM NH4OAc, pH 6.0 using a Bio-rad bio-spin 6 chromatography column and analyzed by MALDI-MS. An identical reaction mixture lacking HcyD was prepared as a control.
WS0269 (MetE)-catalyzed conversion of homocysteine to methionine
100 µL of 169 µM of WS0269 (annotated as MetE) was incubated with 20 µL of 50 mM potassium phosphate, pH 8.0, 1.7 µL of saturated 100 mM 5-DL-methyltetrahydrofolate, 1.7 µL of 10 mM MgSO4 and 1.7 µL of 100 mM DL-homocysteine.21 The sample was incubated at room-temperature for 1 h. The protein was then removed by passing the samples through a millipore ultrafree centrifugal filter device (NMWL = 5000 Da). 150 µL of the flow-through was treated with 50 µL of o-phthalaldehyde reagent (1 mL of 37 mM o-phthalaldehyde in MeOH, 4 mL of 0.1 M boric acid, pH 9.3, 162 µL of 3-mercaptopropionic acid were mixed together and pH was adjusted to 9.3 with NaOH). After 5 min at room-temperature, 20 µL of 1 M potassium phosphate, pH 6.0 were added and the reaction mixture was analyzed by HPLC (Agilent 1200 using Phenomenex Gemini 5µ C18 110A (15 cm × 4.6 mm, 5 µm)) using the following gradient at a flow rate of 1 mL/min: solvent A is water, solvent B is 100 mM potassium phosphate, pH 6.6, solvent C is methanol. 0 min: 100% B; 7 min: 10% A, 90%B; 12 min: 25% A, 60% B, 15% C; 17 min: 25% A, 10% B, 65% C; 19 min: 100% B, 25 min: 100% B.
Results
Growth and over-expression
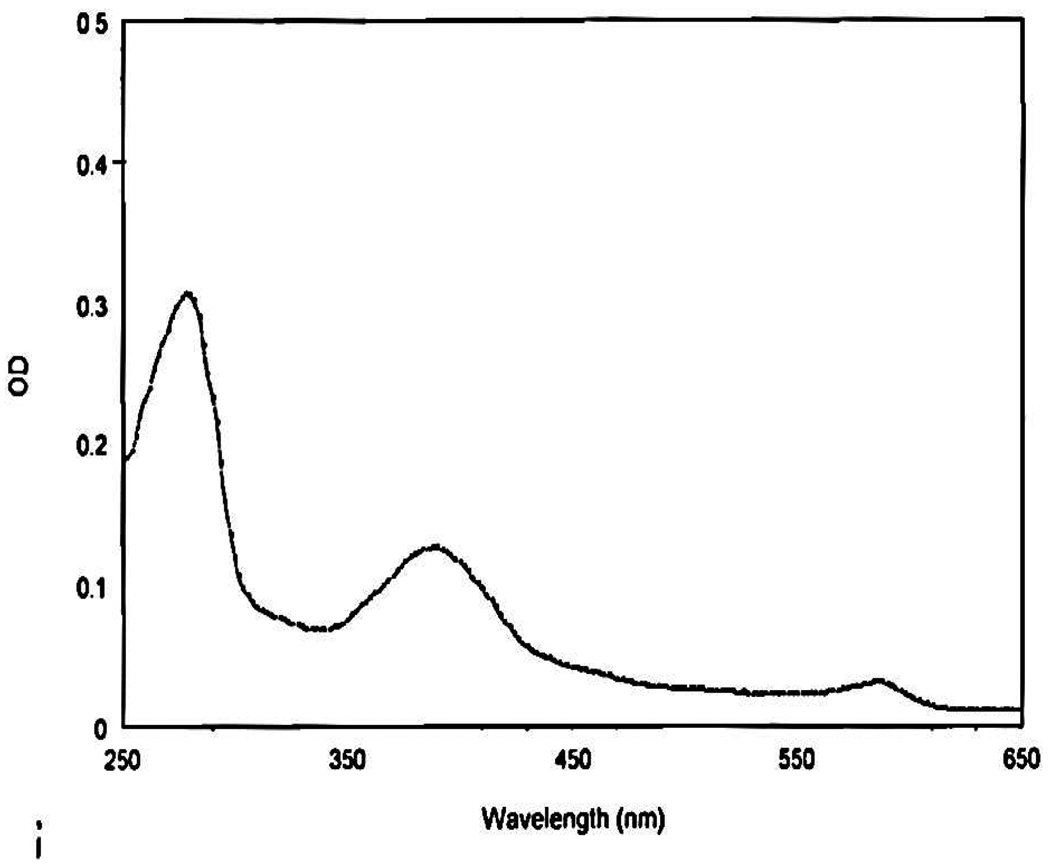
The sir, hcyD, hcyF, hcyS-ala, metY, metZ and WS0269 (metE) genes (Table 1) were cloned into the THT vector. All the proteins, except Sir, were over-expressed in E.coli BL21 (DE3) in LB medium. The sir gene was over-expressed in the methionine-auxotroph E.coli B834 (DE3). In this strain, Sir, annotated as ferredoxin-sulfite reductase, was co-expressed with siroheme synthase (cloned into pACYDuet) and the IscS-cluster assembly proteins (cloned into pDB1282) in M9 medium. Co-expression with these proteins is essential for good reconstitution of active Sir, which utilizes siroheme and [4Fe-4S] cofactors. All proteins were purified on a Ni-NTA affinity column. Sir was purified aerobically (this [4Fe-4S] containing protein did not require anaerobic purification) at 4°C. It had the characteristic absorbance of a siroheme-[4Fe-4S] cluster protein22 (Figure 4). MetY and MetZ were yellow suggestive of bound PLP. SDS-PAGE analysis of all the homocysteine biosynthetic proteins is given in Supplementary material, Figure 1s.
Table 1.
Hypothetical functions of proteins adjacent to the thiocarboxylate-forming protein, HcyS-Ala.
| Protein IDs | Putative function of the proteins |
|---|---|
| WS1004, NP_907206 (sir) | Ferredoxin sulfite reductase* |
| WS1005, NP_907207 (hcyD) | Metalloprotease involved in the removal of C-terminal alanine of HcyS-Ala prior to sulfur transfer and also in the release of homocysteine from HcyS-Homocysteine adduct* |
| WS1006, NP_907208 (hcyF) | ATP-utilizing enzyme involved in adenylating HcyS C-terminal prior to sulfur transfer* |
| WS1007, NP_907209 (hcyS-ala) | Sulfur transfer protein involved in methionine biosynthesis* |
| WS1008 and WS1009, NP_907210 and NP_907211 (cysD and cysN) | Sulfate adenylyltransferase, subunits 1 and 2 |
| WS1010, NP_907212 (cysH) | Phosphoadenylyl-sulfate reductase/Adenylyl-sulfate reductase |
| WS1011, NP_907213 | Conserved hypothetical protein probably involved in assimilatory sulfate reduction |
| WS1012, NP_907214 (metY) | O-acetylhomoserine sulfhydrylase* |
| WS1013, NP_907215 | Methyltransferase (EC 2.1.1.-) |
| WS1014, NP_907216 | Putative efflux protein |
| WS1015, NP_907217 (metZ) | O-acetylhomoserine or O-succinylhomoserine sulfhydrylase* |
Experimentally characterized in this study.
Figure 4.
Absorbance spectrum of Sir with its maxima at 388 nm and 590 nm characteristic of a ferredoxin-sulfite reductase.
HcyD is a metalloprotease involved in C-terminal processing of HcyS-Ala
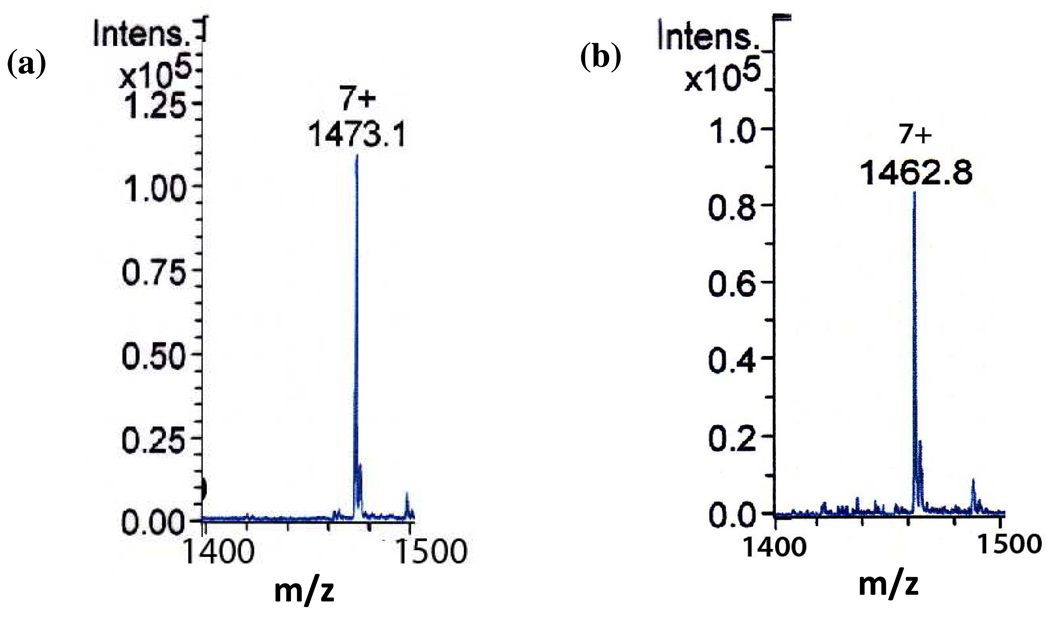
HcyD removes the C-terminal alanine from HcyS-Ala in the presence of exogenous Zn2+. ESI-MS analysis of the reaction mixture demonstrates the conversion of HcyS-Ala to HcyS (observed mass change: 69.37 Da, expected mass change upon removal of alanine: 71.04 Da, Figure 5). 1H-NMR analysis of the small-molecule product confirmed the release of alanine from HcyS-Ala (Supplementary material, Figure 2s).
Figure 5.
ESI-MS analysis of the HcyD-catalyzed removal of alanine from HcyS-Ala (+7 charge state). (a) HcyS-Ala (observed deconvoluted mass: 10304.98 Da, expected mass: 10304 Da, error: 0.01%) (b) HcyS formed after treatment of HcyS-Ala with HcyD. (observed deconvoluted mass: 10235.61 Da, expected mass: 10232.92 Da, error: 0.03%).
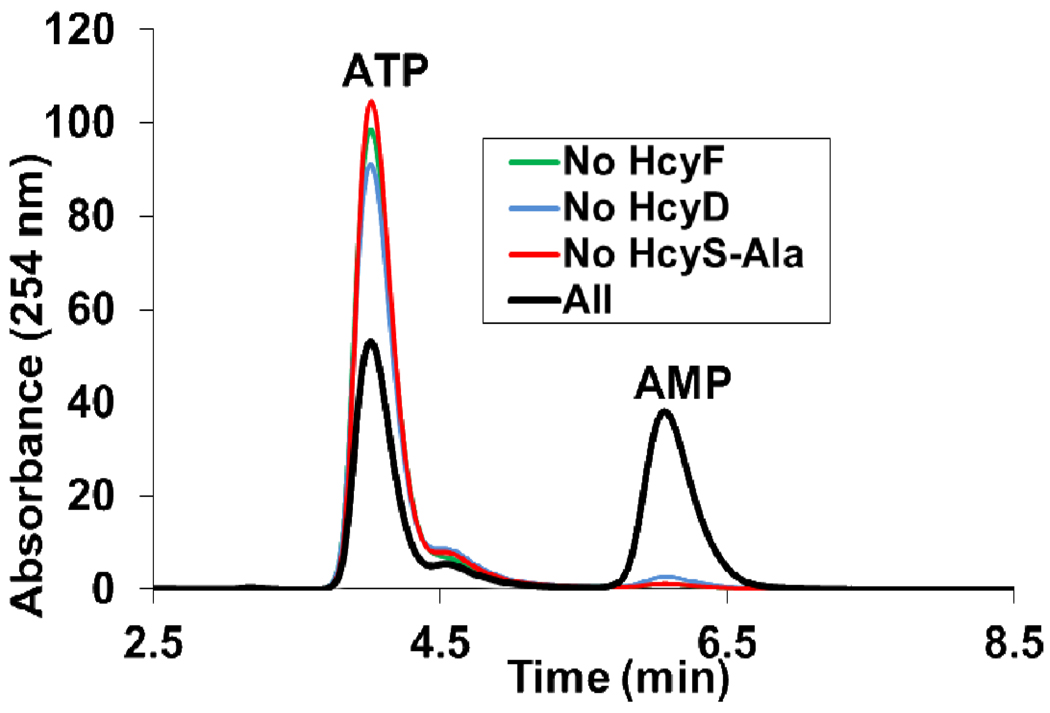
HcyF activates HcyS-COOH by adenylation
HcyF was shown to adenylate HcyS-COOH, generated by HcyD catalyzed hydrolysis of HcyS-Ala. The adenylating reaction was monitored by the release of AMP from the unstable HcyS-COAMP. AMP formation was dependent on HcyF, HcyS-Ala and HcyD (Figure 6).
Figure 6.
Adenylating activity of HcyF. HcyF adenylates HcyS and the resulting acyl-adenylate undergoes hydrolysis in the absence of bisulfide to give AMP.
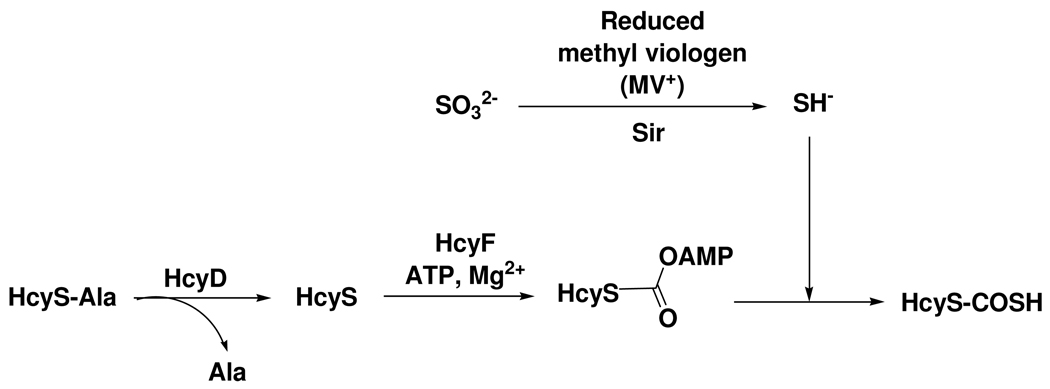
Sir catalyzes the formation of HcyS-COSH from sulfite
The formation of HcyS-COSH from HcyS-Ala requires HcyD catalyzed removal of alanine, HcyF-catalyzed adenylation and AMP displacement by sulfide generated by the Sir-catalyzed reduction of SO32− (Figure 7). LC-MS analysis of the reconstitution reaction mixture demonstrated that this conversion was successfully achieved (Figure 8).
Figure 7.
Reconstitution of HcyS-COSH biosynthesis using methyl viologen as the electron donor and sulfite as the sulfur source.
Figure 8.
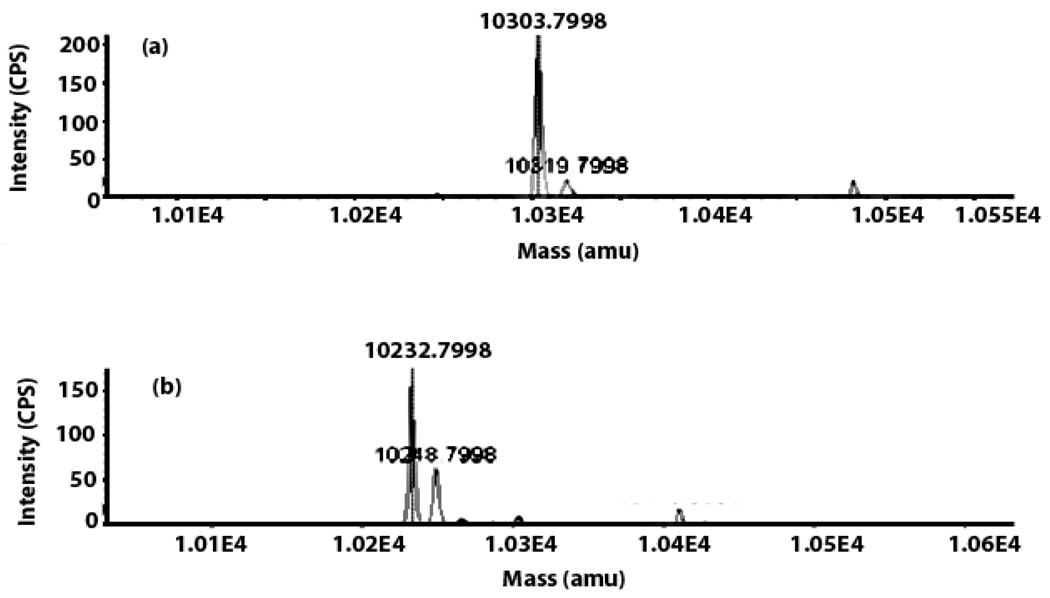
Reconstitution of HcyS-COSH biosynthesis from HcyS-Ala monitored by LC-MS analysis. (a) Reconstitution reaction mixture from which HcyD was omitted showing that HcyS-Ala remains unmodified. (b) The full reaction mixture showing conversion of HcyS-Ala (observed mass: 10303.8 Da, expected mass: 10304 Da, error: 0.002%) to HcyS-COSH (observed mass: 10248.8 Da, expected mass: 10248.92 Da, error: 0.001%) via the intermediacy of HcyS (observed mass: 10232.8, expected mass: 10232.92 Da, error: 0.001%)
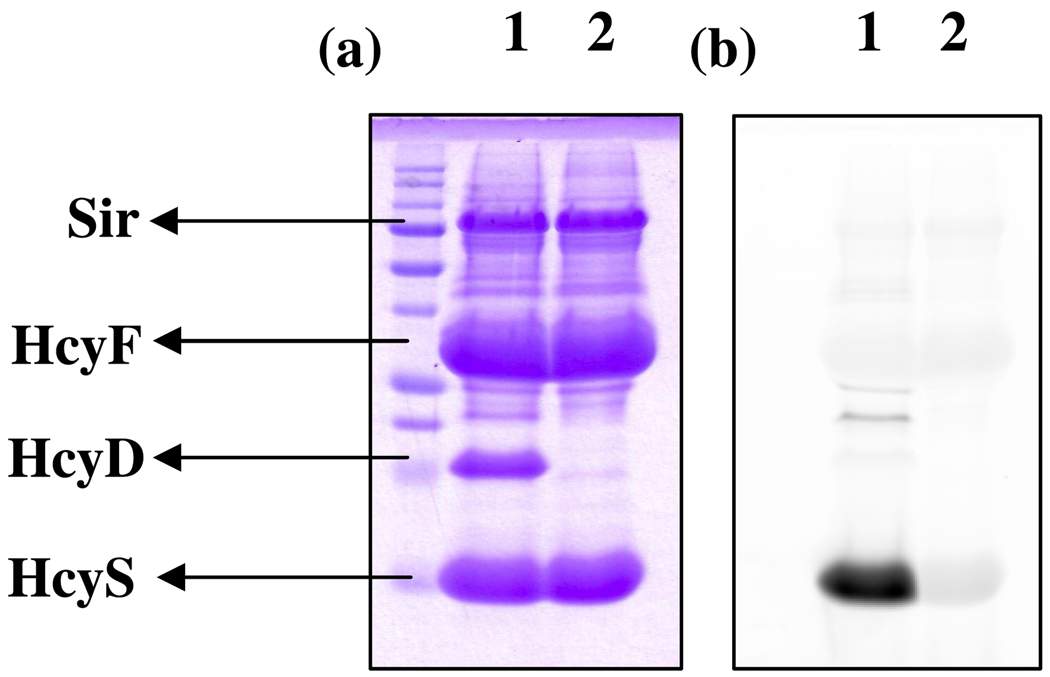
HcyS-COSH formation was also detected using the thiocarboxylate specific fluorescent reagent lissamine rhodamine sulfonyl azide.17 The resulting fluorescent protein was analyzed by SDS-PAGE and scanned for fluorescence using a Typhoon trio (Figure 9).
Figure 9.
SDS-PAGE analysis of HcyS-COSH formation after labeling with lissamine rhodamine sulfonyl azide (a) Coomassie image (b) Fluorescent image. Lane 1: Sample containing all components – HcyD, HcyF, HcyS-Ala, Sir, SO32−, reduced methyl viologen (electron donor), ATP, Mg2+. Lane 2: Same as Lane 1 except HcyD was omitted from the reaction mixture. PMT voltage: 400 V.
HcyS-COSH is the sulfur source for MetY-catalyzed homocysteine biosynthesis
WS1012, annotated as a putative O-acetyl-L-homoserine sulfhydrylase (MetY), catalyzed the reaction of HcyS-COSH with O-acetyl-L-homoserine to form HcyS-Homocysteine (Figure 10 (a)). MetY did not accept O-acetyl-L-serine as a substrate even after extended incubation time (Figure 10 (b)). Longer incubation time or higher concentrations of O-acetyl-L-homoserine resulted in the formation of a higher molecular weight adduct (7911 Da), consistent with homolanthionine formation (Supplementary material, Figure 3s)23. MetY can also use sulfide to convert O-acetyl-L-homoserine to homocysteine (Supplementary material, Figure 4s).
Figure 10.
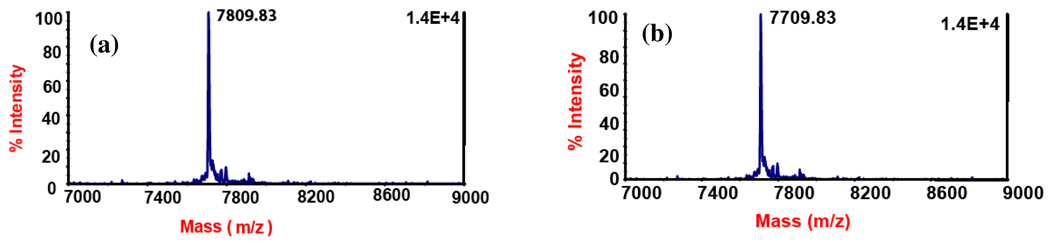
MALDI-MS analysis of the MetY-catalyzed homocysteine biosynthesis (a) HcyS-COSH in the presence of MetY and O-acetyl-L-homoserine, forms HcyS-homocysteine (observed mass: 7809.83 Da, expected mass: 7805.96 Da, error: 0.05%, net mass change of protein: 100 Da). (b) HcyS-COSH (observed mass: 7709.83 Da, expected mass: 7704.82 Da, error: 0.07%) in the presence of MetY and O-acetyl-L-serine, 1 h incubation time, showing that HcyS-COSH cannot transfer sulfide to O-acetyl-L-serine. HcyS-COSH and HcyS-homocysteine (see below) do not have the His6-tag and hence, their masses are different from the His-tagged HcyS used in other experiments in this work.
WS1015, annotated as a putative O-acetyl/succinyl-L-homoserine sulfhydrylase (MetZ), did not catalyze the reaction of HcyS-COSH with O-acetyl-L-homoserine or O-succinyl-L-homoserine to form HcyS-Homocysteine (Supplementary material, Figure 5s).
HcyD catalyzes the release of homocysteine from HcyS-homocysteine
HcyS-homocysteine was prepared by cleaving the HcyS-intein construct with (DL)-homocysteine. HcyD catalyzes the release of homocysteine from HcyS-homocysteine as shown by MALDI-MS analysis of the reaction mixture (Figure 11(a)). This reaction proceeded to greater than 50% indicating that in addition to alanine (see above) HcyD can remove both isomers of homocysteine. HcyD also catalyzes the hydrolysis of HcyS-COSH (Supplementary material, Figure 6s).
Figure 11.
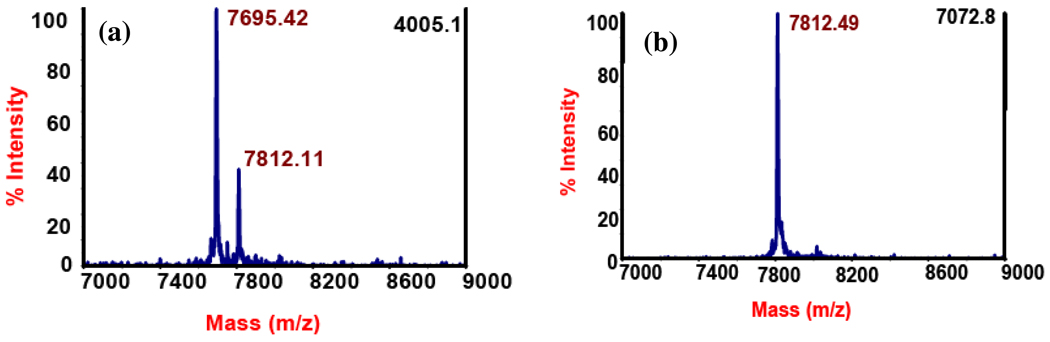
MALDI-MS analysis of HcyD treated HcyS-DL-Homocysteine: (a) HcyS-DL-Homocysteine (observed mass: 7812.11 Da, expected mass: 7805.96 Da, error: 0.08%) in the presence of HcyD is converted to HcyS-homocysteine to HcyS-COOH (observed mass: 7695.42 Da, expected mass: 7688.82 Da, error: 0.09%, net mass change: 116.69 Da) (b) Reference sample of HcyS-DL-homocysteine (observed mass: 7812.49 Da, expected mass: 7805.96 Da, error: 0.08%).
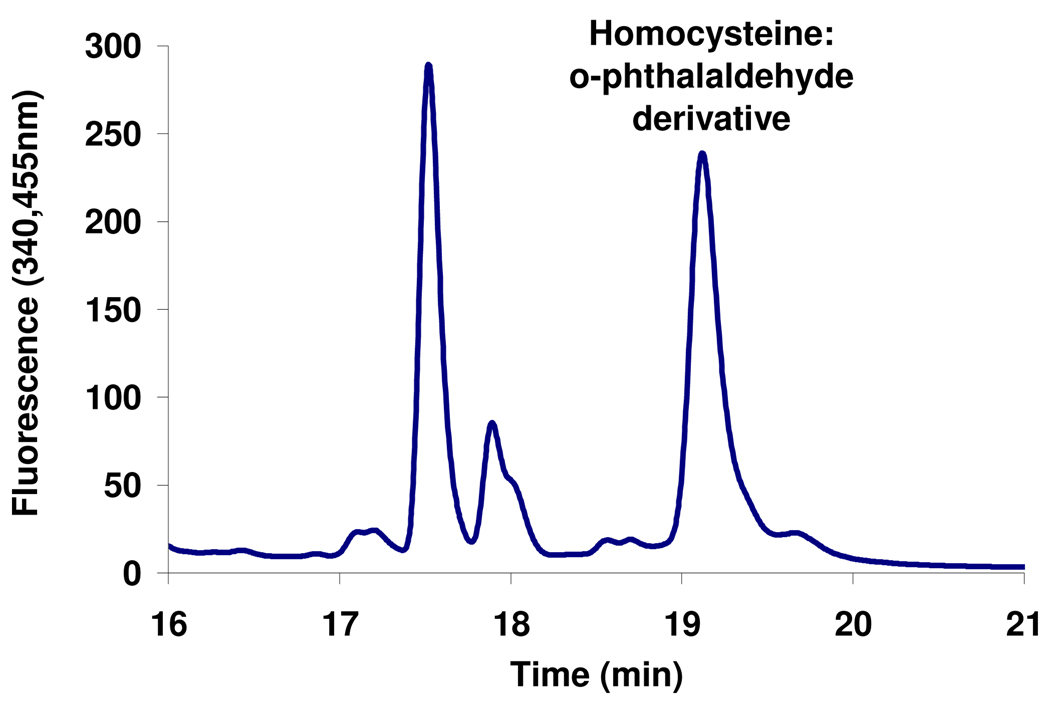
To further characterize the released amino acid, HcyS-L-homocysteine was prepared by reacting HcyS-COSH with O-acetyl-L-homoserine in the presence of MetY and then treated with HcyD. The resulting reaction mixture was then treated with o-phthalaldehyde to yield a fluorescent homocysteine derivative (Supplementary material, Figure 7s) which was analyzed by HPLC18–20 (Figure 12). This detected product was identified as the homocysteine derivative by co-elution with a reference homocysteine:o-phthalaldehyde adduct.
Figure 12.
Characterization of the amino acid released from HcyS-homocysteine by o-phthalaldehyde derivatization. The peak at 19.1 min co-migrated with a reference homocysteine:o-phthalaldehyde adduct.
WS0269 is a methionine synthase
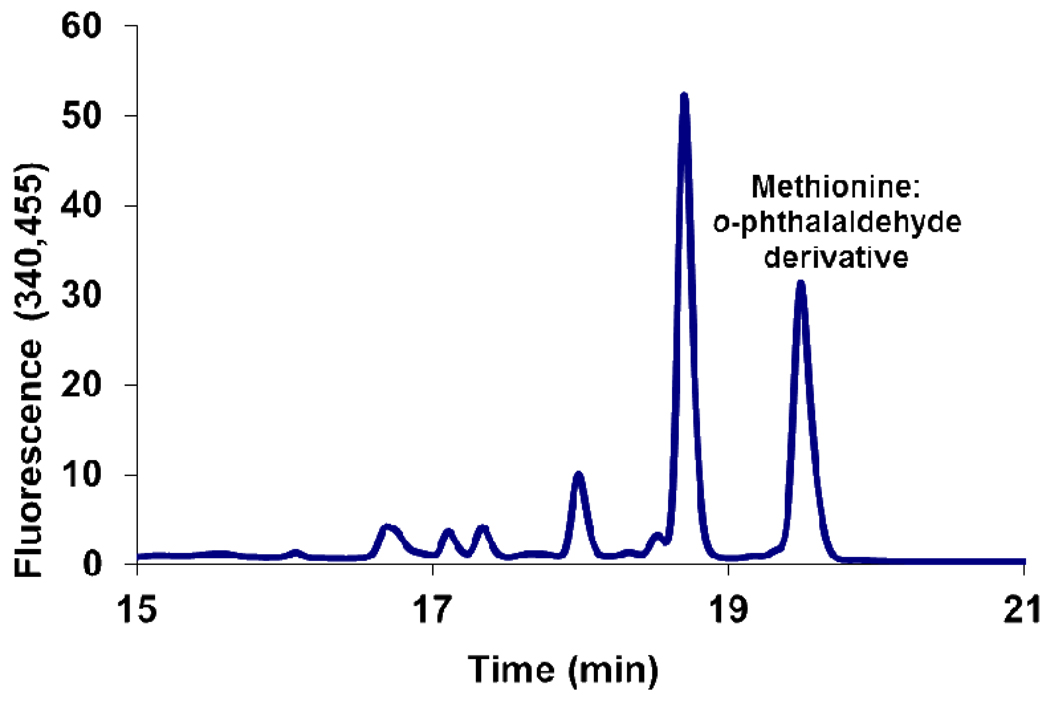
WS0269 (MetE) catalyzes the methyl transfer from methyltetrahydrofolate to homocysteine to form methionine (Figure 13). Methionine production was assayed by HPLC after derivatization with o-phthalaldehyde/mercaptopropionic acid.
Figure 13.
WS0269 is a methyltetrahydrofolate dependent methionine synthase. HPLC analysis of the reaction mixture after derivitization with o-phthalaldehyde. The peak at 19.5 min co-migrated with a reference methionine:o-phthalaldehyde adduct.
Discussion
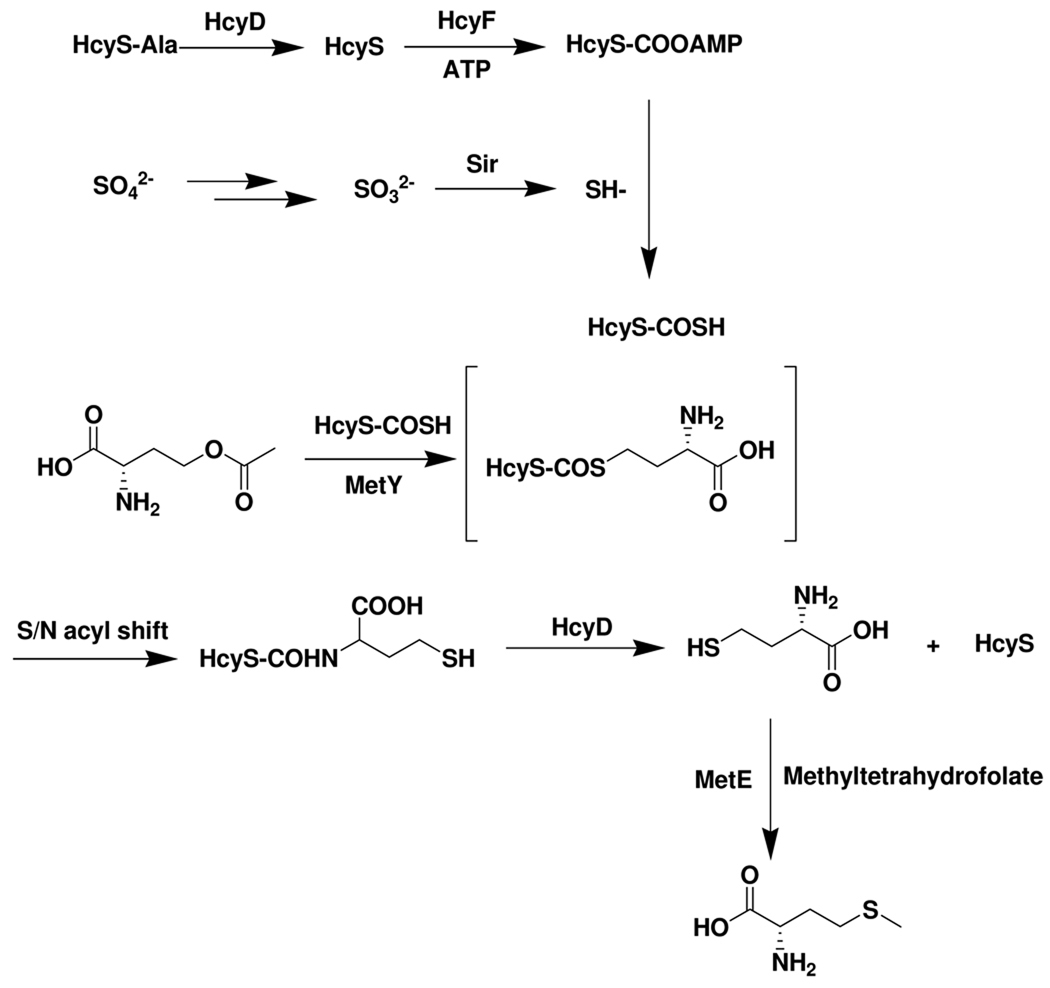
Thiocarboxylated proteins are important intermediates in a variety of biochemical sulfide transfer reactions. For ThiS-COSH, MoaD-COSH, and Urm1p, the thiocarboxylate sulfur is derived from cysteine in a cysteine desulfurase catalyzed reaction4,9,10. The sulfur donor for QbsE, PdtH and CysO is unknown. The identification of a putative protein thiocarboxylate dependent methionine biosynthetic gene cluster in Wolinella succinogenes, also encoding the enzymes involved in the reduction of sulfate to sulfide, suggested that sulfite reductase might function as the sulfide source for protein thiocarboxylate formation. The successful reconstitution of the new methionine biosynthetic pathway described in Figure 14 supports this conclusion. In this pathway, the carboxy terminal alanine is removed from HcyS-Ala in a reaction catalyzed by HcyD. HcyF then catalyzes the adenylation of HcyS. HcyS acyl-adenylate undergoes nucleophilic substitution by bisulfide produced by Sir to form HcyS thiocarboxylate, which adds to O-acetylhomoserine to give HcyS-homocysteine in a reaction catalyzed by MetY. This reaction is likely to proceed via a PLP-mediated substitution followed by an S/N acyl shift analogous to cysteine biosynthesis in M.tuberculosis.5 In that pathway, CysO-thiocarboxylate is converted to CysO-cysteine upon treatment with the PLP-dependent O-phospho-L-serine sulfhydrylase and O-phospho-L-serine5,24. HcyD mediated hydrolysis liberates homocysteine. A final methylation, catalyzed by MetE, completes the methionine biosynthesis.
Figure 14.
A new protein-thiocarboxylate-mediated methionine biosynthesis in Wolinella succinogenes.
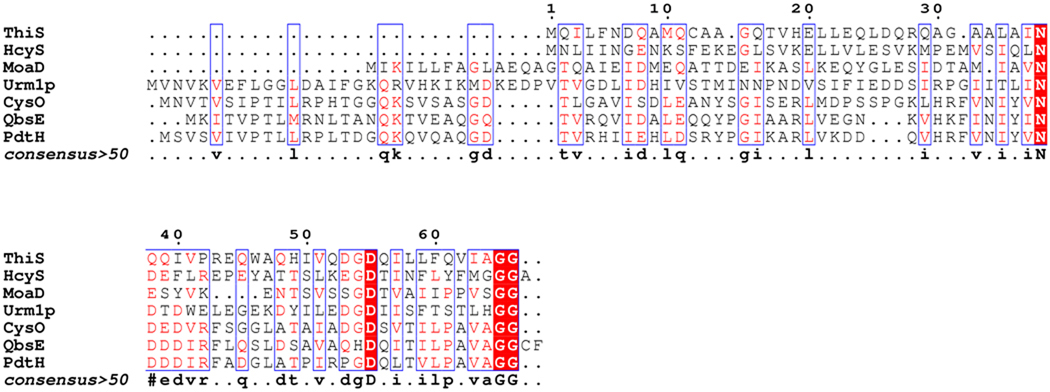
Several features of this pathway merit further comment. All known thiocarboxylate forming proteins have a Gly-Gly C-terminus in their active form (Figure 15). HcyS-Ala ends in a Gly-Gly-Ala sequence that requires processing by the HcyD metalloprotease. The physiological significance of this processing event is unknown. Pathway regulation is one possibility. We have previously observed analogous carboxy terminal processing: QbsD removes Cys-Phe from the carboxy terminal of QbsE during thioquinolobactin biosynthesis in P.fluorescens6 and Mec+ removes cysteine from CysO-Cys during cysteine biosynthesis in M.tuberculosis5. HcyD shows broad substrate tolerance and also catalyzes the removal of both isomers of homocysteine from HcyS-homocysteine as well as sulfide from HcyS-COSH.
Figure 15.
Sequence alignment of biochemically characterized thiocarboxylate-forming proteins (done using Escript 2.2). All have a diglycyl C-terminus in their active form. Both HcyS-Ala and QbsE are activated by carboxy terminal processing. Sequence alignment of putative HcyS-like proteins from various organisms is shown in Supplementary material, Figure 8s.
We assayed for formation of HcyS thiocarboxylate using an MS based assay as well as a recently developed reagent for the detection of protein thiocarboxylates in bacterial proteomes involving a click reaction with lissamine rhodamine B sulfonyl azide.17 The fact that Sir is clustered with the other methionine biosynthetic genes suggests that it is functioning as the sulfur source for methionine biosynthesis in vivo in Wolinella succinogenes. However, we do not have evidence at this time for sulfide channeling between Sir and the HcyF/HcyS complex. Kinetics studies to address this are in progress.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Dr. Cynthia Kinsland at the Protein facility, Cornell University for overexpression of all genes used in this study and Dr. Squire Booker for the methionine-auxotroph E. coli B834(DE3) harboring the iscS-cluster assembly genes. MALDI-MS and LC-MS were obtained by the Laboratory of Biological Mass spectrometry at Texas A&M University. This research was supported by NIH (DK44083) and by the Robert A. Welch Foundation (A-0034).
Footnotes
Supporting Information Available. Additional sequence analysis, SDS-PAGE of purified proteins, reaction schemes, reaction mixture and product spectra and the primers used for cloning are provided as supplementary material.
REFERENCES
- 1.Kessler D. FEMS Microbiol Rev. 2006;30:825. doi: 10.1111/j.1574-6976.2006.00036.x. [DOI] [PubMed] [Google Scholar]
- 2.Mueller EG. Nat Chem Biol. 2006;2:185. doi: 10.1038/nchembio779. [DOI] [PubMed] [Google Scholar]
- 3.Dorrestein PC, Zhai H, McLafferty FW, Begley TP. Chem Biol. 2004;11:1373. doi: 10.1016/j.chembiol.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 4.Leimkuhler S, Wuebbens MM, Rajagopalan KV. J Biol Chem. 2001;276:34695. doi: 10.1074/jbc.M102787200. [DOI] [PubMed] [Google Scholar]
- 5.Burns KE, Baumgart S, Dorrestein PC, Zhai H, McLafferty FW, Begley TP. J Am Chem Soc. 2005;127:11602. doi: 10.1021/ja053476x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Godert AM, Jin M, McLafferty FW, Begley TP. J Bacteriol. 2007;189:2941. doi: 10.1128/JB.01200-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shigi N, Sakaguchi Y, Asai S, Suzuki T, Watanabe K. Embo J. 2008;27:3267. doi: 10.1038/emboj.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leidel S, Pedrioli PG, Bucher T, Brost R, Costanzo M, Schmidt A, Aebersold R, Boone C, Hofmann K, Peter M. Nature. 2009;458:228. doi: 10.1038/nature07643. [DOI] [PubMed] [Google Scholar]
- 9.Begley TP, Xi J, Kinsland C, Taylor S, McLafferty F. Curr Opin Chem Biol. 1999;3:623. doi: 10.1016/s1367-5931(99)00018-6. [DOI] [PubMed] [Google Scholar]
- 10.Leimkuhler S, Rajagopalan KV. J Biol Chem. 2001;276:22024. doi: 10.1074/jbc.M102072200. [DOI] [PubMed] [Google Scholar]
- 11.Noma A, Sakaguchi Y, Suzuki T. Nucleic Acids Res. 2009;37:1335. doi: 10.1093/nar/gkn1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Belfaiza J, Martel A, Margarita D, Saint Girons I. J Bacteriol. 1998;180:250. doi: 10.1128/jb.180.2.250-255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bradford MM. Anal Biochem. 1976;72:248. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 14.Chatterjee A, Li Y, Zhang Y, Grove TL, Lee M, Krebs C, Booker SJ, Begley TP, Ealick SE. Nat Chem Biol. 2008;4:758. doi: 10.1038/nchembio.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kinsland C, Taylor SV, Kelleher NL, McLafferty FW, Begley TP. Protein Sci. 1998;7:1839. doi: 10.1002/pro.5560070821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thorneley RN. Biochim Biophys Acta. 1974;333:487. doi: 10.1016/0005-2728(74)90133-9. [DOI] [PubMed] [Google Scholar]
- 17.Krishnamoorthy K, Begley TP. J Am Chem Soc. 2010;132:11608. doi: 10.1021/ja1034107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benson JR, Hare PE. Proc Natl Acad Sci U S A. 1975;72:619. doi: 10.1073/pnas.72.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okumura HS, Philmus B, Portmann C, Hemscheidt TK. J Nat Prod. 2009;72:172. doi: 10.1021/np800557m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Puri RN, Roskoski R., Jr Anal Biochem. 1988;173:26. doi: 10.1016/0003-2697(88)90153-4. [DOI] [PubMed] [Google Scholar]
- 21.Gonzalez JC, Peariso K, Penner-Hahn JE, Matthews RG. Biochemistry. 1996;35:12228. doi: 10.1021/bi9615452. [DOI] [PubMed] [Google Scholar]
- 22.Koguchi O, Tamura G. Agric. Biol. Chem. 1988;52:373. [Google Scholar]
- 23.Kromer JO, Heinzle E, Schroder H, Wittmann C. J Bacteriol. 2006;188:609. doi: 10.1128/JB.188.2.609-618.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Leary SE, Jurgenson CT, Ealick SE, Begley TP. Biochemistry. 2008;47:11606. doi: 10.1021/bi8013664. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.