Abstract
Fgf signalling is known to play critical roles in tooth development. Twenty-two Fgf ligands have been identified in mammals, but expression of only 10 in molars and three in the incisor loop stem cell region have been documented in murine tooth development. Our understanding of Fgf signalling in tooth development thus remains incomplete and we therefore carried out comparative in situ hybridisation analysis of unexamined Fgf ligands (eight in molars and 15 in cervical loops of incisors; Fgf11–Fgf14 were excluded from this analysis because they are not secreted and do not activate Fgf receptors) during tooth development. To identify where Fgf signalling is activated, we also examined the expression of Etv4 and Etv5, considered to be transcriptional targets of the Fgf signalling pathway. In molar tooth development, the expression of Fgf15 and Fgf20 was restricted to the primary enamel knots, whereas Etv4 and Etv5 were expressed in cells surrounding the primary enamel knots. Fgf20 expression was observed in the secondary enamel knots, whereas Fgf15 showed localised expression in the adjacent mesenchyme. Fgf16, Etv4 and Etv5 were strongly expressed in the ameloblasts of molars. In the incisor cervical loop stem cell region, Fgf17, Fgf18, Etv4 and Etv5 showed a restricted expression pattern. These molecules thus show dynamic temporo-spatial expression in murine tooth development. We also analysed teeth in Fgf15−/− and Fgf15−/−;Fgf8+/− mutant mice. Neither mutant showed significant abnormalities in tooth development, indicating likely functional redundancy.
Keywords: cervical loop, epithelium, Fgf ligand, Fgf15 mutant, Fgf15/Fgf8 mutant, mesenchyme, primary enamel knot, secondary enamel knot, tooth development
Introduction
Fibroblast growth factor (Fgf) signalling has been shown to play essential roles in regulating many biological processes including mitogenic, chemotactic and angiogenic activity and wound healing (Powers et al. 2000; Nie et al. 2006; Mason, 2007). It is known that Fgf signalling controls the development of many craniofacial organs: brain, palate, salivary glands, olfactory, eye and ear (Min et al. 1998; Sekine et al. 1999; De Moerlooze et al. 2000; Mason, 2007).
Teeth are craniofacial organs that are known to be regulated by Fgf signalling during their development (Thesleff et al. 1995; Pispa & Thesleff, 2003; Tucker & Sharpe, 2004; Mikkola, 2007). One of the roles of Fgf signalling in tooth development is for bud formation, since tooth development is arrested as thickening epithelium in Fgf receptor (Fgfr) 2IIIb mutant mice (De Moerlooze et al. 2000; Hosokawa et al. 2009). Fgf signalling is also important for continuous growth of murine incisors, as Fgf10 mutant mice lack cervical loop formation, an epithelial stem-cell niche for providing ameloblast precursors (Harada et al. 2002). Fgf signalling has also been shown to be involved in determining tooth type by inducing homeobox gene expression in presumptive tooth mesenchyme at early stages of development (Tucker et al. 1998). Fgf signalling thus regulates multiple aspects of tooth development (Supporting Information Table S1 summarises the tooth phenotypes caused by targeted disruption of Fgf signalling-related molecules).
Although 22 Fgf ligands have been identified in mammals, only 10 (Fgf1–Fgf10) and three (Fgf3, Fgf9 and Fgf10) Fgf ligands have been documented in murine molar tooth development and murine incisor cervical loop region development, respectively (Kettunen & Thesleff, 1998; Harada et al. 1999; Kettunen et al. 2000; Unda et al. 2001; Wang et al. 2007; Klein et al. 2008). Our understanding of Fgf signalling in tooth development thus remains incomplete.
A comprehensive expression analysis of all unexamined Fgfs in the tooth development was carried out to obtain a more complete understanding of the role of Fgf signalling in tooth development. The expression patterns of unexamined Fgf ligands (eight in the molar and 15 in the cervical loop region) and two Fgf signalling-related molecules (Etv4 and Etv5) were mapped by in situ hybridisation in mouse embryonic tooth germs. Fgf15, Fgf16, Fgf17, Fgf18, Fgf20, Fgf21, Etv4 and Etv5 showed dynamic temporo-spatial expression, whereas the expression of Fgf19 or Fgf22 could not be detected in tooth development. Among the Fgf ligands examined, Fgf15 showed restricted expression patterns in tooth development, and therefore tooth development in Fgf15−/− and Fgf15−/−;Fgf8+/− mutant mice was examined. Neither mutant showed significant abnormalities in tooth development, indicating likely functional redundancy.
Materials and methods
Production and analysis of transgenic mice
Fgf15−/− mice were produced as described by Wright et al. (2004) and Vincentz et al. (2005). Fgf8+/− mice were produced as described by Moon & Capecchi (2000). Mutant and littermate heads were fixed in 4% paraformaldehyde (PFA), wax-embedded and serially sectioned for histological analysis. Eight Fgf15−/− and eight Fgf15−/−;Fgf8+/− mice were examined in this study.
In situ hybridisation
Radioactive in situ hybridisation with [35S]UTP-labeled riboprobes was carried out as described previously (Ohazama et al., 2008). Briefly, CD1 and mutant mouse heads were fixed in 4% PFA, wax-embedded and serially sectioned at 7 μm. Decalcification using 0.5 m EDTA (pH 7.6) was performed after fixation of newborn mice. Sections were split over 5–10 slides and prepared for radioactive in situ hybridisation. Radioactive in situ hybridisation with [35S]UTP-labeled riboprobes was carried out as described by Wilkinson (1995), with modifications. Briefly, sections were floated onto TESPA(3-aminopropyltriethoxysilane)-coated slides. The slides were pretreated with 5 mg mL−1 proteinase K and 0.25% (vol/vol) acetic anhydride to reduce background. Hybridisation was carried out overnight in a humidified chamber at 55 °C. The slides were then washed twice at high stringency in 2 × standard saline citrate (SSC), 50% formamide, 10 mm dithiothreitol (DTT) at 65 °C for 20 min and treated with 40 μg mL−1 RNAse A for 30 min at 37 °C to remove any non-specifically bound probe. The high stringency washes (at 65 °C in 2 × SSC, 50% formamide, 10 mm DTT) were repeated, followed by a further wash at 65 °C in 0.1 × SSC, 10 mm DTT. The sections were then washed in 0.1 × SSC at room temperature and dehydrated through 300 mm ammonium acetate in 70% ethanol, 95% ethanol and absolute ethanol. The slides were air-dried and dipped in Ilford K.5 photographic emulsion. Autoradiography was performed by exposing the sections in a light-tight box at 4 °C for 10–14 days. Slides were developed using Kodak D19, fixed in Kodak UNIFIX, counterstained with malachite green or haematoxylin, and mounted with DePex (BDG). For photography, in some sections the darkfield images were inverted, artificially stained red, and combined with the brightfield image using adobe photoshop. The plasmids used to prepare the radioactive antisense probes in this study have previously been described and were generated from mouse cDNA clones Fgf1, Fgf2, Fgf5, Fgf6, Fgf7, Fgf15, Fgf16, Fgf17, Fgf19, Fgf20, Fgf21 (Yaguchi et al. 2009), Fgf4 (Sun et al. 2000), Fgf8 (Mahmood et al. 1995), Fgf18 (Ohbayashi et al. 2002) and Fgf22 (Umemori et al. 2004).
Results and Discussion
Expression of Fgf ligands and Fgf signalling molecules in tooth development
Teeth develop as a result of sequential and reciprocal interactions between oral epithelium and mesenchyme. The first morphological sign of tooth development is a narrow band of thickened epithelium on the developing jaw primordia. The thickened epithelium progressively takes the form of bud, cap and bell configurations as differentiation proceeds. Subsequently, epithelial cells and mesenchymal cells (dental papilla) differentiate into enamel-producing ameloblasts and dentine-producing odontoblasts, respectively.
Mice have only one incisor and three molars in each jaw quadrant, which are divided by a large edentulous region (diastema). It is known that mice lost ‘diastema’ teeth during evolution and despite the lack of these teeth in the diastema, two rudimentary epithelial buds (called MS and R2) form as vestigial tooth germs in the murine diastema. R2 subsequently integrates into the first molar tooth germs during early stages of development, whereas MS is removed by apoptosis (Ahn et al. 2010; Prochazka et al. 2010).
We carried out comparative in situ hybridisation analysis of molar tooth development at E10.5, E12.5, E13.5, E14.5 and newborn mice. This period encompasses molar tooth development from initiation to the onset of cytodifferentiation.
Fgf ligands can be subdivided broadly into seven subfamilies based on sequence homology. The members of the Fgf11 subfamily (Fgf11–Fgf14) were excluded from this analysis because they are not secreted and do not activate Fgf receptors, but instead are localised to the cell nucleus and have intracellular function (Powers et al. 2000; Zhang et al. 2006; Mason, 2007).
Initiation of tooth development (E10.5)
Initiation begins before the tooth anlagen is morphologically visible. The first signals are derived from tooth presumptive epithelium at E9.5 (Ferguson et al. 2000). The first visible signs of tooth development cannot be recognised at E10.5.
Strong Fgf17 expression was observed in presumptive tooth epithelium (Fig. 1C). Fgf8 and Fgf9 were also expressed in presumptive tooth epithelium, whereas Fgf10 expression was found in both epithelium and mesenchyme at this stage (Kettunen & Thesleff, 1998). Fgf17 was expressed in the presumptive molar region but not in the presumptive incisor region at E10.5, suggesting that Fgf17 expression is very similar to Fgf8 at this stage (data not shown). Both Fgf8 and Fgf17 belong to the Fgf8 subfamily. Fgf17 may thus also regulate the determination of tooth type, as Fgf8 is known to play critical roles in determining tooth type (Neubuser et al. 1997; Tucker et al. 1998). Fgf15, Fgf16, Fgf18, Fgf20, Fgf21, Fgf22 and Fgf23 showed no expression in presumptive tooth regions at E10.5 (Fig. 1A,B,D; data not shown).
Fig. 1.
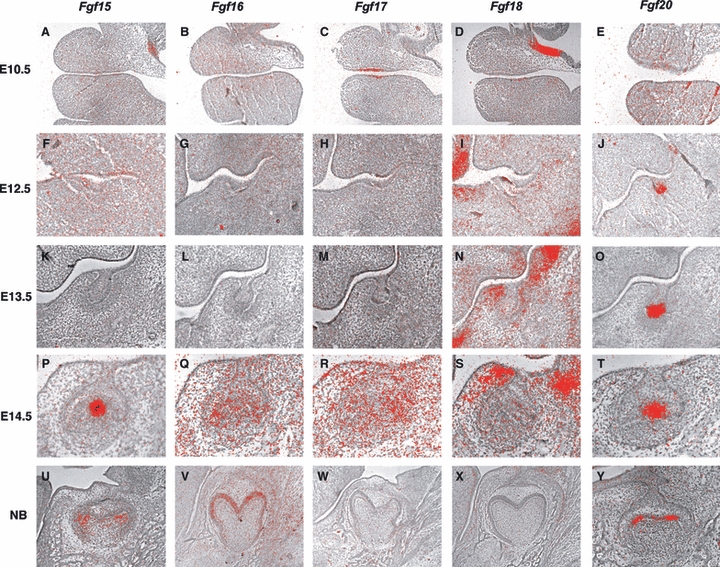
Expression of Fgf ligands in molar tooth development. In situ hybridisation of Fgfs on frontal head sections at E10.5 (A–E), E12.5 (F–J), E13.5 (K–O), E14.5 (P–T) and E18.5 (U–Y).
We next assayed the expression of the ETS-related factor genes Etv4 (Pea3) and Etv5 (Erm), which are considered to be transcriptional targets of the Fgf signalling pathway (O'Hagen & Hassell 1996; Roehl & Nusslein-Volhard, 2001). Expression of both genes was observed in epithelium and mesenchyme (Fig. 2A,B). Fgf signalling is transduced through a family of four transmembrane receptor tyrosine kinases in all vertebrates (Powers et al. 2000; Mason, 2007). It has been shown that Fgfr1IIIc (receptor for Fgf8, Fgf17 and Fgf20) and Fgfr2IIIb (receptor for Fgf10) are expressed in tooth mesenchyme and epithelium, respectively, at this stage (Kettunen et al. 1998; Mason, 2007). It is thus likely that, at this stage, Fgf signalling is activated in epithelium by Fgf10 through Fgfr2IIIb and in mesenchyme by Fgf8, Fgf17 and Fgf20 through Fgfr1IIIc.
Fig. 2.
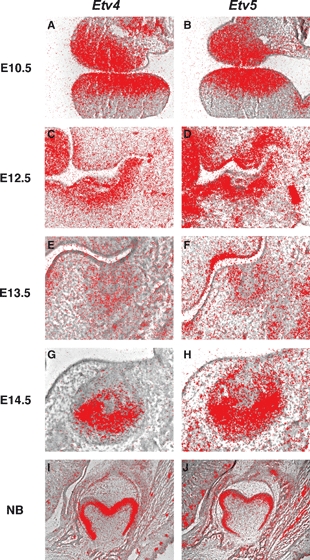
Etv4 and Etv5 expression in molar tooth development. In situ hybridisation of Etv4 and Etv5 on frontal head sections at E10.5 (A,B), E12.5 (C,D), E13.5 (E,F), E14.5 (G,H) and E18.5 (I,J).
Thickening tooth epithelium (E12.5)
Thickening of the oral epithelium takes place from E12. The anterior limit of thickened epithelium is the vestigial tooth germ (MS), which is more prominent than the posterior part of thickened epithelium. Shh expression is only observed in MS epithelium at this stage (Ahn et al. 2010; Prochazka et al. 2010).
Fgf15 was weakly expressed at the lingual side of the thickened tooth epithelium, whereas strong Fgf20 expression was observed at the tip of MS (Fig. 1F,J). Fgf18 was weakly expressed at the buccal side of tooth mesenchyme (Fig. 1I). The expression of Fgf16, Fgf17, Fgf21, Fgf22 and Fgf23 could not be detected in tooth germs at this stage (Fgf1G, 1H; data not shown). Both Etv4 and Etv5 were strongly expressed in mesenchyme, whereas only weak expression was observed in epithelium (Fig. 2C,D). It has been shown that Fgf10 is faintly expressed in the mesenchyme, whereas Fgf8 and Fgf9 expression is observed in epithelium at this stage (Kettunen & Thesleff, 1998; Kettunen et al. 2000). It is also known that Fgfr1IIIc (receptor for Fgf8, Fgf15 and Fgf20) is expressed in mesenchyme and that Fgfr2IIIb (receptor for Fgf10 and Fgf15) expression is found in epithelium at this stage (Kettunen et al. 1998; Mason, 2007). It is likely that Fgf signalling is activated in epithelium by Fgf10 and Fgf15 through Fgfr2IIIb, and in mesenchyme by Fgf8, Fgf15 and Fgf20 through Fgfr1IIIc. In Fgfr2 IIIb mutant mice, tooth development is arrested as thickened epithelium, suggesting that Fgf10 and Fgf15 play critical roles in the transition to the bud stage (De Moerlooze et al. 2000; Hosokawa et al. 2009).
Bud stage (E13.5)
By E13.5 the tooth epithelium invaginates into underlying mesenchyme to form the epithelial bud. R2 buds become prominent posterior to MS expressing Shh, whereas MS loses Shh expression at this stage (Ahn et al. 2010; Prochazka et al. 2010).
Fgf20 expression was found at the posterior part of tooth germs, whereas the anterior part of tooth germs (MS or R2) showed no expression (Fgf 1O; Supporting Information Fig. S1). Expression of Fgf18 (Fig. 1N) was observed at the buccal and lingual sides of tooth mesenchyme but was absent from mesenchyme underneath bud epithelium (Fig. 1N). Fgf15, Fgf16, Fgf17, Fgf21, Fgf22 and Fgf23 showed no expression in tooth germs at E13.5 (Fgf1K,L,M; data not shown). Weak expression of Etv4 and Etv5 was observed in both epithelium and mesenchyme (Fig. 2E,F; Klein et al. 2006). Fgf8 expression was restricted to the oral part of epithelium, whereas Fgf3 was weakly expressed in both epithelium and mesenchyme at this stage (Kettunen & Thesleff, 1998; Kettunen et al. 2000). It has also been shown that strong Fgf9 expression and weak Fgf4 and Fgf3 expression are observed at the tip of bud epithelium, similar to Fgf20 expression (Fig. 1O; Kettunen & Thesleff, 1998). Fgf10 is expressed in the mesenchyme on the buccal and lingual sides of tooth germs at this stage (Kettunen et al. 2000). Fgfr1IIIc (receptor for Fgf4, Fgf8 and Fgf20) and Fgfr2IIIc (receptor for Fgf4, Fgf8, Fgf9, Fgf16, Fgf18 and Fgf20) expression are found in mesenchyme, whereas Fgfr2IIIb (receptor for Fgf3 and Fgf10) is expressed in epithelium at this stage (Kettunen et al. 1998; Mason, 2007). It is likely that Fgf signalling is activated in epithelium by Fgf3 and Fgf10 through Fgfr2IIIb, and in mesenchyme by Fgf4, Fgf8, Fgf9, Fgf18 and Fgf20 through Fgfr1IIIc and Fgfr2IIIc.
Cap stage (E14.5)
By E14.5 the bud basal epithelium develops into the internal and the external (outer) enamel epithelium, and the mesenchyme develops into the dental papilla and the dental follicle. The temporal and spatial folding of the internal enamel epithelium is required for formation of cusps. Primary enamel knots are transient populations of cells in the center of the invaginating dental epithelium originally identified in histological sections of cap-stage tooth germs (MacKenzie et al. 1992; Jernvall et al. 1998). The primary enamel knots are located at the posterior tooth germs that show Shh expression; the anterior part of tooth germs (MS and R2) exhibit no Shh expression at this stage (Ahn et al. 2010; Prochazka et al. 2010).
Fgf15 and Fgf20 expression was found at the primary enamel knots, whereas expression could not be detected in MS or R2 (Fig. 1P,T; Fig. S1). Weak expression of Fgf16 and Fgf17 were observed in cervical loop epithelium and mesenchyme surrounding the cervical loop, whereas MS or R2 showed no expression of these molecules (Fig. 1Q,R). Fgf18 expression was detected in buccal and lingual mesenchyme but was absent from mesenchyme underneath cap epithelium (Fig. 1S). Fgf21, Fgf22 and Fgf23 showed no expression in tooth germs at E14.5 (data not shown). Strong expression of both Etv4 and Etv5 was detected in mesenchyme and epithelium but was absent from the primary enamel knots, whereas both have been shown to be weakly expressed in MS and R2 (Fig. 2G,H; Klein et al. 2006). Fgf3 was expressed in mesenchyme and enamel knots, whereas Fgf4 and Fgf9 showed restricted expression only in primary enamel knots at this stage (Kettunen & Thesleff, 1998; Kettunen et al. 2000). Fgf10 expression was observed in tooth mesenchyme (Kettunen et al. 2000). Fgfr1IIIb (receptor for Fgf3, Fgf10) was expressed in inner enamel epithelium, whereas Fgfr1IIIc (receptor for Fgf4, Fgf15, Fgf17, Fgf20) expression was found in tooth mesenchyme and inner enamel epithelium. Fgfr2IIIb (receptor for Fgf3, Fgf10, Fgf15) was expressed in outer epithelium and the stellate reticulum, whereas weak expression of Fgfr2IIIc (receptor for Fgf4, Fgf9, Fgf15, Fgf16, Fgf17, Fgf18 and Fgf20) was observed in buccal tooth mesenchyme at this stage (Kettunen et al. 1998; Mason, 2007). Fgf signalling is thus activated in inner enamel epithelium by Fgf3, Fgf4, Fgf10, Fgf15, Fgf17 and Fgf20 through Fgfr1IIIb and Fgfr1IIIc, in outer enamel epithelium and the stellate reticulum by Fgf3, Fgf10 and Fgf15 through Fgfr2IIIb, and in mesenchyme by Fgf4, Fgf9, Fgf15, Fgf16, Fgf17, Fgf18 and Fgf20 through Fgfr1IIIc and Fgfr2IIIc.
Fgf signalling was activated in the cells surrounding the primary enamel knot but not in the primary enamel knot cells, although Fgf3, Fgf4, Fgf9, Fgf15 and Fgf20 were expressed in the primary enamel knot at E14.5.
Cytodifferentiation (E18.5)
The terminal differentiation of dentine-forming odontoblasts from dental papilla cells and the enamel-forming ameloblasts from the internal epithelium occurred between E18 to P0. Primary enamel knots were transient and had disappeared by the late cap stage (E15 in mice). In the molars, however, secondary enamel knots developed that are clearly visible from expression of Fgf4 and Slit1 at the bell stage at the tip of the forming cusps (E18; Jernval et al. 1998; Pispa et al. 1999; Loes et al. 2001).
Expression of Fgf20 was found in secondary enamel knots, whereas Fgf15 was expressed in mesenchyme directly underneath the secondary enamel knots (Fig. 1U,Y). Fgf16 expression was observed in ameloblasts (Fig. 1V). Fgf17, Fgf18, Fgf21, Fgf22 and Fgf23 showed no expression in tooth germs at this stage (Fig. 1W,X; data not shown). Strong expression of both Etv4 and Etv5 was observed in ameloblasts, whereas they were weakly expressed in odontoblasts (Fig. 2I,J). Fgf3 was expressed in the dental papilla but was absent from terminally differentiated odontoblasts, whereas Fgf10 was expressed in differentiating odontoblasts at this stage (Kettunen et al. 2000). Fgf4 was also expressed in secondary enamel knots, whereas Fgf9 was expressed in ameloblasts at E18.5 (Kettunen & Thesleff, 1998). Fgfr1IIIb (receptor for Fgf10) expression was observed in both odontoblasts and ameloblasts (Kettunen et al. 1998; Mason, 2007). Fgfr1IIIc (receptor for Fgf4, Fgf15 and Fgf20) was strongly expressed in odontoblasts but weakly expressed in ameloblasts (Kettunen et al. 1998; Mason, 2007). Fgfr2IIIb (receptor for Fgf10 and Fgf15) expression was detected in ameloblasts, and Fgfr2IIIc (receptor for Fgf4, Fgf9, Fgf15, Fgf16 and Fgf20) showed restricted expression in the dental follicle (Kettunen et al. 1998; Mason, 2007). In ameloblast development, Fgf signalling was activated by Fgf4, Fgf10, Fgf15 and Fgf20 through Fgfr1IIIb, Fgfr1IIIc and Fgfr2IIIb, and in odontoblasts by Fgf4, Fgf10, Fgf15 and Fgf20 through Fgfr1IIIb and Fgfr1IIIc. Mice with epithelial conditional mutation of Fgfr1 show abnormal enamel formation due to abnormal enamel-secreting ameloblasts, suggesting that Fgf4, Fgf10, Fgf15 and Fgf20 play critical roles in ameloblast function (Takamori et al. 2008).
The expression of Fgf ligands, Etv4 and Etv5 during murine molar tooth development is summarised in Fig. 3. Etv4 and Etv5 were expressed in an almost identical manner in embryonic molar tooth development.
Fig. 3.
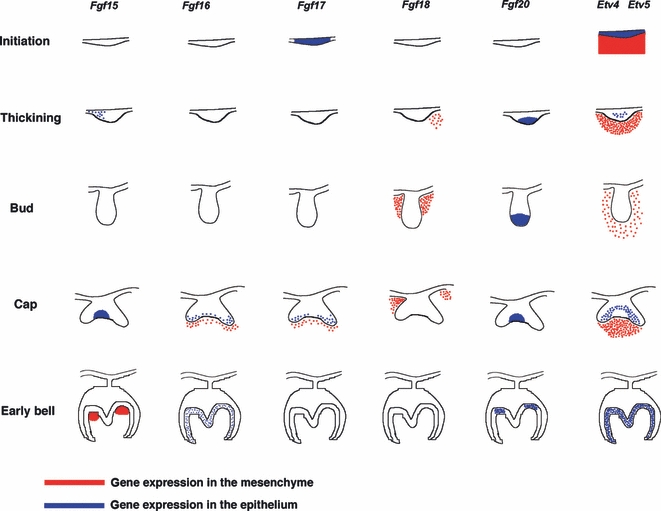
Summary of Fgfs, Etv4 and Etv5 in molar tooth development. Schematic representation of a combination of expression of Fgf ligands, Etv4 and Etv5, in molar tooth development at E10.5, E12.5, E13.5 E14.5 and E18.5. Etv4 and Etv5 were expressed in an identical manner in embryonic tooth development. Expression in epithelium shown in blue and in mesenchyme in red.
Cervical loops of incisors (E16.5)
Murine incisors grow continuously throughout life. The cervical loop region of mouse incisors is an epithelial stem cell niche that provides epithelial cell precursors for continuous growth. Labial and lingual cervical loops are different because rodent incisors have two distinct surfaces – a labial surface of enamel-covered dentine and a lingual surface of dentine only. Fgf10 mutant mice have a compromised cervical loop formation (Harada et al. 2002). Mice with mutations in Sprouty2/4 (antagonists of Fgf signalling) show bilateral enamel deposition due to disrupted inactivation of Fgf signalling in lingual cervical loops (Klein et al. 2008). Fgf signalling thus plays a critical role in the formation and function of cervical loops (Harada et al. 1999, 2002; Wang et al. 2007; Klein et al. 2008). Expression of only three Fgf ligands (Fgf3, Fgf9 and Fgf10) has been described in cervical loops (Harada et al. 2002; Wang et al. 2007; Klein et al. 2008). We therefore examined the expression of other Fgf ligands in incisor cervical loop regions at E16.5 when the cervical loops can be observed as distinct structures.
Fgf1 was expressed in all cervical loop epithelia (Fig. 4A). Weak expression of Fgf7 was found in mesenchyme close to lingual cervical loops (Fig. 4C). Strong Fgf16 expression was observed in lingual mesenchyme close to labial cervical loops, whereas Fgf16 was weakly expressed in labial cervical loop epithelium and labial mesenchyme close to lingual cervical loops (Fig. 4D). Fgf17 expression was localised in labial cervical loop epithelium (Fig. 4D). Fgf18 showed localised expression in mesenchyme between labial and lingual cervical loops (Fig. 4F). Fgf21 was weakly expressed in lingual mesenchyme close to labial cervical loops (Fig. 4G). Fgf2, Fgf4–Fgf6, Fgf8, Fgf15, Fgf19, Fgf20, Fgf22 and Fgf23 expression could not be detected in cervical loop epithelium or incisor mesenchyme (data not shown). Etv4 and Etv5 showed strong expression in both labial and lingual cervical loop epithelia and lingual mesenchyme close to labial cervical loops (Fig. 4H,I; Klein et al. 2008). Fgf3 expression was observed in labial mesenchyme close to cervical loops, whereas Fgf9 expression was localised in a small lingual epithelial domain just anterior to the labial cervical loops (Harada et al. 1999; Wang et al. 2007; Klein et al. 2008). Fgf10 has been shown to be expressed in mesenchyme close to both lingual and labial cervical loops (Harada et al. 1999; Klein et al. 2008). Fgfr1b and Fgfr2b were both expressed in cervical loop epithelium (Harada et al. 1999). It is likely that Fgf signalling is activated in cervical loop epithelium by Fgf1, Fgf3, Fgf7, Fgf10 and Fgf21 through Fgfr1b and Fgfr2b.
Fig. 4.
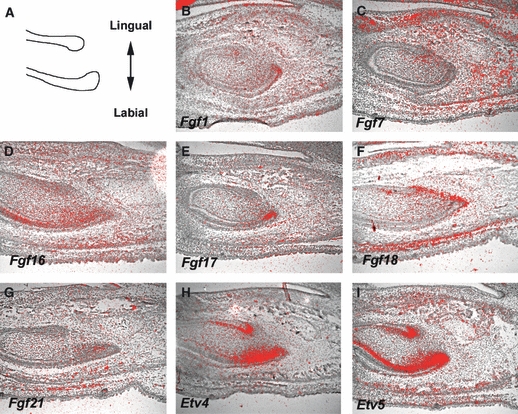
Expression of Fgf ligands Etv4 and Etv5 in cervical loop region at E16.5. (a) Schematic representation of mouse incisor cervical loop at E16.5 and axial definition. (B–I) In situ hybridisation of Fgf ligands Etv4 and Etv5 on sagittal head sections at E16.5.
The expression of Fgf ligands Etv4 and Etv5 in murine embryonic incisor cervical loop regions is summarised in Fig. 5. Etv4 and Etv5 were expressed in an almost identical manner in embryonic cervical loop development.
Fig. 5.
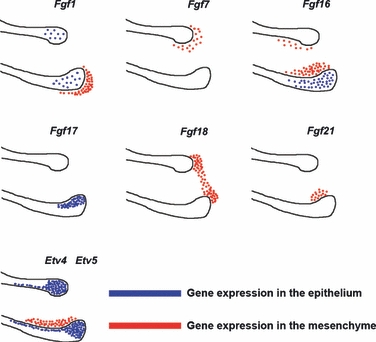
Summary of Fgf ligands Etv4 and Etv5 in embryonic cervical loop region. Schematic drawing of a combination of expression of Fgf ligands Etv4 and Etv5 in cervical loop region at E16.5. Etv4 and Etv5 were expressed in an identical manner in embryonic cervical loops. Expression in epithelium shown in blue and in mesenchyme in red.
Tooth development in Fgf15−/− and Fgf15−/−;Fgf8+/− mice
Fgf ligands show dynamic temporo-spatial expression in embryonic tooth development. Tooth abnormalities are found in mice with mutation in several Fgf signalling-related molecules (De Moerlooze et al. 2000; Harada et al. 2002; Klein et al. 2006, 2008; Hosokawa et al. 2009).
Fgf15 shows restricted expression from the early stages in molar tooth development. Restricted expression pattern of Fgf15 is also observed in incisor tooth development before cervical loop formation (Fig. 6A–C). Fgf15 was co-expressed with Shh in the brain and in primary enamel knots (Dassule et al. 2000; Gritli-Linde et al. 2002; Saitsu et al. 2005; Gimeno & Martinez, 2007; Komada et al. 2008). To investigate the role of Fgf15 in tooth development, we examined teeth from Fgf15 mutant mice (Fgf15−/−). Most Fgf15−/− mice die the first week after birth (Vincentz et al. 2005). No significant abnormalities, however, could be detected in either incisors or molars in the mutant mice at birth (Fig. 7B,D,F,H,J). Fgf15 expression is also observed in the development of other craniofacial organs such as the tongue and whisker follicles (Fig. 6D,E). However, no significant abnormalities were detected in these organs (Fig. 7L,N).
Fig. 6.
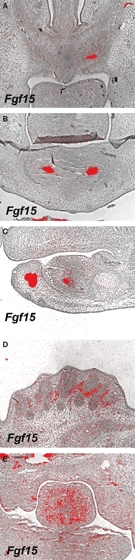
Fgf15 expression in development of craniofacial organs. In situ hybridisation of Fgf15 on frontal head sections (A,B,D,E) and sagittal sections (C) at E13.5 (B,E) and E14.5 (A,D). Upper incisor (A), lower incisor (B,C), whisker hair follicles (D) and tongue (E).
Fig. 7.
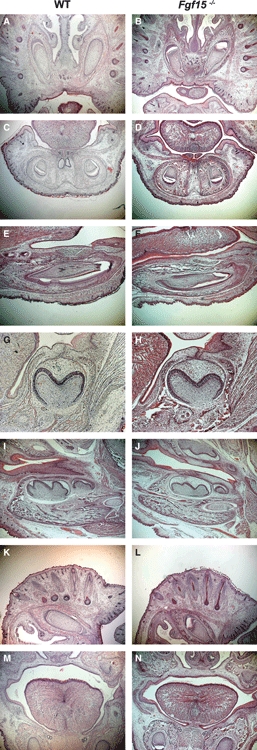
Teeth, whisker hair follicles and tongue in Fgf15−/− mice. No obvious abnormalities are found in upper (B) or lower (D,E) incisors, and molars (H,J) in Fgf15−/− mice. No significant morphological changes are observed in whisker hair follicles (L) and tongue (N) in Fgf15−/− mice. Frontal (A–D,G,H,K–N) and sagittal (E,F,I,J) head sections at birth.
There is an overlap between Fgf8 and Fgf15 expression in tooth development at E12 (Fig. 1; Kettunen et al. 2000) and it is conceivable that Fgf8 compensates for loss of Fgf15 in mutant tooth development. To reveal any genetic interaction between Fgf15 and Fgf18, we analysed teeth in Fgf15−/−;Fgf8+/− mice, but no obvious abnormalities could be detected in either incisors or molars in these mutant mice at birth (Fig. 8).
Fig. 8.
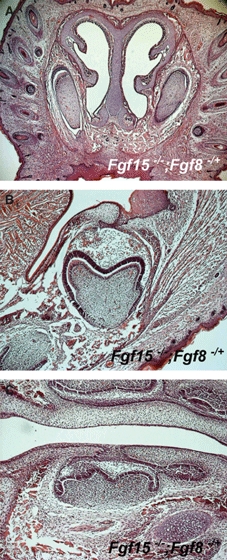
Teeth in Fgf15−/−;Fgf8+/− mice. No obvious abnormalities are found in upper (A) and molars (B,C) in Fgf15−/−;Fgf8+/− mice. Frontal (A,B) and sagittal (C) head sections at birth.
Fgf9 and Fgf20 are expressed in epithelium and Fgf10 and Fgf18 in mesenchyme when Fgf15 expression is found in epithelium at E12.5 (Kettunen & Thesleff, 1998; Kettunen et al. 2000). In common with Fgf15, Fgf3, -4, -9 and -20 are expressed in the primary enamel knots at E14.5 (Kettunen & Thesleff, 1998; Kettunen et al. 2000). It is conceivable that there is multiple functional redundancy between these ligands in tooth development. Fgf8, Fgf10 and Fgf20 can share receptors with Fgf15 at E12.5, whereas Fgf3, Fgf4, Fgf9 and Fgf20 expressed in enamel knots also share receptors with Fgf15 at E14.5.
Expression of all Fgf ligands has been examined at embryonic stages of tooth development, although Fgf5, Fgf6 and Fgf23 could not be detected in tooth development during these embryonic periods (Kettunen & Thesleff, 1998; Kettunen et al. 2000). However, Fgf23 expression was observed at postnatal stages of tooth development (Onishi et al. 2008). Moreover, overexpression of Fgf23 results in the hypo-mineralisation of teeth (Onishi et al. 2008).
The dynamic temporo-spatial expression of Fgf ligands, their receptors and Fgf signalling target molecules shows that Fgf activity regulates multiple aspects of tooth development. There may be multiple functional redundancy between these ligands in tooth development.
Acknowledgments
This work was supported by the MRC. Y.O.-T. is supported by Nihon University. A.O. is an RCUK Fellow. T.P. is supported by W J B Houston Research Scholarship (European Orthodontic Society).
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Fig. S1 Fgf15 and Fgf20 expression in MS or RS at E13.5 or E14.5. Frontal head sections showing no obvious expression of Fgf20 in MS of wild-type at E13.5 (A–C). Frontal head sections showing no obvious expression of Fgf15 (D–F) or Fgf20 (G) in MS or RS of wild-type at E14.5 (D–G).
Table S1 Tooth phenotypes in mice with mutation of Fgf signalling related molecules.
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer-reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
References
- Ahn Y, Sanderson BW, Klein OD, et al. Inhibition of Wnt signaling by Wise (Sostdc1) and negative feedback from Shh controls tooth number and patterning. Development. 2010;137:3221–3231. doi: 10.1242/dev.054668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles C, Lazzari V, Tafforeau P, et al. Modulation of Fgf3 dosage in mouse and men mirrors evolution of mammalian dentition. Proc Natl Acad Sci U S A. 2009;106:22364–22368. doi: 10.1073/pnas.0910086106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassule HR, Lewis P, Bei M, et al. Sonic hedgehog regulates growth and morphogenesis of the tooth. Development. 2000;127:4775–4785. doi: 10.1242/dev.127.22.4775. [DOI] [PubMed] [Google Scholar]
- De Moerlooze L, Spencer-Dene B, Revest JM, et al. An important role for the IIIb isoform of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal-epithelial signalling during mouse organogenesis. Development. 2000;127:483–492. doi: 10.1242/dev.127.3.483. [DOI] [PubMed] [Google Scholar]
- Ferguson CA, Tucker AS, Sharpe PT. Temporospatial cell interactions regulating mandibular and maxillary arch patterning. Development. 2000;127:403–412. doi: 10.1242/dev.127.2.403. [DOI] [PubMed] [Google Scholar]
- Gimeno L, Martinez S. Expression of chick Fgf19 and mouse Fgf15 orthologs is regulated in the developing brain by Fgf8 and Shh. Dev Dyn. 2007;236:2285–2297. doi: 10.1002/dvdy.21237. [DOI] [PubMed] [Google Scholar]
- Gritli-Linde A, Bei M, Maas R, et al. Shh signaling within the dental epithelium is necessary for cell proliferation, growth and polarization. Development. 2002;129:5323–5337. doi: 10.1242/dev.00100. [DOI] [PubMed] [Google Scholar]
- Harada H, Kettunen P, Jung HS, et al. Localization of putative stem cells in dental epithelium and their association with Notch and FGF signaling. J Cell Biol. 1999;147:105–120. doi: 10.1083/jcb.147.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H, Toyono T, Toyoshima K, et al. FGF10 maintains stem cell compartment in developing mouse incisors. Development. 2002;129:1533–1541. doi: 10.1242/dev.129.6.1533. [DOI] [PubMed] [Google Scholar]
- Hosokawa R, Deng X, Takamori K, et al. Epithelial-specific requirement of FGFR2 signaling during tooth and palate development. J Exp Zool B Mol Dev Evol. 2009;312B:343–350. doi: 10.1002/jez.b.21274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jernvall J, Aberg T, Kettunen P, et al. The life history of an embryonic signaling center: BMP-4 induces p21 and is associated with apoptosis in the mouse tooth enamel knot. Development. 1998;125:161–169. doi: 10.1242/dev.125.2.161. [DOI] [PubMed] [Google Scholar]
- Kettunen P, Thesleff I. Expression and function of FGFs-4, -8, and -9 suggest functional redundancy and repetitive use as epithelial signals during tooth morphogenesis. Dev Dyn. 1998;211:256–268. doi: 10.1002/(SICI)1097-0177(199803)211:3<256::AID-AJA7>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Kettunen P, Karavanova I, Thesleff I. Responsiveness of developing dental tissues to fibroblast growth factors: expression of splicing alternatives of FGFR1, -2, -3, and of FGFR4; and stimulation of cell proliferation by FGF-2, -4, -8, and -9. Dev Genet. 1998;22:374–385. doi: 10.1002/(SICI)1520-6408(1998)22:4<374::AID-DVG7>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Kettunen P, Laurikkala J, Itaranta P, et al. Associations of FGF-3 and FGF-10 with signaling networks regulating tooth morphogenesis. Dev Dyn. 2000;219:322–332. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1062>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Klein OD, Minowada G, Peterkova R, et al. Sprouty genes control diastema tooth development via bidirectional antagonism of epithelial-mesenchymal FGF signaling. Dev Cell. 2006;11:181–190. doi: 10.1016/j.devcel.2006.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein OD, Lyons DB, Balooch G, et al. An FGF signaling loop sustains the generation of differentiated progeny from stem cells in mouse incisors. Development. 2008;135:377–385. doi: 10.1242/dev.015081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komada M, Saitsu H, Shiota K, et al. Expression of Fgf15 is regulated by both activator and repressor forms of Gli2 in vitro. Biochem Biophys Res Commun. 2008;369:350–356. doi: 10.1016/j.bbrc.2008.02.015. [DOI] [PubMed] [Google Scholar]
- Lin Y, Cheng Y-SL, Qin C, et al. FGFR2 in the dental epithelium is essential for development and maintenance of the maxillary cervical loop, a stem cell niche in mouse incisors. Dev Dyn. 2009;238:324–330. doi: 10.1002/dvdy.21778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loes S, Luukko K, Kvinnsland IH, et al. Slit1 is specifically expressed in the primary and secondary enamel knots during molar tooth cusp formation. Mech Dev. 2001;107:155–157. doi: 10.1016/s0925-4773(01)00454-3. [DOI] [PubMed] [Google Scholar]
- MacKenzie A, Ferguson MW, Sharpe PT. Expression patterns of the homeobox gene, Hox-8, in the mouse embryo suggest a role in specifying tooth initiation and shape. Development. 1992;115:403–420. doi: 10.1242/dev.115.2.403. [DOI] [PubMed] [Google Scholar]
- Mahmood R, Bresnik J, Hornbruch A, et al. A role for Fgf-8 in the initiation and maintenance of vertebrate limb bud outgrowth. Curr Biol. 1995;5:797–806. doi: 10.1016/s0960-9822(95)00157-6. [DOI] [PubMed] [Google Scholar]
- Mason I. Initiation to end point: the multiple roles of fibroblast growth factors in neural development. Nat Rev Neurosci. 2007;8:583–596. doi: 10.1038/nrn2189. [DOI] [PubMed] [Google Scholar]
- Mikkola ML. Genetic basis of skin appendage development. Semin Cell Dev Biol. 2007;18:225–236. doi: 10.1016/j.semcdb.2007.01.007. [DOI] [PubMed] [Google Scholar]
- Min H, Danilenko DM, Scully SA, et al. Fgf-10 is required for both limb and lung development and exhibits striking functional similarity to Drosophila branchless. Genes Dev. 1998;12:3156–3161. doi: 10.1101/gad.12.20.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon AM, Capecchi MR. Fgf8 is required for outgrowth and patterning of the limbs. Nat Genet. 2000;26:455–459. doi: 10.1038/82601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubuser A, Peters H, Balling R, et al. Antagonistic interactions between FGF and BMP signaling pathways: a mechanism for positioning the sites of tooth formation. Cell. 1997;90:247–255. doi: 10.1016/s0092-8674(00)80333-5. [DOI] [PubMed] [Google Scholar]
- Nie X, Luukko K, Kettunen P. FGF signalling in craniofacial development and developmental disorders. Oral Dis. 2006;12:102–111. doi: 10.1111/j.1601-0825.2005.01176.x. [DOI] [PubMed] [Google Scholar]
- O'Hagan RC, Tozer RG, Symons MS, et al. The activity of the Ets transcription factor PEA3 is regulated by two distinct MAPK cascades. Oncogene. 1996;13:1323–1333. [PubMed] [Google Scholar]
- Ohazama A, Johnson EB, Ota MS, et al. Lrp4 modulates extracellular integration of cell signaling pathways in development. PLoS ONE. 2008;3:e4092. doi: 10.1371/journal.pone.0004092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohbayashi N, Shibayama M, Kurotaki Y, et al. Fgf18 is required for normal cell proliferation and differentiation during osteogenesis and chondrogenesis. Genes Dev. 2002;16:870–879. doi: 10.1101/gad.965702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi T, Umemura S, Shintani S, et al. Phex mutation causes overexpression of FGF23 in teeth. Arch Oral Biol. 2008;53:99–104. doi: 10.1016/j.archoralbio.2007.08.009. [DOI] [PubMed] [Google Scholar]
- Pispa J, Thesleff I. Mechanisms of ectodermal organogenesis. Dev Biol. 2003;262:195–205. doi: 10.1016/s0012-1606(03)00325-7. [DOI] [PubMed] [Google Scholar]
- Pispa J, Jung HS, Jernvall J, et al. Cusp patterning defect in Tabby mouse teeth and its partial rescue by FGF. Dev Biol. 1999;216:521–534. doi: 10.1006/dbio.1999.9514. [DOI] [PubMed] [Google Scholar]
- Powers CJ, McLeskey SW, Wellstein A. Fibroblast growth factors, their receptors and signaling. Endocr Relat Cancer. 2000;7:165–197. doi: 10.1677/erc.0.0070165. [DOI] [PubMed] [Google Scholar]
- Prochazka J, Pantalacci S, Churava S, et al. Patterning by heritage in mouse molar row development. Proc Natl Acad Sci U S A. 2010;107:15497–15502. doi: 10.1073/pnas.1002784107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roehl H, Nusslein-Volhard C. Zebrafish pea3 and erm are general targets of FGF8 signaling. Curr Biol. 2001;11:503–507. doi: 10.1016/s0960-9822(01)00143-9. [DOI] [PubMed] [Google Scholar]
- Saitsu H, Komada M, Suzuki M, et al. Expression of the mouse Fgf15 gene is directly initiated by Sonic hedgehog signaling in the diencephalon and midbrain. Dev Dyn. 2005;232:282–292. doi: 10.1002/dvdy.20236. [DOI] [PubMed] [Google Scholar]
- Sekine K, Ohuchi H, Fujiwara M, et al. Fgf10 is essential for limb and lung formation. Nat Genet. 1999;21:138–141. doi: 10.1038/5096. [DOI] [PubMed] [Google Scholar]
- Sun X, Lewandoski M, Meyers EN, et al. Conditional inactivation of Fgf4 reveals complexity of signalling during limb bud development. Nat Genet. 2000;25:83–86. doi: 10.1038/75644. [DOI] [PubMed] [Google Scholar]
- Takamori K, Hosokawa R, Xu X, et al. Epithelial fibroblast growth factor receptor 1 regulates enamel formation. J Dent Res. 2008;87:238–243. doi: 10.1177/154405910808700307. [DOI] [PubMed] [Google Scholar]
- Thesleff I, Vaahtokari A, Partanen AM. Regulation of organogenesis. Common molecular mechanisms regulating the development of teeth and other organs. Int J Dev Biol. 1995;39:35–50. [PubMed] [Google Scholar]
- Tucker A, Sharpe P. The cutting-edge of mammalian development; how the embryo makes teeth. Nat Rev Genet. 2004;5:499–508. doi: 10.1038/nrg1380. [DOI] [PubMed] [Google Scholar]
- Tucker AS, Al Khamis A, Sharpe PT. Interactions between Bmp-4 and Msx-1 act to restrict gene expression to odontogenic mesenchyme. Dev Dyn. 1998;212:533–539. doi: 10.1002/(SICI)1097-0177(199808)212:4<533::AID-AJA6>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Umemori H, Linhoff MW, Sanes JR. Fgf22 and its close relatives are presynaptic organizing molecules in the mammalian brain. Cell. 2004;118:257–270. doi: 10.1016/j.cell.2004.06.025. [DOI] [PubMed] [Google Scholar]
- Unda FJ, Martin A, Hernandez C, et al. FGFs-1 and -2, and TGF beta 1 as inductive signals modulating in vitro odontoblast differentiation. Adv Dent Res. 2001;15:34–37. doi: 10.1177/08959374010150010801. [DOI] [PubMed] [Google Scholar]
- Vincentz JW, McWhirter JR, Murre C, et al. Fgf15 is required for proper morphogenesis of the mouse cardiac outflow tract. Genesis. 2005;41:192–201. doi: 10.1002/gene.20114. [DOI] [PubMed] [Google Scholar]
- Wang XP, Suomalainen M, Felszeghy S, et al. An integrated gene regulatory network controls stem cell proliferation in teeth. PLoS Biol. 2007;5:e159. doi: 10.1371/journal.pbio.0050159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson DG. In Situ Hybridization: A Practical Approach. Oxford: IRL Press; 1995. [Google Scholar]
- Wright TJ, Ladher R, McWhirter J, et al. Mouse FGF15 is the ortholog of human and chick FGF19, but is not uniquely required for otic induction. Dev Biol. 2004;269:264–275. doi: 10.1016/j.ydbio.2004.02.003. [DOI] [PubMed] [Google Scholar]
- Yaguchi Y, Yu T, Ahmed MU, et al. Fibroblast growth factor (FGF) gene expression in the developing cerebellum suggests multiple roles for FGF signaling during cerebellar morphogenesis and development. Dev Biol. 2009;238:2058–2072. doi: 10.1002/dvdy.22013. [DOI] [PubMed] [Google Scholar]
- Zhang X, Ibrahimi OA, Olson SK, et al. Receptor specificity of the fibroblast growth factor family The complete mammalian fgf family. J Biol Chem. 2006;281:15694–15700. doi: 10.1074/jbc.M601252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


