Abstract
BACKGROUND
It is thought that HLA antibodies might contribute to the pathogenesis of transfusion-related acute lung injury (TRALI). Many methods exist to detect HLA antibodies, including ELISA, flow cytometry, and multiplex bead-based assays, as well as the older lymphocytotoxicity assay. The sensitivity of each of these testing platforms varies, and it is not obvious how to compare HLA antibody results obtained on different platforms. This issue has become increasingly important as some blood centers have begun HLA antibody apheresis donor screening as a TRALI risk-reduction measure.
STUDY DESIGN AND METHODS
525 serum samples were selected from 7,841 donors in the Leukocyte Antibody Prevalence Study (LAPS) repository based on risk for the development of HLA antibodies, using the number of pregnancies as the risk factor. There were 81 males, and the pregnancy history of the 444 female donors was 0 (n=187), 1 (n=67), or 2+ pregnancies (n=190). Replicate frozen serum aliquots were sent to four different assay manufacturers blinded for HLA Ab detection using five different assays.
RESULTS
Among the assays with different manufacturer designated cutoffs, the flow cytometry and multiplex bead based-assays (Luminex) typically resulted in a larger proportion of HLA Ab positive samples compared with ELISA based assays. Capitalizing upon having quantitative results from five different assays on the same group of samples, latent variable analysis was used to derive a new set of cutoffs. These cutoffs, termed consensus cutoffs, yielded similar sensitivities across test platforms, thereby increasing concordance amongst assays. Assay agreement was higher in ever pregnant females than in males and never pregnant females.
CONCLUSION
Different assays resulted in varied positivity rates when the manufacturer’s suggested cutoffs were used, demonstrating that care needs to be taken when comparing clinical outcomes data generated using different HLA antibody assays and testing platforms. The method used here, involving latent variable analysis, presents one possible approach to calculating comparable cutoffs that result in broad agreement across assays with respect to positivity designation.
INTRODUCTION
Antibodies generated against human leukocyte antigens (HLA) have long been recognized in subjects who have been exposed to alloantigens, such as previously pregnant women 1 and transfusion recipients 2. These antibodies play a role in rejection of transplanted organs 3 and in development of refractoriness to platelet transfusions 4. It has been also been proposed that HLA antibodies in blood donors can cause transfusion related lung injury (TRALI), a leading cause of transfusion related mortality reported to the FDA in recent years 5,6. The cause of TRALI is an area of active research, with possible causes including soluble inflammatory mediators in transfused plasma 7, human neutrophil antigen (HNA) antibodies 8,9, as well as HLA antibodies 10. Blood banks have taken active steps to reduce TRALI incidence, including deferral of donors whose products were associated with TRALI cases and screening platelet and plasma apheresis donors for HLA antibodies 11
The classic method for detection of HLA antibodies is the lymphocytotoxicity assay, which relies on lysis of cells expressing the HLA antigen of interest if the corresponding matching HLA antibody is present in the test serum 12,13. More recently developed techniques with increased sensitivity include ELISA 14, multiplex bead-based assays using the Luminex platform 15, and flow cytometry assays 16. Each of the assay formats has its relative strengths and weaknesses, such as assay throughput, sensitivity, dynamic range and cost. All three assay types have been developed as potential testing modalities for blood bank use in screening donors as a TRALI risk reduction measure. However, while many blood centers have already, or are in the process of implementing HLA antibody screening of donors at risk for alloimmunization, there is no industry standard as to which assay platform should be used. Given that results are being generated with different assays, it would be useful to know how these results can be compared. The current analysis utilized a well characterized panel of blood donor specimens to compare a number of commercially available and prototype HLA antibody detection assays. Results were analyzed using manufacturers’ suggested cutoffs and using a “consensus” cutoff designed to maximize agreement amongst assays.
MATERIALS AND METHODS
Subject and sample selection
LAPS (Leukocyte Antibody Prevalence Study) was a prospective cross-sectional six-center study conducted by the Retrovirus Epidemiology Donor Study – II (REDS-II) program of the National Heart, Lung, and Blood Institute. All six REDS-II blood centers participated in the study. These included: American Red Cross New England region (Dedham, MA), American Red Cross Southern Region (Douglasville, GA), BloodCenter of Wisconsin (Milwaukee, WI), Blood Centers of the Pacific (San Francisco, CA), Hoxworth Blood Center/University of Cincinnati Academic Health Center (Cincinnati, OH) and the Institute for Transfusion Medicine (Pittsburgh, PA). The REDS-II Coordinating Center is Westat (Rockville, MD) and the REDS-II central laboratory is Blood Systems Research Institute (San Francisco, CA). LAPS enrollment and study design have been previously described in detail (Triulzi et al 2009). Donors consenting to the study provided a blood sample for HLA Class I and II antibody testing and a detailed history of pregnancy and transfusion. A total of 8171 (6011 females, 2160 males) donors were enrolled. Females and transfused males were intentionally oversampled.
We used a stratified random design to select 525 REDS-II donors and their serum samples from the LAPS repository of 7,841 donors previously tested for HLA antibodies 17. Donors were selected based on the availability of adequate serum samples and risk for the development of HLA-antibodies, specifically using gender, transfusion status, and the number of pregnancies as strata. Blinded serum samples from 268 never pregnant donors (81 non-transfused males and 187 nulliparous women) and from 257 ever pregnant donors (67 women with one pregnancy and 190 women with more than one pregnancy) were selected. A total of 525 donors was selected due to budgetary constraints. There were only 81 non-transfused males and 67 women with one pregnancy that had adequate serum samples for this study, and thus, all were selected. Roughly equal numbers of donors within the strata of nulliparous women and women with more than one pregnancy were then randomly selected. The samples were sent to the sent to the following manufacturers for determination of the presence of HLA Class I or Class II antibodies: GTI Diagnostics (Waukesha, WI) for analysis using the DonorScreen, QuickStep ELISA assay, One Lambda (Canoga Park, CA) for analysis using the Lambda Antigen Tray, LATM, ELISA assay, Tepnel Lifecodes (Stamford, CT) for analysis using the Lifescreen Luminex assay, and Transfusion & Transplantation Technologies (3Ti; Atlanta, GA) for analysis using the Aegis System flow cytometry assay. Replicate serum aliquots with the same freeze-thaw history were used, with the exception of the samples for 3Ti testing, which had undergone one additional freeze thaw cycle. Previous HLA antibody test results had been obtained at the REDS-II Central Laboratory using the One Lambda LabScreen mixed Luminex assay.
HLA antibody assays
GTI Diagnostics QuickStep (ELISA)
Samples were assayed by GTI using the QuickStep assay according to the product insert. GTI determined a sample to be positive for Class I or Class II, if the Optical Density (O.D.) value was greater than or equal to the cutoff, where the cutoff was calculated as 2x the mean of the O.D. values of the negative control, as described in the product insert. Normalized O.D. values were also calculated to account for assay run variability by expressing the results for a given sample as a ratio of the sample to cutoff O.D. values (sample O.D. value/cutoff O.D. value).
One Lambda LATM20 (ELISA)
Samples were assayed by One Lambda using the LATM20 assay according to the product insert. As recommended in the product insert, samples were determined to be positive for Class I if their averaged values obtained in the Class I wells were greater than the calculated 20% cutoff (calculated according to the LATM product insert); if the values were less than or equal to the 20% cutoff, they were determined to be negative. Class II positive and negative determinations were made in a similar fashion, using values obtained from the wells containing Class II antigens. Normalized test values were also calculated to account for assay run variability by expressing values for each sample (Average Test Value – Average Negative Control) as a percent of the total range (Average Positive controls – Average Negative controls).
One Lambda LABScreen Mixed (Luminex)
Previously, LAPS samples were screened for the presence of HLA antibodies using the One Lambda LABScreen Mixed (LSM12) assay on the Luminex flow cytometry platform 17. The normalized background (NBG) ratio cutoff value of 2.2 was used for both Class I and Class II assays because at the time of this study, NBG ratios of 2.2 were the manufacturer’s suggested cutoffs for the organ transplantation field.
Tepnel Lifescreen (Luminex)
Samples were assayed by Tepnel using the Lifescreen Luminex assay according to the product insert. Tepnel determined a sample to be positive for Class I or Class II, by first calculating three adjusted MFI (median fluorescence index) ratios for each bead as described in the product insert, then determining whether at least one of the class I HLA beads or class II HLA beads was positive. Normalized MFI values for each sample were recorded by taking the maximum adjusted MFI ratio according to the following algorithm for Class I: maximum MFI ratio (max of Adj. Ratios for Bead 1 or median of Adj. Ratios for each of Beads 2 through 7). For Class II: maximum MFI ratio (max of Adj. Ratios for Bead 1 or median of Adj. Ratios for each of Beads 2 through 5).
3Ti Aegis (Flow cytometry)
Samples were assayed by 3Ti using the Aegis assay (One Lambda Flow PRA reagents) according to the manufacturer’s instructions. 3Ti determined a sample to be positive for Class I or Class II if the percent of beads was above a calculated cutoff (2 × negative control); the sample was determined to be negative if the percentage of beads were below the negative cutoff (0.75 × negative control). Results from samples with values in-between the cutoffs were examined manually to make the final call of positive or negative. The reported percentage of gated cells was used as the normalized value.
Derivation of consensus cutoffs
Taking advantage of the fact that we had five measures for each sample, we conducted a statistical approach called latent variable analysis 18, which is particularly well suited for situations where there are multiple measures of a variable which is latent. We describe the HLA antibodies as latent variables because there is no direct means to observe the amount, or potency, of HLA antibodies. Latent variable analysis was used to derive a set of cutoffs (termed the consensus cutoffs) for the HLA Class I and Class II assays that maximized concordance when we used the 99th percentile cutoffs as HLA positive. It is important to note that latent variable analysis is most effective with three or more measures (analysis with one or two measures is possible with certain parameters assumptions). A separate analysis was performed for HLA class I and class II using the same methodology and the statistical software LISREL (from Scientific Software International, Lincolnwood, IL). The normalized values were analyzed using latent variable analysis, modeling a single latent variable for each class. One option in the LISREL software, as well as with other statistical software, is to produce a latent variable score for each observation 19. The latent variable score is an estimated value, or realization, of the unobserved latent variable. The latent variable score is calculated as a linear combination of the observed (normalized) values which have been centered. A univariate analysis was performed on the latent variable score for each class using data from subjects who were never pregnant (i.e. males and never-pregnant females). The 99th percentiles from the univariate analyses were calculated as cutoffs. The latent variable score at a given percentile was used to calculate consensus cutoffs for each assay using coefficients derived in the latent variables analysis and the assay means (to adjust for the centering), where
yi: Consensus cutoff for assay i
X̄i: Sample mean of assay i
λi: Coefficient from latent variable analysis
z: Latent variable score at selected percentile
The coefficients derived in the latent variable analysis are analogous to a regression coefficient that would be derived if it were possible to perform a regression analysis of the observed (normalized) value regressed on the latent variable.
Interassay agreement
In order to determine the extent of agreement between assays with respect to a positive or negative designation (i.e. concordance), the Kendall Tau statistic was calculated using different combinations of cutoffs (SAS 9.1.3 (2004) SAS Institute Inc., Cary NC). The Kendall Tau statistics were calculated among the manufacturer’s designations of positive or negative, as well as the designations of positive or negative as determined by the consensus cutoffs established via the latent variable analyses. The Pearson procedure (SAS 9.1.3) was used to calculate correlation coefficients and p-values as a measure of correlation among assay values and associated significance levels.
RESULTS
HLA Class I and Class II antibody positivity rates varied depending upon the assay
One of the unique features of this study is the fact that 525 samples were analyzed by five different HLA antibody assays by the assay’s manufacturer. The assay’s manufacturer provided either a designation of positivity, or a suggested cutoff from which we determined positivity status. The positivity rates varied greatly across assays when using the manufacturer’s cutoffs (Table 1) as one might infer from the absence of a gold standard. The two Luminex based assays (Tepnel Lifescreen and One Lambda LABScreen Mixed) and the flow cytometry based assay (3Ti Aegis) resulted in the highest proportions of positive samples in the never pregnant group (i.e. males and never-pregnant females). The One Lambda LABScreen Mixed resulted in 29.1%, Tepnel Lifescreen 13.1%, and 3Ti Aegis 6.3% of all never pregnant donors being designated as having any HLA antibody. The two ELISA assays resulted in only 3% (One Lambda LATM20) and 1.9% (GTI QuickStep) of all never pregnant donors having any HLA antibody. A similar trend with varying positivity rates across the assays was observed in the ever pregnant group. However, the positivity rates were higher in the ever pregnant group compared with the never pregnant group. The two Luminex assays resulted in over one-third of the ever pregnant group having any HLA antibody (52.5% for One Lambda LABScreen Mixed and 37.9% for Tepnel Lifescreen). The flow cytometry assay resulted in slightly fewer positive samples (29.3% of the ever pregnant group had any HLA antibody), and the two ELISAs resulted in a 19 – 20% positivity rate in the ever pregnant group.
Table 1.
Proportion of samples positive using manufacturers’ designations or suggested assay cutoffs
| Never pregnant (n=268)a | Ever pregnant (n=257) a | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Assay | Method | Class I and II positive | Class I only positive | Class II only positive | Any HLA Ab positive | Class I and II positive | Class I only positive | Class II only positive | Any HLA Ab positive |
| GTI QuickStep | ELISA | 1 (0.4%) | 2 (0.7%) | 2 (0.7%) | 5 (1.9%) | 14 (5.4%) | 24 (9.3%) | 11 (4.3%) | 49 (19.1%) |
| One Lambda LATM20 | ELISA | 1 (0.4%) | 6 (2.2%) | 1 (0.4%) | 8 (3.0%) | 11 (4.3%) | 22 (8.6%) | 17 (6.6%) | 50 (19.5%) |
| 3Ti Aegis | Flow cytometry | 1 (0.4%) | 14 (5.2%) | 2 (0.7%) | 17 (6.3%) | 29 (11.3%) | 27 (10.5%) | 19 (7.4%) | 75 (29.3%) |
| Tepnel Lifescreen | Luminex | 3 (1.2%) | 9 (3.5%) | 22 (8.5%) | 34 (13.1%) | 39 (15.7%) | 30 (12.1%) | 25 (10.1%) | 94 (37.9%) |
| One Lambda LABScreen Mixedb | Luminex | 20 (7.5%) | 39 (14.6%) | 19 (7.1%) | 78 (29.1%) | 64 (19.2%) | 54 (16.2%) | 17 (5.1%) | 135 (52.5%) |
Samples missing HLA antibody screen data: GTI class I and II, never pregnant = 1; Tepnel class I and II, ever pregnant= 9 (high background signal was obtained); 3Ti class I and II, ever pregnant= 1 (QNS).
NBG ratio of 2.2 was the manufacturer’s suggested cutoffs for the organ transplantation field.
Consensus cutoffs were derived which resulted in more consistent proportions of positive samples across the five assays
Using latent variable analysis, we established consensus cutoffs at the 99th percentile of the never pregnant donors. Table 2 gives the equivalent assay cutoff values at 99th percentiles based on normalized values, the manufacturers’ cutoff values based on normalized values, and the respective positivity rates for each assay at each of the analyzed cutoff values per assay among ever pregnant and never pregnant donors. The 99th percentile cutoffs resulted in more consistency among positivity rates in ever pregnant females compared with the manufacturers’ designations, as exemplified in Figure 1. Among the ever pregnant females, the positivity rates were between 22 and 26% for any HLA antibody when the 99th percentile cutoffs were implemented.
Table 2.
HLA antibody Class I and II positivity at selected consensus cutoffs and manufacturers’ cutoffs
| 99th percentile Consensus Cutoffsa | Manufacturer’s Designations | ||||||
|---|---|---|---|---|---|---|---|
| Assay (Assay Methodology) | Cutoff values | % never pregnant above cutoff | % ever pregnant above cutoff | Cutoff values | % never pregnant above cutoff | % ever pregnant above cutoff | |
| Class I (Class I only, or Class I and II) | GTI QuickStep (ELISA) | 0.8 | 2.2% | 17.2% | 1.0 | 1.1% | 14.8% |
| One Lambda LATM20 (ELISA) | 12.5 | 3.7% | 19.5% | N/A b | 2.6% | 12.8% | |
| 3Ti Aegis (Flow cytometry) | 9.3 | 3.4% | 16.0% | N/A b | 5.6% | 21.9% | |
| Tepnel Lifescreen (Luminex) | 7.6 | 0.4% | 16.3% | N/A b | 4.6% | 27.8% | |
| One Lambda LABScreen Mixed (Luminex) | 17.8 | 0.4% | 16.3% | 2.2c | 22% | 45.9% | |
| Class II (Class II only, or Class I and II) | GTI QuickStep (ELISA) | 0.7 | 4.1% | 14.5% | 1.0 | 1.1% | 9.7% |
| One Lambda LATM20 (ELISA) | 13.3 | 2.2% | 14.8% | N/A b | 0.7% | 10.9% | |
| 3Ti Aegis (Flow cytometry) | 10.1 | 0.4% | 17.5% | N/A b | 1.1% | 18.8% | |
| Tepnel Lifescreen (Luminex) | 8.7 | 0.8% | 14.0% | N/A b | 9.7% | 25.8% | |
| One Lambda LABScreen Mixed (Luminex) | 16.2 | 0.7% | 13.2% | 2.2c | 14.6% | 31.5% | |
| Any HLA Ab (Class I and/or Class II) | GTI QuickStep (ELISA) | See Above | 5.6% | 24.6% | See Above | 1.9% | 19.1% |
| One Lambda LATM20 (ELISA) | 5.2% | 25.7% | 3.0% | 19.5% | |||
| 3Ti Aegis (Flow cytometry) | 3.4% | 25.3% | 6.3% | 29.3% | |||
| Tepnel Lifescreen (Luminex) | 1.2% | 22.6% | 13.1% | 37.9% | |||
| One Lambda LABScreen Mixed (Luminex) | 1.1% | 21.8% | 29.1% | 52.5% | |||
Percentage of never pregnant donors tested that would fall below the consensus cutoffs, where the 99th percentile consensus cutoff means that approximately 99% of the never pregnant donors would be designated as negative using the indicated cutoff values. The 99th percentile cutoff was selected for the concordance analyses in this study based on the assumption that 1% of never pregnant donations (from males and never pregnant females) would be anticipated to have naturally occurring HLA antibodies, which is consistent with what was detected in the entire LAPS cohort, using the One Lambda Luminex screening assay 17.
No manufacturer’s cutoff value was provided for 3Ti Aegis or Tepnel Lifescreen assays because the algorithm that was used to determine whether a sample was positive or negative for class I and/or II HLA antibodies was more complicated than simply applying a cutoff. No manufacturer’s cutoff value was provided for One Lambda LATM20 because the adjusted 20% cutoff values were determined on a plate-by-plate basis.
NBG ratio of 2.2 was the manufacturer’s suggested cutoffs for the organ transplantation field.
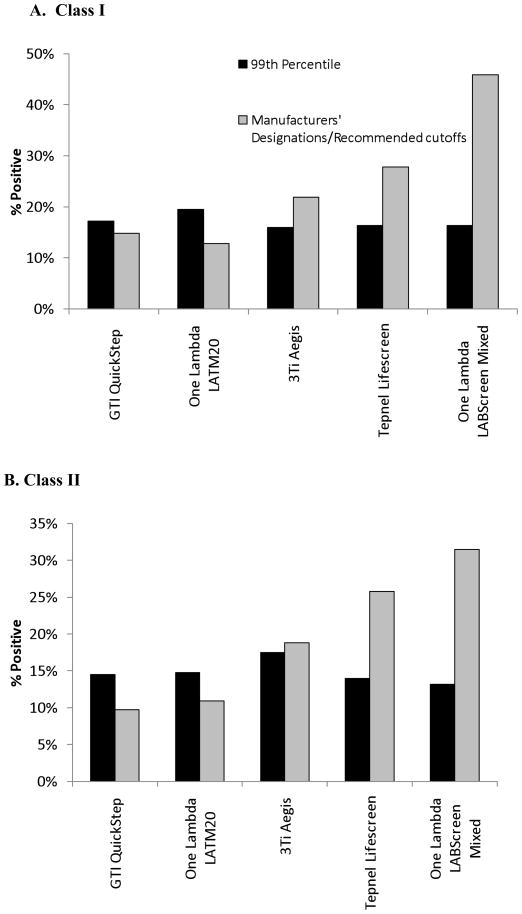
Fig. 1. Proportion of HLA Class I (panel A) and Class II (panel B) positive ever pregnant donors using three different designations of positivity.
Shown here is a graphical representation of the proportion of assayed samples determined to be positive using the 99th percentile consensus cutoff (black bars) and the manufacturer’s designation (white bars). The 99th percentile cutoffs give more consistent proportions of positive samples among the five assays for both Class I and II.
Subsequent analyses for concordance and correlations analyses were limited to the 99th percentile cutoff values for two reasons: 1) prior research suggested that approximately 1% of never pregnant donors would have HLA antibodies 20 and 2) use of the 99th percentile for consensus cutoffs resulted in more consistency among the ever pregnant samples (Figure 1). One explanation for this may be that the precision of the assays increases with increasing levels of HLA antibodies. This claim is supported below by the concordance and correlation analyses (Tables 3 and 4) showing higher correlation among assays when comparing groups with higher levels of HLA antibodies relative to groups with lower levels of HLA antibodies.
Table 3.
Concordance among the five HLA antibody assays (using 99th percentile consensus cutoffs)
| Class I | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Never pregnant (n=268) | Ever pregnant (n=257) | |||||||||
| GTI QuickStep | One Lambda LABScreen Mixed | One Lambda LATM20 | Tepnel Lifescreen | 3Ti Aegis | GTI QuickStep | One Lambda LABScreen Mixed | One Lambda LATM20 | Tepnel Lifescreen | 3Ti Aegis | |
| GTI QuickStep | N/A | 0.40* | 0.10 | 0.40* | 0.11 | N/A | 0.80* | 0.64* | 0.69* | 0.70* |
| One Lambda LABScreen Mixed | N/A | 0.31* | 1.00* | 0.33* | N/A | 0.77* | 0.83* | 0.84* | ||
| One Lambda LATM20 | N/A | 0.31* | 0.40* | N/A | 0.63* | 0.67* | ||||
| Tepnel Lifescreen | N/A | 0.33* | N/A | 0.73* | ||||||
| 3Ti Aegis | N/A | N/A | ||||||||
| Class II | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Never pregnant (n=268) | Ever pregnant (n=257) | |||||||||
| GTI QuickStep | One Lambda LABScreen Mixed | One Lambda LATM20 | Tepnel Lifescreen | 3Ti Aegis | GTI QuickStep | One Lambda LABScreen Mixed | One Lambda LATM20 | Tepnel Lifescreen | 3Ti Aegis | |
| GTI QuickStep | N/A | 0.18 | 0.32* | −0.02 | −0.01 | N/A | 0.78* | 0.72* | 0.78* | 0.73* |
| One Lambda LABScreen Mixed | N/A | 0.57* | 0.50* | −0.01 | N/A | 0.87* | 0.90* | 0.79* | ||
| One Lambda LATM20 | N/A | 0.28* | 0.40* | N/A | 0.78* | 0.76* | ||||
| Tepnel Lifescreen | N/A | −0.01 | N/A | 0.85* | ||||||
| 3Ti Aegis | N/A | N/A | ||||||||
Kendall’s Tau coefficients are reported in the tables.
Concordance between the two assays was statistically significant at the p<0.0001 level.
Table 4.
Correlation coefficients among the five HLA antibody assays
| Class I | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Never pregnant (n=268) | Ever pregnant (n=257) | |||||||||
| GTI QuickStep | One Lambda LABScreen Mixed | One Lambda LATM20 | Tepnel Lifescreen | 3Ti Aegis | GTI QuickStep | One Lambda LABScreen Mixed | One Lambda LATM20 | Tepnel Lifescreen | 3Ti Aegis | |
| GTI QuickStep | N/A | 0.44* | 0.30* | 0.66* | 0.57* | N/A | 0.85* | 0.86* | 0.79* | 0.77* |
| One Lambda LABScreen Mixed | N/A | 0.32* | 0.48* | 0.39* | N/A | 0.89* | 0.78* | 0.87* | ||
| One Lambda LATM20 | N/A | 0.31* | 0.47* | N/A | 0.76* | 0.79* | ||||
| Tepnel Lifescreen | N/A | 0.55* | N/A | 0.78* | ||||||
| 3Ti Aegis | N/A | N/A | ||||||||
| Class II | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Never pregnant (n=268) | Ever pregnant (n=257) | |||||||||
| GTI QuickStep | One Lambda LABScreen Mixed | One Lambda LATM20 | Tepnel Lifescreen | 3Ti Aegis | GTI QuickStep | One Lambda LABScreen Mixed | One Lambda LATM20 | Tepnel Lifescreen | 3Ti Aegis | |
| GTI QuickStep | N/A | 0.09 | 0.40* | 0.16 | 0.16 | N/A | 0.89* | 0.89* | 0.89* | 0.76* |
| One Lambda LABScreen Mixed | N/A | 0.21 | 0.41* | 0.19 | N/A | 0.90* | 0.90* | 0.82* | ||
| One Lambda LATM20 | N/A | 0.18 | 0.26* | N/A | 0.89* | 0.84* | ||||
| Tepnel Lifescreen | N/A | 0.25* | N/A | 0.80* | ||||||
| 3Ti Aegis | N/A | N/A | ||||||||
Pearson correlation coefficients are reported in the tables.
Concordance between the two assays was statistically significant at the p<0.0001 level.
Concordance of assays was higher with the ever pregnant donors
As a statistical measure of the extent to which any two assays agreed on positivity status, we used the Kendall Tau statistic to measure concordance when the 99th percentile consensus cutoff was implemented (Table 3). This statistic is a measure of correlation between binary variables, with low values between 0 and 1 indicating low concordance and high values indicating high concordance (negative values indicate discordance). Concordance estimates for Class I antibodies in the never pregnant donors ranged from 0.10 (One Lambda LATM versus GTI) to 0.40 (GTI versus One Lambda LabScreen and Tepnel; One Lambda LATM20 versus 3Ti), with the exception of One Lambda LabScreen versus Tepnel, which yielded a concordance estimate of 1.0. The latter observation indicated that for never pregnant donors, both the One Lambda LabScreen and Tepnel assays made the same designations of whether a sample was positive or negative. Concordance estimates for Class II antibodies in the non-alloexposed donors ranged from negative 0.01 to 0.57 (One Lambda LabScreen versus One Lambda LATM20). Overall, the low concordances observed within the never pregnant samples indicates that there was wide variability in designating a sample as positive or negative when the expectation of finding a true positive was low (with the exception being One Lambda LabScreen and Tepnel Class I assays).
In the ever pregnant donors, the Class I antibody concordance estimates range from 0.63 (One Lambda LATM versus Tepnel) to 0.84 (One Lambda LabScreen versus 3Ti Aegis). Class II antibody concordance estimates were higher, ranging from 0.72 (GTI versus One Lambda LATM20) to 0.90 (One Lambda LabScreen versus Tepnel). This indicates that all of the assays performed similarly in terms of designating whether a sample was positive or negative when restricted to the ever pregnant group. Therefore, it seems that in a population where HLA antibodies are anticipated to be present, all of the assays performed similarly provided that the consensus cutoff was used.
Correlation of assay values was higher with the ever pregnant donors
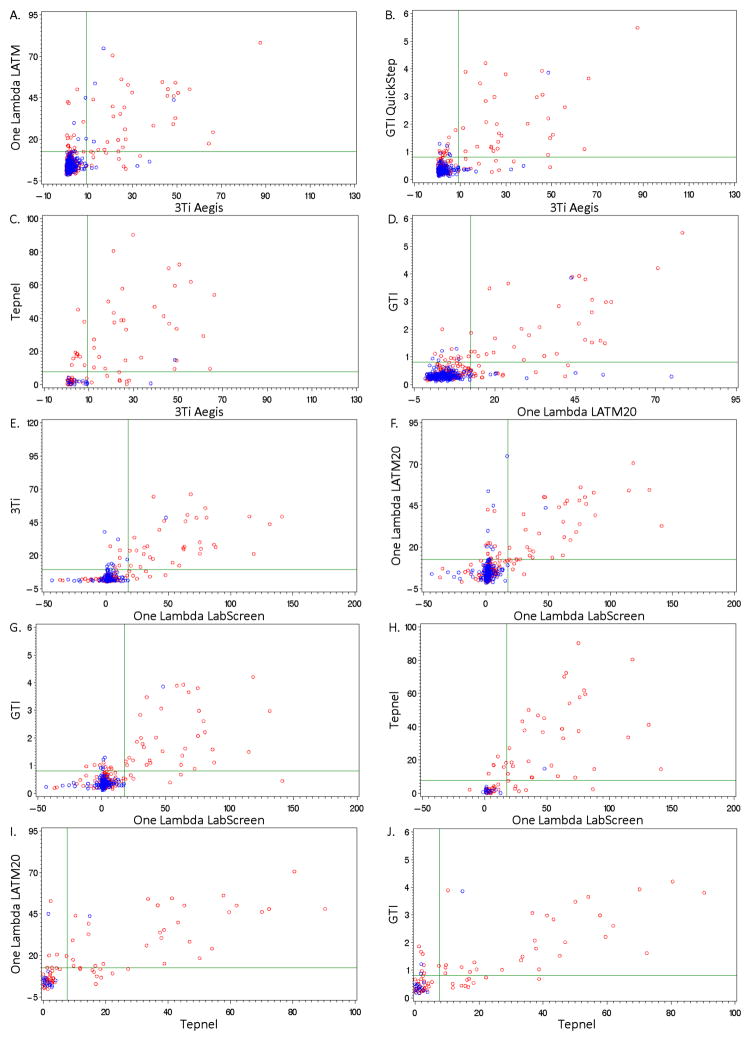
In addition to measuring whether assays agreed using a consensus cutoff, we measured the correlation of normalized values (see Methods for normalized value derivations) amongst assays in the ever pregnant sample and never pregnant sample. An intuitive way to analyze the data is to examine the scatter plots for Class I in Figure 2 (Class II data not shown because of space limitations) which show all pairs of assays, the 99th percentile consensus cutoffs, and classification of pregnancy history. As a statistical measure of how well the normalized assay values correlated with each other, we calculated the Pearson correlation coefficients (Table 4). Correlation coefficients for Class I antibody assays ranged from 0.31 (Tepnel versus GTI) to 0.67 (One Lambda LATM versus GTI) in the never pregnant donors. In the ever pregnant donors, the Class I correlation coefficients were all above 0.70. The highest correlation coefficients for Class I values in the ever pregnant donors were between the two One Lambda assays. The lowest correlation coefficient was between the One Lambda LATM20 ELISA assay and the Tepnel Luminex assay.
Fig. 2. Normalized values obtained from the five assays for Class I antibodies are higher for ever pregnant donors.
Red circles, ever pregnant donors; blue circles, never pregnant donors; green horizontal and vertical lines, 99th percentile consensus cutoffs for the given assay.
Correlations of Class II values were similar to those for Class I (Table 4). In the never pregnant donors, the correlation coefficients ranged from 0.01 (One Lambda LATM versus GTI and 3Ti) to 0.66 (One Lambda LabScreen versus 3Ti). As with Class I, the Class II correlation coefficients were higher in the ever pregnant donors (all were above 0.76). These findings suggest that at lower assay values, there may be less accuracy in the assays. However, at the higher levels of the assay, such as obtained with the ever pregnant donors, the assays likely produce more precise estimates.
Positivity rates using One Lambda LABScreen Mixed cutoffs reported in the literature and their relation to other assay cutoffs
One assay that is currently used in the field by several blood bank laboratories is the One Lambda LABScreen Mixed 21. The product insert for this assay recommends that HLA Ab screening laboratories establish their own cutoffs. As such, cutoffs implemented by various laboratories vary. In order to relate the data from this manuscript to the broader scientific community, we reanalyzed the positivity rates from One Lambda LABScreen Mixed in the subset of ever pregnant donors using a variety of different NBG cutoffs and report the corresponding cutoffs for the other assays (Table 5); values greater than the cutoffs would be considered positive. For example, using the highest One Lambda LABScreen assay cutoffs reportedly used by others (Class I, 59.3; Class II, 27.5) 21, we estimate positivity rates of 7.4% Class I and 11.3% Class II positive ever pregnant females. These rates correspond to estimated cutoffs of 2.1 for Class I and 0.9 for Class II in the GTI assay.
Table 5.
Equivalent Cutoffs by Assay
| Corresponding Cutoffs* | ||||||||
|---|---|---|---|---|---|---|---|---|
| Class I (Class I only or Class I and II) | One Lambda LABScreen Mixed NBG cutoff | 2.2a | 10.0 b | 12.0 c | 14.0 d | 22.9 c | 30.0 b | 59.3 b |
| GTI QuickStep cutoff | 0.4 | 0.6 | 0.7 | 0.7 | 1.0 | 1.2 | 2.1 | |
| One Lambda LATM20 | 5.3 | 8.9 | 9.8 | 10.7 | 14.8 | 18.1 | 31.5 | |
| Tepnel Lifescreen | −0.3 | 3.6 | 4.7 | 5.7 | 10.2 | 13.8 | 28.8 | |
| 3Ti Aegis | 3.1 | 6.2 | 7.0 | 7.8 | 11.4 | 14.2 | 25.9 | |
| % of Ever Pregnant Females Positive for HLA Ab | 44.7% e | 19.8% e | 19.5% e | 17.5% e | 15.2% e | 12.5% e | 7.4% e | |
| Class II (Class II only or Class I and II) | One Lambda LABScreen Mixed NBG cutoff | 2.2a | 9.0 c, d | 10.0 b | 15.0 b | 16 c | 27.5 b | |
| GTI QuickStep cutoff | 0.4 | 0.5 | 0.6 | 0.7 | 0.7 | 0.9 | ||
| One Lambda LATM20 | 5.3 | 9.2 | 9.8 | 12.6 | 13.2 | 19.7 | ||
| Tepnel Lifescreen | 0.4 | 4.4 | 5.0 | 8.0 | 8.6 | 15.4 | ||
| 3Ti Aegis | 3.4 | 6.6 | 7.1 | 9.5 | 10.0 | 15.5 | ||
| % of Ever Pregnant Females Positive for HLA Ab | 28.0% e | 16.3% e | 16.3% e | 14.4% e | 14.4% e | 11.3% e | ||
The data presented show cutoffs that are roughly equivalent to each other in terms of positivity rates in ever pregnant donors. The data is organized based on increasing One Lambda LABScreen Mixed NBG cutoff values that are currently in use by laboratories across the U.S.
Manufacturer’s suggested cutoff for organ transplantation;
Reported cutoff used by blood bank laboratories 21;
Cutoff used by Vassallo et al. 25;
Percent positive may differ from that reported in Table 2 as calculations are based on the latent variable score and not observed values.
DISCUSSION
As infectious risks from transfusion in the developed world have decreased, the threat posed by non-infectious transfusion complications has gained increasing attention. In particular, TRALI risk-reduction procedures have been implemented at US blood banks, and in many cases these procedures involve testing donors at risk for alloimmunization for the presence of HLA antibodies.
A number of different assay platforms are available for HLA antibody testing, and not all blood collection centers will opt to use the same platform. The ability to properly compare results obtained across testing platforms will be key to understanding future studies of TRALI and the role HLA antibodies might play. Our analysis suggests that there is a high level of concordance among HLA antibody testing platforms when comparable cutoffs are used. Furthermore, concordance is higher for samples obtained from subjects with a history of pregnancy and with samples containing higher levels of HLA antibodies.
The flow cytometry and multi-analyte bead based assays yielded higher positivity rates overall than the ELISAs when using the manufacturer’s specifications for assessing positivity status, which is consistent with prior work 22–24. While some assays yielded a higher proportion of positive designations, concordance amongst assays could be increased by adjusting the cutoffs. This implies that while samples will not have a one-to-one concordance using different HLA antibody testing platforms, one can better compare results obtained using different platforms if the data are analyzed using a derived consensus cutoff.
The positivity rates for any HLA antibody in ever pregnant females (i.e. exposed females) using our 99th percentile consensus cutoffs were between 22% and 26%. This is consistent with our previously reported positivity rate of 24% for any HLA antibody in the cohort of LAPS all ever pregnant females in using the One Lambda LABScreen Mixed assay 17. Additionally, the rates we observed are consistent with the 21% positivity rates obtained with One Lambda LABScreen and the GTI Quick Step assays by Vassallo et al. in a cohort of American Red Cross ever pregnant female donors 25. While the positivity rates are consistent with previous reports, the cutoff values we implemented (99th percentile consensus cutoffs) are slightly different from those previous analyses. One reason for this difference may be lot-to-lot variation in the assays.
A comparison of assays requires consideration of several caveats. First, the concordance amongst assays depends on the pre-test probability of obtaining a positive result. Screening a low-risk population for HLA antibodies would yield lower agreement amongst assays, so it is important in any analysis to take into account the demographic history of test subjects when evaluating results of HLA antibody testing. “Natural” HLA antibodies, likely induced by environmental antigens, have been described in never pregnant populations 26, and while our data suggest concordance is low among never pregnant donors, it is not clear how the concordance of different HLA antibody assays would be in a population with “natural” HLA antibodies. A second major limitation of comparing results across different testing platforms is that assay sensitivity may change from lot-to-lot, so the consensus cutoffs derived here may not apply to newer lots of commercial reagents (e.g. the bead format for LSM12 was recently changed). A third limitation is that using the manufacturer’s recommended designations of positivity may not be sufficient. When the manufacturers’ recommended designations of positivity were analyzed, there was a wide range of positivity observed amongst the assays. This observation indicates that comparing results among assays is difficult. If different assays are utilized, it is likely that varying outcomes will result. Therefore, in the blood banking setting, one needs to make sure to evaluate individual assays and determine appropriate cutoffs, particularly for the more sensitive assays (Luminex and flow cytometry).
Additionally, the choice of anticoagulant may affect assay results 27. Finally, newer methods for HLA antibody detection are continuously being developed 28, further adding to the complexity of data available in the field.
One solution to allow comparison of results across assays and from lot-to-lot of a given assay would be to develop calibration and proficiency panels composed of samples of known HLA antibody specificities and strength including negative control sera. These panels could be employed in routine assay runs to allow derivation of consistent cut-off values for determination of the reactivity status of unknown (donor) samples and to evaluate new instruments, reagent lots and technologists to assure optimally consistent performance of HLA antibody screening. The current study was not intended to address the blood bank HLA testing laboratories operational issue of where to set a cutoff value. REDS-II is undertaking other studies that will evaluate the potential trade-offs made when selecting various cutoffs, including examining the impact different cutoffs have on donation loss and loss of detection of donations with antibodies that are likely to cause TRALI.
In summary, while none of the available HLA antibody tests showed complete concordance among all groups, there was reasonable agreement amongst assays. Multi-analyte bead based assays and flow cytometry platforms showed higher sensitivity than ELISA platforms when using the manufacturer’s cutoffs. Concordance was higher among samples with a history of pregnancy and when assay cutoffs were set at higher levels, and particularly when empirically derived consensus cutoffs were employed.
Acknowledgments
The authors thank the staff at all six participating blood centers. Without their help, this study would not have been possible.
This work was supported by NHLBI contracts N01-HB-47170, -47175 and –57181
Appendix
The Retrovirus Epidemiology Donor Study - II (REDS-II Study Group) is the responsibility of the following people:
Blood Centers
American Red Cross Blood Services, New England Region
R. Cable, J. Rios and R. Benjamin
American Red Cross Blood Services, Southern Region/Department of Pathology and Laboratory Medicine, Emory University School of Medicine
J.D. Roback
Hoxworth Blood Center, University of Cincinnati Academic Health Center
R.A. Sacher, S.L. Wilkinson and P.M. Carey
Blood Centers of the Pacific, University of California San Francisco, Blood Systems Research Institute
E.L. Murphy, B. Custer and N. Hirschler
The Institute for Transfusion Medicine
D. Triulzi, R. Kakaiya and J. Kiss
Blood Center of Wisconsin
J. Gottschall and A. Mast
Coordinating Center: Westat, Inc
J. Schulman and M. King
National Heart, Lung, and Blood Institute, NIH
G.J. Nemo
Central Laboratory: Blood Systems Research Institute
M.P. Busch and P.J. Norris
Footnotes
Conflicts of interest: PJN received a speaker’s honorarium from One Lambda. The other authors state no conflicts.
References
- 1.Payne R. The development and persistence of leukoagglutinins in parous women. Blood. 1962;19:411–24. [PubMed] [Google Scholar]
- 2.Howard JE, Perkins HA. The natural history of alloimmunization to platelets. Transfusion. 1978;18:496–503. doi: 10.1046/j.1537-2995.1978.18478251250.x. [DOI] [PubMed] [Google Scholar]
- 3.Reed E, Hardy M, Benvenisty A, et al. Effect of antiidiotypic antibodies to HLA on graft survival in renal-allograft recipients. N Engl J Med. 1987;316:1450–5. doi: 10.1056/NEJM198706043162305. [DOI] [PubMed] [Google Scholar]
- 4.Slichter SJ, Davis K, Enright H, et al. Factors affecting posttransfusion platelet increments, platelet refractoriness, and platelet transfusion intervals in thrombocytopenic patients. Blood. 2005;105:4106–14. doi: 10.1182/blood-2003-08-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holness L, Knippen MA, Simmons L, Lachenbruch PA. Fatalities caused by TRALI. Transfus Med Rev. 2004;18:184–8. doi: 10.1016/j.tmrv.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 6.Starkey J, Kapler R. Transfusion Recipient Fatalities Reported to the Food and Drug Administration, FY2004 – 2006. ABC Newsletter; Washington, DC: 2007. p. 21. [Google Scholar]
- 7.Silliman CC, Boshkov LK, Mehdizadehkashi Z, et al. Transfusion-related acute lung injury: epidemiology and a prospective analysis of etiologic factors. Blood. 2003;101:454–62. doi: 10.1182/blood-2002-03-0958. [DOI] [PubMed] [Google Scholar]
- 8.Davoren A, Curtis BR, Shulman IA, et al. TRALI due to granulocyte-agglutinating human neutrophil antigen-3a (5b) alloantibodies in donor plasma: a report of 2 fatalities. Transfusion. 2003;43:641–5. doi: 10.1046/j.1537-2995.2003.00374.x. [DOI] [PubMed] [Google Scholar]
- 9.Silliman CC, Curtis BR, Kopko PM, et al. Donor antibodies to HNA-3a implicated in TRALI reactions prime neutrophils and cause PMN-mediated damage to human pulmonary microvascular endothelial cells in a two-event in vitro model. Blood. 2007;109:1752–5. doi: 10.1182/blood-2006-05-025106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Popovsky MA, Moore SB. Diagnostic and pathogenetic considerations in transfusion-related acute lung injury. Transfusion. 1985;25:573–7. doi: 10.1046/j.1537-2995.1985.25686071434.x. [DOI] [PubMed] [Google Scholar]
- 11.Kopko P, Silva M, Shulman I, Kleinman S. AABB survey of transfusion-related acute lung injury policies and practices in the United States. Transfusion. 2007;47:1679–85. doi: 10.1111/j.1537-2995.2007.01403.x. [DOI] [PubMed] [Google Scholar]
- 12.Terasaki PI, McClelland JD. Microdroplet Assay of Human Serum Cytotoxins. Nature. 1964;204:998–1000. doi: 10.1038/204998b0. [DOI] [PubMed] [Google Scholar]
- 13.Phelan DL, Rodey GE, Anderson CB. The development and specificity of antiidiotypic antibodies in renal transplant recipients receiving single-donor blood transfusions. Transplantation. 1989;48:57–60. doi: 10.1097/00007890-198907000-00013. [DOI] [PubMed] [Google Scholar]
- 14.Uboldi de Capei M, Pratico L, Curtoni ES. Comparison of different techniques for detection of anti-HLA antibodies in sera from patients awaiting kidney transplantation. Eur J Immunogenet. 2002;29:379–82. doi: 10.1046/j.1365-2370.2002.00334.x. [DOI] [PubMed] [Google Scholar]
- 15.Pei R, Lee J, Chen T, et al. Flow cytometric detection of HLA antibodies using a spectrum of microbeads. Hum Immunol. 1999;60:1293–302. doi: 10.1016/s0198-8859(99)00121-4. [DOI] [PubMed] [Google Scholar]
- 16.Wahrmann M, Exner M, Haidbauer B, et al. [C4d] FlowPRA screening--a specific assay for selective detection of complement-activating anti-HLA alloantibodies. Hum Immunol. 2005;66:526–34. doi: 10.1016/j.humimm.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 17.Triulzi DJ, Kleinman S, Kakaiya RM, et al. The effect of previous pregnancy and transfusion on HLA alloimmunization in blood donors: implications for a transfusion-related acute lung injury risk reduction strategy. Transfusion. 2009;49:1825–35. doi: 10.1111/j.1537-2995.2009.02206.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bollen KA. Structural Equations wtih Latent Variables. New York: John Wiley & Sons; 1989. [Google Scholar]
- 19.Joreskog KG, Sorbom D, Wallentin FY. Latent Variable Scores and Observational Residuals [monograph on the internet] 2006 Available from: http://www.ssicentral.com/lisrel/techdocs/obsres.pdf.
- 20.Powers A, Stowell CP, Dzik WH, et al. Testing only donors with a prior history of pregnancy or transfusion is a logical and cost-effective transfusion-related acute lung injury prevention strategy. Transfusion. 2008;48:2549–58. doi: 10.1111/j.1537-2995.2008.01902.x. [DOI] [PubMed] [Google Scholar]
- 21.Kleinman S, Grossman B, Kopko P. A national survey of transfusion-related acute lung injury risk reduction policies for platelets and plasma in the United States. Transfusion. doi: 10.1111/j.1537-2995.2010.02659.x. in press. [DOI] [PubMed] [Google Scholar]
- 22.Worthington JE, Robson AJ, Sheldon S, et al. A comparison of enzyme-linked immunoabsorbent assays and flow cytometry techniques for the detection of HLA specific antibodies. Hum Immunol. 2001;62:1178–84. doi: 10.1016/s0198-8859(01)00282-8. [DOI] [PubMed] [Google Scholar]
- 23.Lopes LB, Fabron A, Jr, Chiba AK, et al. Impact of using different laboratory assays to detect human leukocyte antigen antibodies in female blood donors. Transfusion. 2010;50:902–8. doi: 10.1111/j.1537-2995.2009.02523.x. [DOI] [PubMed] [Google Scholar]
- 24.Fadeyi E, Adams S, Peterson B, et al. Analysis of a high-throughput HLA antibody screening assay for use with platelet donors. Transfusion. 2008;48:1174–9. doi: 10.1111/j.1537-2995.2008.01684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vassallo RR, Hsu S, Einarson M, et al. A comparison of two robotic platforms to screen plateletpheresis donors for HLA antibodies as part of a transfusion-related acute lung injury mitigation strategy. Transfusion. doi: 10.1111/j.1537-2995.2010.02626.x. in press. [DOI] [PubMed] [Google Scholar]
- 26.Morales-Buenrostro LE, Terasaki PI, Marino-Vazquez LA, et al. “Natural” human leukocyte antigen antibodies found in nonalloimmunized healthy males. Transplantation. 2008;86:1111–5. doi: 10.1097/TP.0b013e318186d87b. [DOI] [PubMed] [Google Scholar]
- 27.Book BK, Higgins NG, Rosner KM, Pescovitz MD. Impact of anticoagulant in samples for HLA antibody screening by Luminex. Hum Immunol. 2008;69:S35. [Google Scholar]
- 28.Heinold A, Kuehl B, Brenner-Weiss G, et al. Sequential analysis by immunoprecipitation-MALDI-TOF: A novel method for detection and identification of alloantibody specificities. Hum Immunol. 2010 doi: 10.1016/j.humimm.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 29.Norris PJ, Lee JH, Carrick DM, et al. Long-term in vitro reactivity for human leukocyte antigen antibodies and comparison of detection using serum versus plasma. Transfusion. 2009;49:243–51. doi: 10.1111/j.1537-2995.2008.01955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]