Abstract
Monitoring the kinetics of protein interactions on a high density sensor array is vital to drug development and proteomic analysis. Label-free kinetic assays based on surface plasmon resonance are the current gold standard, but they have poor detection limits, suffer from non-specific binding, and are not amenable to high throughput analyses. Here we show that magnetically responsive nanosensors that have been scaled to over 100,000 sensors/cm2 can be used to measure the binding kinetics of various proteins with high spatial and temporal resolution. We present an analytical model that describes the binding of magnetically labeled antibodies to proteins that are immobilized on the sensor surface. This model is able to quantify the kinetics of antibody-antigen binding at sensitivities as low as 20 zeptomoles of solute.
Affinity-based sensing of DNA hybridization, antigen-antibody binding, and DNA-protein interactions play a vital role in basic science research, clinical diagnostics, biomolecular engineering, and drug design1,2,3,4,5,6,7,8,9,10. As the state of the art advances, demand for accurate, sensitive, specific, high-throughput, and rapid methods for the determination of molecular identities and reaction details places increasing pressure on evolving analytical methods11,12,13,14,15,16,17. To meet these pressing needs, researchers have turned to nano-scale labels in order to enhance the limit of detection (LOD) and specificity for detecting low abundance molecules. Such labels, however, can alter diffusion and steric phenomena. In addition, high-throughput, or speed requirements often prohibit the use of classical equilibrium methods, so a precise understanding of reaction kinetics, transport phenomena, and the implications of surface immobilization becomes critical for extracting meaningful molecular reaction parameters for nanoparticle labeling methodologies. This report addresses these issues and demonstrates that nanoparticle labeled proteins offer unique advantages over label-free methods, making this system very effective for modeling and extracting binding kinetics and analyte transport.
Extant modeling of molecular interactions has predominantly been restricted to label-free binding in solution. Early work by Berg and Stenberg proposed some of the first kinetic models of surface antigen-antibody interactions that explained the new restrictions that labeled reagents introduce on surface reaction kinetics by altering rotational and translational motion18,19,20. Furthermore, they argued that the use of targets immobilized on sensor surfaces implies that diffusion can become problematic due to the existence of long range concentration gradients, which can require ligands to traverse macroscopic distances (>100 μm) prior to reaction. Though many of these details are elaborated by Waite21 and Sheehan22, their emphasis on numerical methods precludes the derivation of semi-analytical expressions. While binding kinetics of quantum-dot-labeled macromolecules in liquid phase has been studied with fluorescence cross-correlation spectroscopy23,24, we found no similar literature describing reactions on a sensor surface. Our investigation provides new quantitative insight into the binding kinetics of labeled macromolecules interacting with targets immobilized on a sensor surface, addressing this gap in the literature.
GMR nanosensor platform and magnetic nanoparticle tags
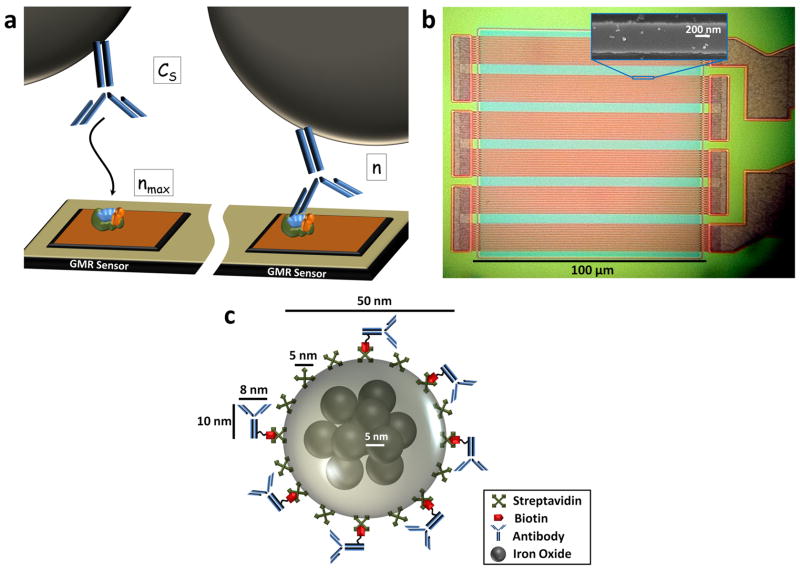
Our approach utilizes giant magnetoresistive (GMR) biosensors, an emerging tool for both basic science research and clinical diagnostics. Their superior LOD, multiplex capacity, broad linear dynamic range, and real-time readout capabilities make them ideal for kinetic analysis measurements25,26,27. GMR nanosensors, initially utilized as read head elements in computer hard drives, operate by changing their electrical resistance in response to changes in the local magnetic field28,29,30,31. Recent work has adapted GMR sensors for detection of biological species in solution by implementing a traditional sandwich assay directly on GMR nanosensors. If a magnetic particle is introduced to label the biomolecule of interest, GMR sensors are capable of highly sensitive DNA and protein detection32,33,34,35,36. This prior work25,26 has involved quantifying the amount of protein, but has provided little information about the kinetics of the biomolecular reaction. In the current research, we pre-label the soluble ligand with a magnetic nanoparticle (MNP) in order to monitor the real-time binding kinetics of the ligand-MNP complex to antigens immobilized on sensor surfaces (Fig. 1a). As the antibody-MNP complexes are captured, their magnetic fields induce changes in electrical resistance in the underlying GMR sensor. Using the rapid, real-time readout of our GMR sensor array25, we can monitor and quantify the kinetics of binding, thus determining the associated kinetic rate constants. Each GMR sensor in the array covers a total area of 100 μm × 100 μm and is comprised of twelve parallel GMR sensor stripes that are connected in series six times, producing a total of 72 stripes per sensor (Fig. 1b). Each stripe is 750 nm wide, approximately 20 nm thick, and spans 100 μm in length. Using scanning electron microscopy, it is possible to resolve nanoparticles bound over each sensor stripe (Fig. 1b insert).
Figure 1. GMR nanosensor and nanoparticle system for kinetic analysis.
a, Schematic representation of antibody-antigen binding. On the left, antibody labeled with a magnetic nanoparticle tag in solution at concentration Cs approaches the GMR sensor surface. When not bound, most diffusing magnetically labeled antibodies are too far from the GMR sensor to be detected. Antigens are immobilized on the sensor surface at an initial surface concentration of nmax. Once the magnetically labeled antibody binds to the antigen, as depicted on the right, the magnetic field from the magnetic tag is detected by the underlying, proximity-based GMR nanosensor. The captured antibody-antigen complex surface concentration is n. b, Optical micrograph showing the GMR sensor architecture comprising 72 stripes connected in parallel and in series. Insert: scanning electron microscopy image of one stripe of the GMR sensor with several bound magnetic nanoparticle tags. c, Schematic representation of a magnetically labeled antibody drawn to scale. The magnetic tag is comprised of a dozen iron oxide cores embedded in a dextran polymer and then functionalized with antibody or receptor.
The MNPs that label the protein or antibody of interest are comprised of approximately twelve 10 nm iron oxide cores embedded in a dextran polymer (Fig. 1c), as determined by TEM analysis37. The entire nanoparticle averages 46 ± 13 nm in diameter (from number weighted Dynamic Light Scattering). Based on the Stokes-Einstein relation, these particles have a translational diffusion coefficient of approximately 9.3×10-12 m2 s-1. The MNPs have a reported zeta potential of -11 mV38. These particles are superparamagnetic and colloidally stable, so they do not aggregate or precipitate during the reaction. Importantly, the GMR sensors operate as proximity-based detectors of the dipole fields from the magnetic tags; thus, only tags within ∼150 nm of the sensor surface are detected25. Therefore, unbound MNP tags contribute negligible signal in the absence of binding, making this unique nanosensor-MNP system ideal for real-time analysis of kinetic binding process at the sensor surface.
Our GMR assay using MNP labels has several advantages over surface plasmon resonance (SPR) - the standard method of monitoring protein binding interactions, which operates by measuring changes in the refractive index of a thin film when unlabeled solute molecules bind to the surface39,40. While recent studies have attempted to design higher density SPR instrumentation41,42,43, their methods typically monitor only a few reactions in parallel, and their LOD and dynamic range are limited to ∼ 25 ng mL-1 and ∼ 2 logs, respectively. In contrast, the GMR biosensor array can simultaneously monitor hundreds to thousands of sensors at sensitivities of ∼ 1 pg mL-1 or below and dynamic ranges of 6 logs or more26. Furthermore, we have fabricated GMR sensor arrays with 1,008 sensors on a 1 mm2 chip area44. The calculated feature density is now over 100,000 GMR sensors per cm2, the highest density reported to date for biosensor technologies. Each sensor within a sub-array is individually addressable by row and column decoders via a shared 6-bit control bus fabricated with state of the art VLSI technology. Such highly integrated GMR sensor arrays allow for exceptionally sensitive, massively parallel multiplex monitoring of protein binding kinetics.
Mathematical model of binding kinetics
Given our ability to monitor real-time binding kinetics using highly sensitive and multiplexed nanosensors, we first tested the widely used Langmuir absorption isotherm model, which assumes that there is negligible depletion of reactants and that the concentration of reactants near the surface is the same as that in the bulk. However, this did not adequately describe the binding kinetics in our system (Supplementary Fig. 1). A new model that would accurately describe the binding rates and would give an analytical solution is needed to explicitly describe the microscopic processes and offer insights into the mechanism of the overall reaction. We devised an analytical model capable of fitting the data with a high degree of generality across a variety of reaction conditions. This new model is capable of fitting real-time binding kinetics data for nanoparticle labeled macromolecules so that one can calculate the molecular association and dissociation rate constants, kon and koff.
Traditionally, protein binding is envisioned as a two-step process in which the target protein or antibody in the bulk solution first enters a surface compartment (reaction zone) via diffusion and flow, and then binds to or escapes from the immobilized targets via the chemical processes of association and dissociation (Supplementary Fig. 2)45. If the soluble ligand diffuses slowly, as for a large MNP tag, the diffusion into the reaction zone becomes negligible (see materials and methods and Supplementary Figs. 2-3 for complete derivation). Therefore, replenishment of soluble antibody-MNP via diffusion from the bulk compartment (bulk zone) to the surface compartment can be assumed to be zero in our model. Accordingly, the rate equation for the traditional two-compartment model can be reduced to the following (see materials and methods for derivation):
| (1) |
where n is the surface concentration of bound MNP-antibody-antigen complexes that have formed over the sensor, kon is the association rate constant, koff is the dissociation rate constant, C0 is the bulk concentration of magnetically tagged antibody, nmax is the maximum moles of surface bound complexes per area, V is the volume of the surface compartment and A is the reaction area (in this case it refers to the surface area of each sensor). The quantity of nA/V is equivalent to the bulk concentration in the reaction zone which would be consumed to produce the surface concentration, n, according to mass conservation. Conceptually, the two terms within the parentheses represent the depletion of tagged antibody in the reaction zone and reduction of available surface sites due to binding, respectively.
As both volume and surface concentrations are discussed, we must be aware of the different dimensions: n and nmax are expressed in mol m-2 whereas Cs and C0 are expressed in mol m-3. Note that nmax is limited by the maximum concentration of close packed MNPs, not by the maximum concentration of analyte on the surface. Therefore, in order to exclude steric effects related to MNP crowding on the sensor surfaces, the highest analyte surface concentration tested was significantly lower than the surface concentration of the close packed antibody-MNP complexes. In addition, by restricting the amount of protein deposited on the sensor surface to be under this limit, the proteins will be spaced across the sensor at distances greater than one MNP diameter. Therefore, each binding event monitored will be from one antibody on a MNP binding to one antigen on the sensor, and not from multiple antibodies binding to multiple antigens, mitigating avidity issues that may complicate the model.
We further simplify Equation 1 by assuming that koff is zero, since antibody-antigen dissociation is typically negligible on the 400-1,000 second time scales of our experiments (Supplemental Fig. 3). Equation 1 now has the following analytical solution (derivation in online materials and methods):
| (2) |
If C0V ≫ nmaxA, which implies a vast excess of solution molecules over available surface sites, the kinetics reduce to Langmuir absorption dynamics (derivation in online materials and methods), demonstrating that the Langmuir model is merely a special case of our more general analytical model.
Experimental and theoretical binding kinetics measurements
To test this solution, we examined antibody-antigen binding kinetics for epithelial cell adhesion molecule (EpCAM) using this analytical model, and compared our experimental results to literature values. Anti-EpCAM antibody was selected because it was formulated into a chemotherapeutic drug, edrecolomab46.
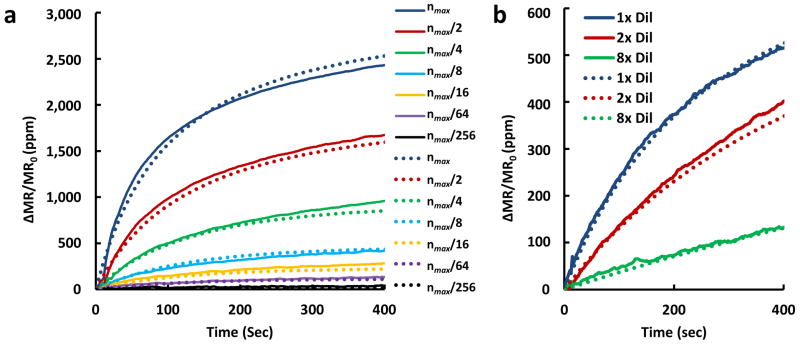
In the first set of binding experiments, presented in Figure 2a, we performed a binding assay of MNP-labeled anti-EpCAM antibody to surface-bound EpCAM protein. C0, nmax, V, and A in the model were fixed values determined from dimensions and concentrations, and kon was determined via best fit of the predicted binding curves to experimental data. We first performed a binding assay for varying concentrations of surface bound EpCAM protein at a fixed concentration of MNP-anti-EpCAM antibody complexes. Twofold dilutions were used to prepare a series of sensor surfaces, beginning at a loading mass (i.e., the amount of protein that is actually bound to the sensor surface and functional) of 5 attomoles of EpCAM and diluting sequentially down to 20 zeptomoles. Therefore, the only parameter that varied between binding curves was nmax; all other parameters were unchanged. When this one parameter variation is implemented in the model, each experimental binding curve was fitted accurately (Fig. 2a). The values of the parameters (the undiluted case) were nmax = 9.5 × 10-10 mol m-2, C0 = 6.8 × 10-7 M, A = 5.4 × 10-9 m2 and V = 5.5 × 10-12 m3. Accordingly, kon = 2.5 × 104 M-1 s-1 fit the data best (R2 = 0.98). In addition, after the MNP-antibody solution was washed away and replaced by antigen-loaded buffer, dissociation constants were calculated by fitting the subsequent data to a basic exponential decay model, nRelease(t) = n0e−kofft, where n0 is the surface concentration of bound MNPs at the time of washing (derivation in online materials and methods). Accordingly, the anti-EpCAM antibody-antigen dissociation constant, koff, was determined to be 2.0 × 10-6 s-1, supporting our assumption that koff is essentially negligible compared to kon.
Figure 2. Comparison of experimentally generated binding curves to kinetic model predictions.
a, Binding curves for anti-EpCAM antibody binding to EpCAM antigens immobilized on the surface. Dotted lines are predictions using the analytical model in equation 2 (•••) and full lines are experimental data (–) obtained for surface loading amounts that vary from 5 attomoles (nmax) to 20 zeptomoles (nmax /256) in serial dilutions of 2×. The fitting error of all the curves in this experiment to the curves predicted by the model is R2=0.98. b, Binding curves for MNP-anti-EpCAM antibody binding to 833 zeptomoles (nmax/6) of EpCAM antigen immobilized on the sensor surface. Dotted lines show prediction using the analytical model and full lines are experimental data obtained for MNP-anti-EpCAM that are undiluted, diluted 2× and diluted 8×. The fitting error of all the curves to the model is R2=0.96. The y-axis is presented as changes in magnetoresistance (MR) normalized to the initial MR in parts per million (ppm).
In a second set of binding experiments, presented in Figure 2b, each sensor was immobilized with a constant load mass of 833 zeptomoles of EpCAM protein (1/6 of the maximum amount used in Figure 2a). The concentration of antibody-MNP complex applied to the sensors was varied between undiluted, twofold diluted, and eightfold diluted solutions of antibody-MNP complexes (corresponding to C0, C0/2 and C0/8 in the model). Since the antibody-antigen interaction should remain the same whether C0 or nmax is altered, the rate constants describing the interaction above should remain the same across these diverse experiments, as was observed. In fitting the model, we obtained the same kon of 2.5 × 104 M-1 s-1 (R2=0.96), supporting the validity of our analytical model. Furthermore, these results lie within the expected range reported in the literature, confirming both the validity of our kinetic model to predict binding and the accuracy of the results derived (Table 1).
Table 1.
Comparison of kon for binding of biotin to streptavidin, binding of EpCAM antibody to EpCAM antigen, binding of CEA antibody to CEA antigen and binding of VEGF antibody to VEGF antigen when using the GMR sensor array and SPR. For both SPR and GMR sensor experiments, the exact same antibody pairs were used. In addition, each method of kinetic analysis was consistent with the literature.
| GMR sensor (×104 M-1 s-1) |
SPR (×104 M-1 s-1) |
Literature (×104 M-1 s-1) |
|
|---|---|---|---|
| Biotinylated DNA and streptavidin | 467 | 550 | 300 – 4500[48] |
| EpCAM antigen and antibody | 2.5 | N/A | 3.2 – 40[49] |
| CEA antigen and antibody | 5.0 | 5.2 | 3.7 – 11[50] |
| VEGF antigen and antibody | 1.6 | N/A | 0.5 – 7[51] |
Similar real-time experiments were performed in order to quantify binding kinetics for MNP-anti-carcinoembryonic antigen (CEA) antibody to CEA, MNP-anti-vascular endothelial growth factor (VEGF) antibody to VEGF, and MNP-streptavidin to biotin binding kinetics (Supplementary Fig. 4). CEA and VEGF were chosen because they are among the most well-known clinical tumor markers, and anti-VEGF antibody drugs, such as bevacizumab (Avastin; Genentech/Roche), are highly effective anti-cancer drugs46,47. For anti-CEA antibody, binding and dissociation constants were monitored with the same reagents on both GMR sensors and SPR instruments, and the results were compared (Supplementary Figs. 4c,d). The GMR sensor array and SPR measurements yielded kinetic constants that were consistent with one another and with literature values (Table 1).
Scanning electron microscopy (SEM) confirmed that our analytical system also has the ability to quantify the precise number of proteins captured on each sensor. Thus, by calibrating GMR signal to an absolute number of magnetic tags bound to the sensor surface, we can derive the mass of protein bound and the signal generated per MNP. To accomplish this, a combination of the prior two experiments was performed and accurately modeled where EpCAM protein was serially diluted in twofold increments, starting at 2.5 attomoles and ending at 78 zeptomoles, on three to eight replica sensors (Fig. 3 left panel). A 20-fold dilution of the MNP-antibody complexes was subsequently added to all the sensors and the binding kinetics were monitored. After 20 minutes of incubation time, the solution of magnetically labeled antibody was washed away to terminate the binding reaction, at which point the sensors were imaged by SEM (Fig. 3 right panel). By normalizing the real-time experimental data and fitting it to the model, we were able to convert the sensor signal, measured as a change in magnetoresistance (MR) normalized to the initial MR (ΔMR/MR0), into the number of magnetic tags bound to each sensor. For example, the sensor functionalized with 2.5 attomoles of EpCAM protein captured 190,000 MNP tags within the time of the experiment. SEM imaging reveals that experimental results match with predictions from the kinetic model, thus extending the validity of the model for precisely quantifying the number of tags bound per sensor and determining the number of proteins that bind during a given reaction.
Figure 3. The kinetic model can predict the number of protein binding events.
By fitting real-time binding curves to the model, it is possible to convert the signal generated from the GMR sensor into an absolute number of magnetic tags bound to the sensor surface. Here, EpCAM protein was loaded onto the sensors at a mass of 2.5 attomoles (nmax) and at masses serially diluted in twofold increments down to 78 zeptomoles (nmax/64). At least three replica sensors were used for each dilution. After 20 mins incubation with the 20-fold diluted solution of MNP-anti-EpCAM antibody (C0/20), the solution was washed away to terminate the binding reaction. Subsequently, a small section of the sensors was imaged with SEM (right panel; colour-coded boxes represent the different loading masses) to compare the number of MNPs bound in the experiment with that predicted by the model (left panel). The number of MNPs presented above each binding curve in the left panel represents the number of MNPs predicted to have bound to each corresponding sensor. Dotted lines are predictions using the analytical model in equation 2 (•••) and full lines are experimental data (–). SEM image of nmax/64 was not shown due to the low surface coverage of MNPs.
Furthermore, using our models, we estimate that every 150 MNPs produce ∼1 ppm of the normalized signal. The LOD of our sensors is ∼20 ppm (defined by the average background signal of a non-complementary antibody coated sensor plus two standard deviations). Therefore, we can detect as few as 0.6 particles μm-2. In addition, the model is capable of explaining when saturation of the sensor surface will occur. For example, when 10 attomoles of protein are deposited on the sensor surface, experimental data shows that the sensor surface is approaching saturation as judged by comparing signals at 5 attomoles and lower (Supplementary Fig. 5). A loading mass of 10 attomoles is described in the model as nmax being equal to 1.9×10-9 mol m-2. The saturation value predicted by the model is thus in close agreement with the maximum surface concentration of the MNPs in a close packed single layer, 1.0 × 10-9 mol m-2.
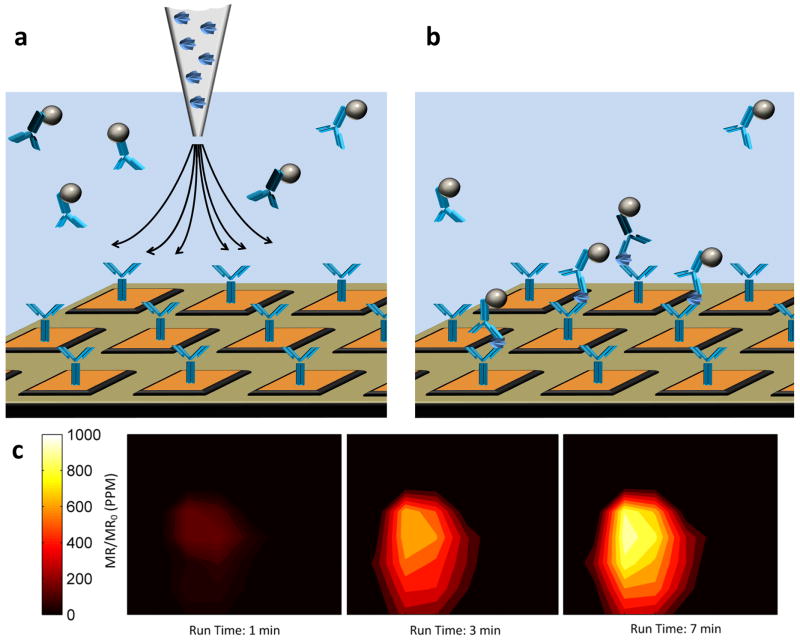
Our real-time binding assay and kinetic model has been extended to visualize protein binding events in both space (due to the high density of the array architecture) and time (due to the rapid and real-time readout). As an example, we first immobilized the same anti-CEA capture antibody across a sensor array. We then added MNP-anti CEA conjugates in solution to the sensor well. Upon delivery of the CEA antigen, we were able to monitor the movement of CEA protein across the sensor array (Fig. 4a). With this method, we can visualize the binding of soluble CEA protein to each sensor, which is spatially distributed in the array, by means of the MNP-anti-CEA antibody binding (Fig. 4b and Supplementary Fig. 6). In this manner, each sensor monitors the reaction zone above it, and thus we can investigate protein binding, protein transport, and protein dynamics with high spatial and temporal resolution--an important and unique development. Such techniques will be applied to monitor localized cell-cell communications via cellular protein secretome analysis in future works.
Figure 4. Visualization of spatiotemporal resolution of the sensor array.
a, Schematic depicting the GMR sensor array functionalized with monoclonal anti-CEA capture antibody (not to scale). The solution above the sensor array is comprised of magnetically labeled anti-CEA detection antibodies. The schematic includes a pipette tip containing a solution of CEA protein prior to injection. b, Once the CEA antigen is introduced into the solution above the sensor array, the transport of CEA antigen from the near center of the array radially was monitored in real-time as magnetically labeled detection antibodies bound to captured CEA protein, forming detectable sandwich structures. c, Visualization of CEA protein surface concentration at different times using a high density GMR sensor array. The units of the y-axis are presented in changes in MR normalized to the initial MR in parts per million (ppm).
Discussion and conclusions
We have presented a high-density, highly sensitive, real-time binding assay for quantifying protein binding kinetics and analyte transport at the surface of a biosensor array. Notably, we have also developed a novel analytical kinetics model that provides a precise and physically intuitive description of the dominant processes involved in labeled protein-protein interactions. We proved that the reduced rate of diffusion, that is consistent with the addition of nano-scale labels, can be exploited to derive generalizable kinetic binding models with analytical solutions. The combination of the GMR sensor technology and analytical model has enabled us to measure protein binding constants and quantify the number of proteins bound to a given sensor with unprecedented LOD. Furthermore, we have demonstrated the unique benefits of combining our GMR sensor array, kinetic binding assay, and analytical model to visualize protein transport in a reaction well.
The tools we have developed can be used broadly in basic science research for understanding receptor-ligand binding interactions involved in signal transduction in cell biology or for profiling the affinity of specific compounds of interest against an entire proteome on a high density array. In addition, applications to pharmacodynamics via receptor occupancy assays for drug development would open an entirely new and broad segment for our technology. MHC-peptide and T-cell receptor interactions could also be studied with exceptional speed and accuracy.
Further, the potential clinical applications of our method are vast, ranging from in-vitro clinical assay development52, to targeted molecular imaging for early cancer diagnostics53, to investigating drug on-target and off-target cross-reaction54 binding kinetics. In each case, the affinity and cross-reactivity of reagents and targeting probes are critical issues to clinical adoption and can be easily addressed with our platform. In short, our method represents a significant advance in this domain as it has the potential to provide fundamentally important solutions for both cutting-edge basic science research and clinical practice.
Materials and Methods
GMR nanosensor array architecture
The giant magnetoresistive (GMR) sensor used in our experiment has a bottom spin valve structure of the type: Si/Ta(5)/seed layer/IrMn(8)/CoFe(2)/Ru/(0.8)/CoFe(2)/Cu(2.3)/ CoFe(1.5)/Ta(3), all numbers in parenthesis are in nanometers. Each chip contains an array of GMR sensors, which are connected to peripheral bonding pads by a 300 nm thick Ta/Au/Ta lead. To protect the sensors and leads from corrosion, two passivation layers were deposited by ion beam sputtering: first, a thin passivation layer of SiO2(10 nm)/Si3N4(20 nm)/ SiO2(10nm) was deposited above all sensors and leads, excluding only the bonding pad area; then a thick passivation layer of SiO2(100 nm)/Si3N4(150 nm)/SiO2(100 nm) was deposited on top of the reference sensors and leads, but excluding the active sensors and bonding pad area. The magnetoresistive ratio is approximately 12% after patterning. The pinning direction of the spin valve is in-plane and perpendicular to the sensor strip. The easy axis of the free layer is set by the shape anisotropy to be parallel with the sensor strip. This configuration allows the GMR sensors to work at the most sensitive region of their MR transfer curves.
Due to the GMR effect, the resistance of the sensor changes with the orientation of the magnetization of the two magnetic layers, which are separated by a copper spacer layer:
| (3) |
Here R0 is the resistance under zero magnetic field, δRmax is the maximum resistance change and θ is the angle between the magnetization of the two magnetic layers. In the bottom spin valve structure, the magnetization of the bottom magnetic layer (pinned layer) is pinned to a fixed direction, while the magnetic orientation of the top magnetic layer (free layer) can freely rotate with the external magnetic field. As a result, the stray field from magnetic labels can change the orientation of the free layer magnetization and therefore change the resistance of the sensor.
Magnetic labels
The magnetic labels we used are commercial magnetic nanoparticles from Miltenyi Biotech Inc., referred to as “MACS”. Each MACS particle is a cluster of 10nm Fe2O3 nanoparticles held together by a matrix of dextran. Due to the small size of the Fe2O3 nanoparticles, the MACS particle is superparamagnetic, with a hydrodynamic diameter of ∼46 nm, and contains ∼10% magnetic material (wt/wt). MACS particles can be functionalized with specific affinity molecules corresponding to the analyte being studied. For EpCAM (CEA, VEGF) experiments, the MACS particles are functionalized with anti-EpCAM (CEA, VEGF) antibodies; for biotin-streptavidin experiments, the MACS particles are functionalized with streptavidin.
Surface chemistry
The sensor surface was first cleaned using acetone, methanol and isopropanol and subsequently exposed to oxygen plasma for three minutes. A 2% (w/v) polyallylamine solution in Milli-Q water was applied to the sensor for 5 minutes. The chips were then rinsed with Milli-Q water and baked at 150°C for 45 minutes. A 10% (w/v) solution of EDC and 10% (w/v) solution of NHS were then added to the sensor surface at room temperature for 1 hour, after which the sensor was rinsed and dried. Capture protein EpCAM (960-EP-050 from R&D Systems), CEA (4128-CM-050 from RD Systems) or VEGF (293-VE165 from R&D Systems) or capture antibody (antibody to EpCAM (ab20160 from Abcam or 960 from R&D), CEA (5910 from BiosPacific) or VEGF (ab69479 from Abcam) was robotically deposited (Scienion sciFlexarrayer from BioDot) over each sensor in 360 picoliter droplets 3 times (total volume of ∼1 nanoliter). In order to monitor reproducibility, from 3 to 8 sensors, randomly distributed across the GMR sensor array, were incubated with the same capture protein. The control sensors were immobilized in a similar fashion with either BSA at 1 mg/mL or a non-complementary antibody (typically anti-survivin antibody (H00000332-P01 from Novus Biologicals, LLC) at 500 μg/mL. Finally, the entire surface of the sensor array was blocked with 1 mg/mL of BSA in phosphate buffered saline for 30 minutes.
Kinetic Assay
After the sensor surface is functionalized with the appropriate capture protein, the GMR sensor array was placed in the test station. The BSA blocking buffer was washed away and a 50 μL solution of the magnetically labeled detection antibody (described above) was added to the reaction well. The GMR sensor array was monitored in real-time as the magnetically labeled detection antibodies bound to the corresponding surface-immobilized proteins. The binding curves, unique to each protein, were then plotted and the binding constants could be determined. An assay is typically run for approximately 5 minutes.
Supplementary Material
Acknowledgments
This work was supported in part by United States National Cancer Institute grants 1U54CA119367, 1U54CA143907, 1U54CA151459 and N44CM–2009-00011, United States National Science Foundation grant ECCS-0801385-000, the United States Defense Advanced Research Projects Agency/Navy Grant N00014–02-1–0807, Gates Foundation Grand Challenge Exploration Award and The National Semiconductor Corporation. R.S.G. acknowledges financial support from the Stanford Medical School Medical Scientist Training Program and a National Science Foundation graduate research fellowship. The authors thank Mark Hammer and Amit Bhattacharjee for editing the manuscript.
Footnotes
Author contributions: R.S.G. and S.X.W designed research; R.S.G. performed research; R.S.G., R.J.W., and S.X.W developed the model; R.S.G., L.X., S.H., R.J.W., D.A.H., S.J.O., H.Y. and S.X.W. contributed analytical tools; R.S.G., R.J.W. and S.X.W analyzed data; S.J.O., L.X., S.H., and S.X.W. designed the magnetic sensor arrays; R.S.G. and H.Y. developed the biochemistry; and R.S.G. and S.X.W. wrote the paper.
Supplementary information accompanies this paper at www.nature.com/naturenanotechnology.
Reprints and permission information is available online at http://npg.nature.com/reprintsandpermissions/.
Competing financial interests: Stanford University has licensed part of the magnetic bioassay chip technology contained in this publication to MagArray Inc., an early stage startup company in Silicon Valley. The authors S.X.W, H.Y., and S.J.O. hold financial interests in MagArray in the form of stock options.
References
- 1.Schena M, Shalon D, Davis RW, Brown PO. Quantitative Monitoring of Gene Expression Patterns with a Complementary DNA Microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 2.MacBeath G, Schreiber SL. Printing Proteins as Microarrays for High-Throughput Function Determination. Science. 2000;289:1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- 3.Zheng G, Patolsky F, Cui Y, Wang WU, Lieber CM. Multiplexed electrical detection of cancer markers with nanowire sensor arrays. Nature Biotech. 2005;23:1294–1301. doi: 10.1038/nbt1138. [DOI] [PubMed] [Google Scholar]
- 4.James LC, Tawfik DS. Structure and kinetics of a transient antibody binding intermediate reveal a kinetic discrimination mechanism in antigen recognition. Proc Natl Acad Sci USA. 2005;102:12730–12735. doi: 10.1073/pnas.0500909102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.LaBaer J, Ramachandran N. Protein microarrays as tools for functional proteomics. Curr Opin Chem Biol. 2005;9:14–19. doi: 10.1016/j.cbpa.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 6.Park J, et al. A highly sensitive and selective diagnostic assay based on virus nanoparticles. Nature Nanotech. 2009;4:259–264. doi: 10.1038/nnano.2009.38. [DOI] [PubMed] [Google Scholar]
- 7.Hudson PJ, Souriau C. Engineered antibodies. Nature Med. 2003;9:129–134. doi: 10.1038/nm0103-129. [DOI] [PubMed] [Google Scholar]
- 8.Schrama D, Reisfeld RA, Becker JC. Antibody targeted drugs as cancer therapeutics. Nature Rev Drug Discov. 2006;5:147–159. doi: 10.1038/nrd1957. [DOI] [PubMed] [Google Scholar]
- 9.Sinensky AK, Belcher AM. Label-free and high-resolution protein//DNA nanoarray analysis using Kelvin probe force microscopy. Nature Nanotech. 2007;2:653–659. doi: 10.1038/nnano.2007.293. [DOI] [PubMed] [Google Scholar]
- 10.Haab BB, Dunham MJ, Brown PO. Protein microarrays for highly parallel detection and quantitation of specific proteins and antibodies in complex solutions. Genome Biol. 2001;2:research0004.1–research0004.13. doi: 10.1186/gb-2001-2-2-research0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilson WD. Analyzing Biomolecular Interactions. Science. 2002;295:2103–2105. doi: 10.1126/science.295.5562.2103. [DOI] [PubMed] [Google Scholar]
- 12.Bornhop DJ, et al. Free-Solution, Label-Free Molecular Interactions Studied by Back-Scattering Interferometry. Science. 2007;317:1732–1736. doi: 10.1126/science.1146559. [DOI] [PubMed] [Google Scholar]
- 13.Stern E, et al. Label-free biomarker detection from whole blood. Nature Nanotech. 2010;5:138–142. doi: 10.1038/nnano.2009.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Squires TM, Messinger RJ, Manalis SR. Making it stick: convection, reaction and diffusion in surface-based biosensors. Nature Biotech. 2008;26:417–426. doi: 10.1038/nbt1388. [DOI] [PubMed] [Google Scholar]
- 15.Ramachandran N, et al. Self-Assembling Protein Microarrays. Science. 2004;305:86–90. doi: 10.1126/science.1097639. [DOI] [PubMed] [Google Scholar]
- 16.Patolsky F, et al. Electrical detection of single viruses. Proc Natl Acad Sci USA. 2004;101:14017–14022. doi: 10.1073/pnas.0406159101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patolsky F, Lieber CM. Nanowire nanosensors. Materials Today. 2005;8:20–28. [Google Scholar]
- 18.Berg H, Purcell E. Physics of chemoreception. Biophys J. 1977;20:193–219. doi: 10.1016/S0006-3495(77)85544-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berg OG, von Hippel PH. Diffusion-controlled macromolecular interactions. Annu Rev Biophys Biophys Chem. 1985;14:131–160. doi: 10.1146/annurev.bb.14.060185.001023. [DOI] [PubMed] [Google Scholar]
- 20.Stenberg M, Nygren H. Kinetics of antigen-antibody reactions at solid-liquid interfaces. J Immunol Methods. 1988;113:3–15. doi: 10.1016/0022-1759(88)90376-6. [DOI] [PubMed] [Google Scholar]
- 21.Waite BA, Stewart JD. An idealized dynamical model of simple diffusional interactions between macromolecules and between macromolecules and surfaces. Math Biosci. 1993;114:173–213. doi: 10.1016/0025-5564(93)90077-n. [DOI] [PubMed] [Google Scholar]
- 22.Sheehan PE, Whitman LJ. Detection Limits for Nanoscale Biosensors. Nano Lett. 2005;5:803–807. doi: 10.1021/nl050298x. [DOI] [PubMed] [Google Scholar]
- 23.Swift JL, Cramb DT. Nanoparticles as fluorescence labels: is size all that matters? Biophys J. 2008;95:865–876. doi: 10.1529/biophysj.107.127688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Röcker C, Pötzl M, Zhang F, Parak WJ, Nienhaus GU. A quantitative fluorescence study of protein monolayer formation on colloidal nanoparticles. Nature Nanotech. 2009;4:577–580. doi: 10.1038/nnano.2009.195. [DOI] [PubMed] [Google Scholar]
- 25.Osterfeld SJ, et al. Multiplex protein assays based on real-time magnetic nanotag sensing. Proc Natl Acad Sci USA. 2008;105:20637–20640. doi: 10.1073/pnas.0810822105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaster RS, et al. Matrix-insensitive protein assays push the limits of biosensors in medicine. Nature Med. 2009;15:1327–1332. doi: 10.1038/nm.2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaster RS, Hall DA, Wang SX. nanoLAB: An ultraportable, handheld diagnostic laboratory for global health. Lab Chip. 2011;11:950–956. doi: 10.1039/c0lc00534g. [DOI] [PubMed] [Google Scholar]
- 28.Baibich MN, et al. Giant Magnetoresistance of (001)Fe/(001)Cr Magnetic Superlattices. Phys Rev Lett. 1988;61:2472. doi: 10.1103/PhysRevLett.61.2472. [DOI] [PubMed] [Google Scholar]
- 29.Barnas J, Fuss A, Camley R, Grünberg P, Zinn W. Novel magnetoresistance effect in layered magnetic structures: Theory and experiment. Phys Rev B, Condensed Matter. 1990;42:8110–8120. doi: 10.1103/physrevb.42.8110. [DOI] [PubMed] [Google Scholar]
- 30.Prinz GA. Magnetoelectronics. Science. 1998;282:1660–1663. doi: 10.1126/science.282.5394.1660. [DOI] [PubMed] [Google Scholar]
- 31.Wolf SA, et al. Spintronics: A Spin-Based Electronics Vision for the Future. Science. 2001;294:1488–1495. doi: 10.1126/science.1065389. [DOI] [PubMed] [Google Scholar]
- 32.Mulvaney S, Myers K, Sheehan P, Whitman L. Attomolar protein detection in complex sample matrices with semi-homogeneous fluidic force discrimination assays. Biosens Bioelectron. 2009;24:1109–1115. doi: 10.1016/j.bios.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 33.Baselt DR, et al. A biosensor based on magnetoresistance technology. Biosens Bioelectron. 1998;13:731–9. doi: 10.1016/s0956-5663(98)00037-2. [DOI] [PubMed] [Google Scholar]
- 34.Sandhu A. Biosensing: New probes offer much faster results. Nature Nanotech. 2007;2:746–748. doi: 10.1038/nnano.2007.398. [DOI] [PubMed] [Google Scholar]
- 35.Koets M, van der Wijk T, van Eemeren J, van Amerongen A, Prins M. Rapid DNA multi-analyte immunoassay on a magneto-resistance biosensor. Biosens Bioelectron. 2009;24:1893–1898. doi: 10.1016/j.bios.2008.09.023. [DOI] [PubMed] [Google Scholar]
- 36.Xu L, et al. Giant magnetoresistive biochip for DNA detection and HPV genotyping. Biosens Bioelectron. 2008;24:99–103. doi: 10.1016/j.bios.2008.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koh AL, Sinclair R. TEM Studies of Iron Oxide Nanoparticles for Cell Labeling and Magnetic Separation. Technical Proceedings of the 2007 NSTI Nanotechnology Conference and Trade Show; pp. 101–104. [Google Scholar]
- 38.De Palma R, et al. Magnetic Particles as Labels in Bioassays: Interactions between a Biotinylated Gold Substrate and Streptavidin Magnetic Particles. J Phys Chem C. 2007;111:12227–12235. [Google Scholar]
- 39.Malmqvist M. Biospecific interaction analysis using biosensor technology. Nature. 1993;361:186–187. doi: 10.1038/361186a0. [DOI] [PubMed] [Google Scholar]
- 40.Katsamba PS, et al. Kinetic analysis of a high-affinity antibody/antigen interaction performed by multiple Biacore users. Anal Biochem. 2006;352:208–221. doi: 10.1016/j.ab.2006.01.034. [DOI] [PubMed] [Google Scholar]
- 41.Lausted C, Hu Z, Hood L. Quantitative Serum Proteomics from Surface Plasmon Resonance Imaging. Mol Cell Proteomics. 2008;7:2464–2474. doi: 10.1074/mcp.M800121-MCP200. [DOI] [PubMed] [Google Scholar]
- 42.Rich RL, Myszka DG. Higher-throughput, label-free, real-time molecular interaction analysis. Anal Biochem. 2007;361:1–6. doi: 10.1016/j.ab.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 43.Campbell CT, Kim G. SPR microscopy and its applications to high-throughput analyses of biomolecular binding events and their kinetics. Biomaterials. 2007;28:2380–2392. doi: 10.1016/j.biomaterials.2007.01.047. [DOI] [PubMed] [Google Scholar]
- 44.Wang S, Guanxiong Li. Advances in Giant Magnetoresistance Biosensors With Magnetic Nanoparticle Tags: Review and Outlook. IEEE Transactions on Magnetics. 2008;44:1687–1702. [Google Scholar]
- 45.Myszka DG, He X, Dembo M, Morton TA, Goldstein B. Extending the range of rate constants available from BIACORE: interpreting mass transport-influenced binding data. Biophys J. 1998;75:583–594. doi: 10.1016/S0006-3495(98)77549-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reichert JM, Valge-Archer VE. Development trends for monoclonal antibody cancer therapeutics. Nature Rev Drug Discov. 2007;6:349–356. doi: 10.1038/nrd2241. [DOI] [PubMed] [Google Scholar]
- 47.Willett CG, et al. Direct evidence that the VEGF-specific antibody bevacizumab has antivascular effects in human rectal cancer. Nature Med. 2004;10:145–147. doi: 10.1038/nm988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Srisa-Art M, Dyson EC, deMello AJ, Edel JB. Monitoring of Real-Time Streptavidin–Biotin Binding Kinetics Using Droplet Microfluidics. Anal Chem. 2008;80:7063–7067. doi: 10.1021/ac801199k. [DOI] [PubMed] [Google Scholar]
- 49.Barbosa MD, Chamberlain AK, Desjarlais JR. United States Patent: 7557190 - Optimized proteins that target Ep-CAM. 2009 http://patft.uspto.gov/netacgi/nph-Parser?Sect1=PTO2&Sect2=HITOFF&p=1&u=%2Fnetahtml%2FPTO%2Fsearch-bool.html&r=1&f=G&l=50&co1=AND&d=PTXT&s1=7557190.PN.&OS=PN/7557190&RS=PN/7557190.
- 50.Hefta LJF, Neumaier M, Shively JE. Kinetic and affinity constants of epitope specific anti-carcinoembryonic antigen (CEA) monoclonal antibodies for CEA and engineered CEA domain constructs. Immunotechnology. 1998;4:49–57. doi: 10.1016/s1380-2933(98)00004-9. [DOI] [PubMed] [Google Scholar]
- 51.Chen Y, et al. Selection and analysis of an optimized anti-VEGF antibody: crystal structure of an affinity-matured fab in complex with antigen. J Mol Bio. 1999;293:865–881. doi: 10.1006/jmbi.1999.3192. [DOI] [PubMed] [Google Scholar]
- 52.Anderson NL. The Clinical Plasma Proteome: A Survey of Clinical Assays for Proteins in Plasma and Serum. Clin Chem. 2010;56:177–185. doi: 10.1373/clinchem.2009.126706. [DOI] [PubMed] [Google Scholar]
- 53.Cai W, et al. Peptide-Labeled Near-Infrared Quantum Dots for Imaging Tumor Vasculature in Living Subjects. Nano Lett. 2006;6:669–676. doi: 10.1021/nl052405t. [DOI] [PubMed] [Google Scholar]
- 54.Gaster RS, Hall DA, Wang SX. Autoassembly Protein Arrays for Analyzing Antibody Cross-Reactivity. Nano Lett. doi: 10.1021/nl1026056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.