Abstract
Amplification of the 8p11-12 region has been found in about 15% of human breast cancer and is associated with poor prognosis. Earlier, we used genomic analysis of copy number and gene expression to perform a detailed analysis of the 8p11-12 amplicon to identify candidate oncogenes in breast cancer. We identified 21 candidate genes and provided evidence that three genes; LSM-1, TC-1 and BAG4 have transforming properties when over expressed. In the present study, we systematically investigated the transforming properties of thirteen newly identified 8p11-12 candidate oncogenes in vitro. We found that WHSC1L1, DDHD2 and ERLIN2 are the most potently transforming oncogenes we tested from the 8p11-12 region based on the number of altered phenotypes expressed by the cells. WHSC1L1 contains a PWWP-domain that is a methyl-lysine recognition motif involved in histone code modification and epigenetic regulation of gene expression. Knock-down of WHSC1L1 in 8p11-12 amplified breast cancer cells resulted in profound loss of growth and survival of these cells. Further, we identified several WHSC1L1 target genes, one of which is iroquois homeobox 3 gene (IRX3), a member of the Iroquois homeobox transcription factor family.
Keywords: gene amplification, breast cancer, WHSC1L1
Introduction
An important mechanism for the activation of oncogenes in human cancers is gene amplification, which results in gene over expression at both the message and protein levels (1, 2). Oncogenes, such as ERBB2 at 17q12, CCND1 at 11q13 and C-MYC at 8p24, have previously been identified as amplification targets linked to the development, progression, or metastasis of human cancers, including breast, prostate, lung and other cancers (2, 3). ERBB2 is the most frequently amplified oncogene in breast cancer, and its over expression is associated with poor clinical outcomes. The prognostic and predictive values of ERBB2 amplification and over expression have been used to guide treatment decisions for patients with both lymph node-positive and negative diseases. More significantly, recognition of the mechanistic roles of ERBB2 in breast cancer has led to the development of ERBB2-targeting drugs such as Trastuzumab to treat breast cancer (4–6).
Amplification of 8p11-12 occurs in approximately 15% of human breast cancer (HBC), and this region of amplification is significantly associated with disease-specific survival and distant recurrence in breast cancer patients (7–11). Chin et al. performed an analysis of the association of gene amplification and disease-free survival and distant relapse in human breast cancer specimens (12). They identified 23 genes from the 8p11-12 region as being correlated with progression. Recently, our laboratory published results of a detailed analysis of copy number and gene expression in the 8p11-12 region in a panel of breast cancer cell lines and primary human breast cancers (13). We identified 21 genes that are over expressed when their copy number is increased (10). Furthermore, we directly tested the transforming function of eight 8p11-12 amplified genes in human mammary epithelial cells. From these experiments, we identified several genes including LSM1, BAG4 and C8orf4 (TC-1) as having the transforming properties in vitro (10, 14, 15). Accumulating evidence suggests that the 8p11-12 amplicon contains multiple candidate oncogenes that could play a role in breast cancer development (7–11).
Recent extensive genomic analyses and siRNA knock-down studies have identified the Wolf-Hirschhorn syndrome candidate 1-like 1 gene (WHSC1L1, also known as NSD3) as one of major candidate oncogenes of the 8p11-12 amplicon in breast cancer (7–11). WHSC1L1 is the third member of a gene family including NSD1 and WHSC1 (NSD2) (16, 17). De novo translocation of NSD1 genes causes the childhood overgrowth syndrome, Sotos syndrome that is associated with elevated risks of cancer; while de novo deletion of NSD2/WHSC1 causes the Wolf-Hirschhorn syndrome that displays growth retardation (18, 19). WHSC1L1/NSD3, NSD1 and WHSC1/NSD2 show strong sequence similarity and share multiple functional domains (16). WHSC1L1 has two isoforms that are derived from alternative splicing of exon 10, and both WHSC1L1 protein isoforms contain a PWWP domain. The PWWP domain belongs to the royal superfamily which includes chromodomain, tudor, malignant brain tumor (MBT) and plant agent motifs, and these domains exist in multiple histone modifying proteins. The N-terminal half of the PWWP domain exhibits a beta-barrel structure that resembles a SAND domain, while the C-terminal portion is made up of a five-helix bundle. Both the crystal and MMR solution structures of the superfamily complexes show that the beta-barrel structure recognizes and binds the histone lysine pocket (20, 21). A study on PWWP function in the DNA methyltransferase DNMT3B demonstrated that the PWWP domain binds methylated DNA (22). Recently, Wang et al. demonstrated that a PWWP domain protein binds to histone lysine in vitro and in vivo, and regulates Set9-mediated H4K20 methylation (23). Their results demonstrated that the PWWP domain is a methyl-lysine recognition motif that plays important roles in epigenetic regulation.
In the present study, we systematically investigated the transforming properties of thirteen newly identified 8p11-12 candidate oncogenes in vitro. We found that WHSC1L1, DDHD2 and ERLIN2 are the most potently transforming oncogenes we tested from the 8p11-12 region based on the number of altered phenotypes expressed by the cells. Knock-down of WHSC1L1 in 8p11-12 amplified breast cancer cells resulted in profound loss of growth and survival of these cells. Further, we identified several WHSC1L1 target genes, one of which is iroquois homeobox 3 gene (IRX3), a member of the Iroquois homeobox transcription factor family.
Materials and Methods
Genomic array CGH
The isolation and culture of the SUM series of human breast cancer cell lines and MCF10A cells have been described in the supplementary Materials and Methods (10, 24). Genomic array CGH experiments were performed using the Agilent 44K human genome CGH microarray chip (Agilent Technologies, Palo Alto, CA). Agilent's CGH Analytics software was used to calculate various measurement parameters, including log2 ratio of total integrated Cy-5 and Cy-3 intensities for each probe.
Semiquantitative RT-PCR reactions
Total RNA was prepared from human breast cancer cell lines and the MCF10A cell line by standard methods (10, 25). For RT-PCR reactions, RNA was converted into cDNA via a reverse transcription reaction using random hexamer primers. Primers were ordered from Invitrogen (Carlsbad, CA) and all the relevant primer sequences are available on request. A GAPDH primer set was used as a control. Semiquantitative RT-PCR was done using the iQSYBR Green Supermix (Bio-Rad, Hercules, CA).
Lentivirus construction and transduction of cells
The lentiviral expression constructs containing the 13 genes tested in the present experiments, stared in Table 1, were established as previously described (10). Briefly, we first created entry clones from cDNA of SUM-44 cells using the pENTR directional TOPO cloning kit and then performed the LR recombination reaction to transfer the gene into the Gateway destination vector, pLenti6/V5-DEST. Specifically, the pLenti-WHSC1L1 construct was established from the full-length short isoform which only contained the PWWP domain. The lentivirus for each construct was generated and used to infect immortalized, nontransformed mammary epithelial MCF10A cells. Control infections with pLenti-LacZ virus were performed in parallel. Selection began 48 hours after infection in growth medium with 10 µg/mL blasticidin in the absence of insulin. Upon confluence, selected cells were passaged and serially cultured.
Table 1.
List of the 21 candidate genes of 8p11-12 region
| Gene | Description | |
|---|---|---|
| ZNF703 | zinc finger protein 703 | |
| * | ERLIN2 | ER lipid raft associated 2 |
| * | PROSC | proline synthetase co-transcribed homolog (bacterial) |
| * | BRF2 | BRF2, subunit of RNA polymerase III transcription initiation factor, BRF1-like |
| RAB11FIP1 | Rab coupling protein = RCP | |
| EIF4EBP1 | elongation factor 4 binding protein 1 | |
| * | ASH2L | ash2 (absent, small, or homeotic)-like (Drosophila) |
| LSM1 | LSM1 homolog, U6 small nuclear RNA associated (S. cerevisiae) | |
| BAG4 | BCL2-associated athanogene 4 | |
| * | DDHD2 | DDHD domain containing 2 |
| * | PPAPDC1B | phosphatidic acid phosphatase type 2 domain containing 1B |
| * | WHSC1L1 | Wolf-Hirschhorn syndrome candidate 1-like 1 |
| * | LETM2 | leucine zipper-EF-hand containing transmembrane protein 2 |
| FGFR1 | fibroblast growth factor receptor 1 (fms-related tyrosine kinase 2, Pfeiffer syndrome) | |
| TACC1 | Transforming acidic coiled-coil | |
| * | TM2D2 | TM2 domain containing 2 |
| C8orf4 | Chromosome 8 open reading frame 4 = TC-1 | |
| * | AP3M2 | adaptor-related protein complex 3, mu 2 subunit |
| * | POLB | polymerase (DNA directed), beta |
| * | VDAC3 | voltage-dependent anion channel 3 |
| * | HOOK3 | hook homolog 3 (Drosophila) |
Growth in soft agar and Matrigel
Soft agar assays were performed as previously described (10). For three-dimensional morphogenesis assays in Matrigel, cells grown in monolayer culture were detached by trypsin/EDTA treatment and seeded in Matrigel (BD Biosciences, San Jose, CA) precoated 8-well chamber slides. The appropriate volume of medium was added and maintained in culture for 10–18 days. Phase-contrast images and immunostained images were photographed with bright-field and confocal microscopy (26).
Lentivirus-mediated shRNA knock-down of gene expression
We knocked down the expression of the human WHSC1L1 gene in breast cancer cell lines, SUM-44 and SUM-52, and in the MCF10A cell line using the Expression Arrest GIPZ lentiviral shRNAmir system (OpenBiosystems, Huntsville, AL). Lentivirus was produced by transfecting 293FT cells with the combination of the lentiviral expression plasmid DNA and Trans-Lentiviral packaging mix (OpenBiosystems. Huntsville, AL). For cell infection, viral supernatants were supplemented with 6 µg/ml polybrene and incubated with cells for 24 h. Cells expressing shRNA were selected with puromycin for 2–3 weeks for functional studies (cell proliferation and colony formation assays) and for 4 to 10 days after infection for RNA extraction.
Results
The effect of different 8p11-12 genes on growth factor independent proliferation
Recently, our group identified 21 candidate oncogenes within the 8p11-12 amplicon in breast cancer based on statistical analysis of copy number increase and gene over expression. We tested eight of the 21 candidate oncogenes for transforming function in vitro and identified three genes, LSM1, BAG4 and C8orf4 (TC-1), that could induce transformed phenotypes (10). In the present report, we expanded our analysis to the remaining 13 candidate oncogenes. Table 1 shows the original 21 gene list with the 13 genes tested in the present experiments highlighted with an asterisk. Details on the origins and sequence validations of each clone are given in Materials and Methods and in supplementary data.
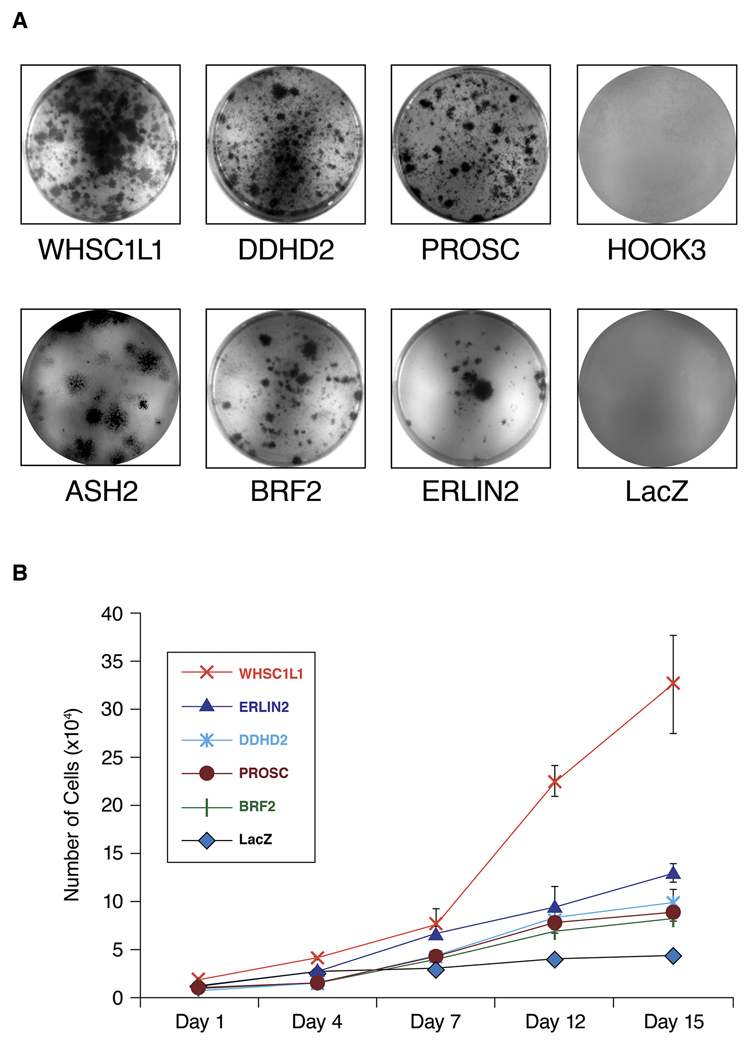
To systematically investigate the transforming properties of thirteen 8p11-12 candidate breast cancer oncogenes, we transduced MCF10A cells, which are highly growth factor dependent, with individual lentiviral expression vectors for each gene. Growth factor independent proliferation of MCF10A cells transduced with each candidate gene was investigated. RT-PCR was performed to confirm the expression of the gene using primers specific for the gene and for the vector. Over expression of WHSC1L1 protein in MCF10A-WHSC1L1 cells was further confirmed by western blot (Supplementary Figure S1). As shown in Figure 1, MCF10A cells expressing 6 genes WHSC1L1, DDHD2, PROSC, BRF2, ASH2L and ERLIN2 formed expanding colonies in insulin-free medium, and then grew continuously in the absence of insulin-like growth factors. Colony formation assays in MCF10A cells with equalized viral titer of the tested genes indicated that over expression of WHSC1L1 and DDHD2 resulted in the highest number of insulin-independent colonies. Growth curves of MCF10A cells over expressing the 5 genes (WHSC1L1, DDHD2, PROSC BRF2, and ERLIN2) were performed within two passages of isolation in insulin-free medium. Data in Figure 1B shows that over expression of WHSC1L1 not only resulted in the largest number of colonies emerging in insulin-free medium, but also gave rise to cells with the most rapid proliferation rate under these conditions. These results extend our previous findings and indicate that a total of 9 genes from the 8p11-12 have ability to induce insulin-like growth factor independent proliferation when over expressed in MCF10A cells.
Figure 1.
(A) MCF10A cells expressing 6 genes WHSC1L1, BRF2, DDHD2, PROSC, ERLIN2 and ASH2L formed expanding colonies in insulin-free medium, while MCF10A cells expressing HOOK3 and control LacZ did not form colonies. (B) In vitro growth rate of the MCF10A cells that stably over express the 5 genes (ERLIN2, WHSC1L1, DDHD2, PROSC and BRF2) relative to MCF10A-LacZ control cells in insulin deficient media. Cells were seeded into 35-mm culture wells and grown in the absence of insulin-like growth factors.
Transforming properties of 8p11-12 candidate genes
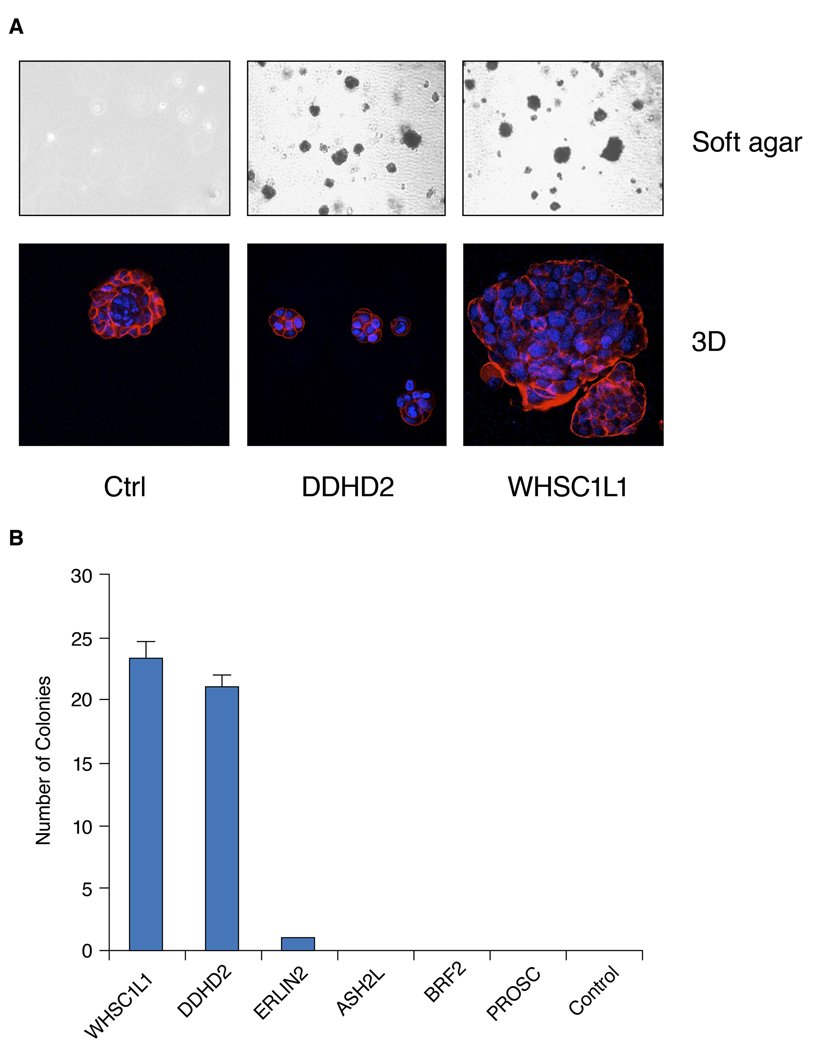
To assess the expression of other transformed phenotypes of MCF10A cells over expressing the newly identified candidate oncogenes, we evaluated each of them for their ability to form colonies in soft agar and for altered morphogenesis in Matrigel. Figure 2 shows that after 3 weeks in culture, MCF10A cells over expressing WHSC1L1, DDHD2, and ERLIN2 formed colonies in soft agar. MCF10A cells over expressing WHSC1L1 and DDHD2 had the highest soft-agar colony forming efficiency (Figure 2B). By contrast, MCF10A cells over expressing ASH2, BRF2, and PROSC did not form soft agar colonies. We also examined whether these six genes affect the growth or morphology of colonies in 3D Matrigel culture, as aberrant behavior in this environment is frequently associated with transformation and/or tumor progression (27). In 3D basement membrane cultures, the immortalized, nontransformed mammary epithelial cells, MCF10A, formed acinar-like structures consisting of a single cell layer of polarized, growth-arrested mammary epithelial cells surrounding a hollow lumen (Figure 2A). As shown in Figure 2A, MCF10A cells over expressing WHSC1L1 formed strikingly abnormal acini that were enlarged, disorganized, and contained filled lumens. In contrast, MCF10A cells over expressing DDHD2 formed disorganized, small abnormal acini. MCF10A cells over expressing ERLIN2 also formed large, highly-proliferative colonies, while insulin-independent MCF10A cells over expressing the other three candidate oncogenes formed polarized, growth-arrested acinar structures with hollow lumens similar to MCF10A parental cells (Data not shown). These experiments demonstrate that three of the transforming genes, PROSC, ASH2L and BRF2, induced insulin-independent growth and no other altered phenotypes. By contrast, WHSC1L1, DDHD2, and ERLIN2 were the most transforming oncogenes based on the number of altered phenotypes expressed by the cells.
Figure 2.
(A) Top: Representative pictures of MCF10A cells that stably over express DDHD2 and WHSC1L1 genes and control cell soft agar colonies. Cells were grown for 3 weeks in soft agar and stained with the vital dye p-iodonitrotetrazolium violet. Bottom: Effects of DDHD2 and WHSC1L1 on mammary acinar morphogenesis. MCF10A-DDHD2, WHSC1L1 and control cells were cultured on a bed of Matrigel as described in Materials and Methods. Representative images of structures with staining for actin with phalloidin conjugated to Alexa Fluor-568 (red), and DAPI as a marker of nuclei (blue). (B) Soft agar colony forming efficiency in MCF10A cells stably over expressing the 6 genes (WHSC1L1, BRF2, DDHD2, PROSC, ERLIN2 and ASH2L) and control cell soft agar colonies. Data represent the mean number of colonies per low power field three weeks after seeding 105 cells per well.
Amplification of WHSC1L1 isoforms in breast cancer
In our analysis of the transforming properties of the 8p11 candidate oncogenes, we were surprised by the potency of WHSC1L1 for transforming MCF-10A cells. As described above, WHSC1L1 over expressing MCF-10A cells exhibited the highest transforming efficiency. The cells had the highest growth rate in insulin-free medium, and the cells grew with high efficiency in soft agar, while forming very abnormal colonies in Matigel. Because of the extrodinary transforming potency of WHSC1L1, the role of this gene as a driver oncogene in breast cancer cell lines and specimens with the amplicon was examined further.
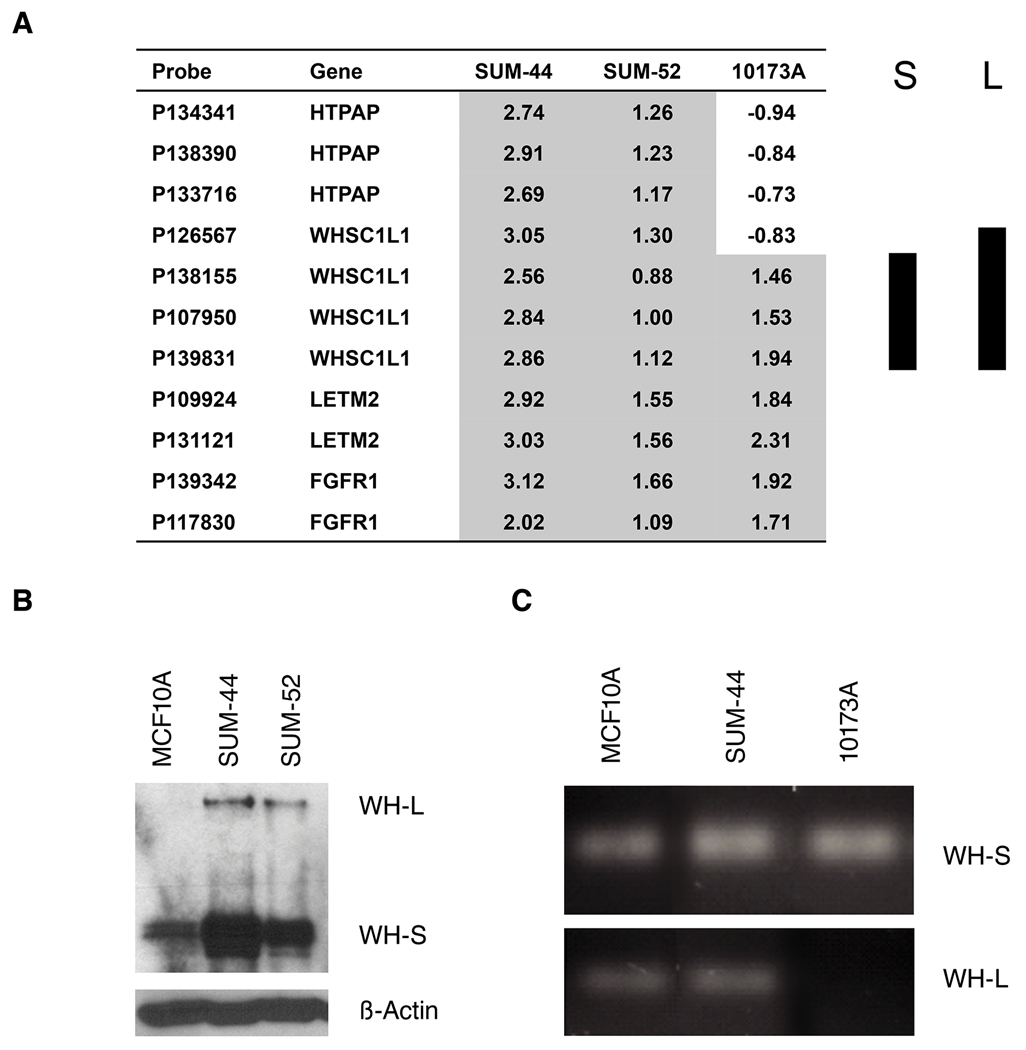
Expression of the WHSC1L1 gene results in two alternatively spliced variants, a long isoform and a short isoform that are derived from alternative splicing of exon 10. The WHSC1L1 long isoform encodes a 1437 amino acid protein containing 2 PWWP domains, 2 PHD-type zinc finger motifs, a TANG2 domain, an AWS domain and a SET domain. The short isoform encodes a 645 amino acid protein containing a PWWP domain only (Supplement Figure S2). Data shown in figure 3 demonstrates that both SUM-44 and SUM-52 cells have amplifications of the full-length gene, but at the protein level expression of the short isoform predominates. The transformation data for WHSC1L1 over expressing MCF10A cells shown above was obtained using an expression construct coding for the short isoform, and similar results were obtained when we transduced MCF10A cells with a vector coding for full length WHSC1L1 (data not shown). Alternative splicing in cancer is an important mechanism for gene regulation and for generating proteomic diversity. Interestingly, we identified one primary breast cancer specimen (10173A) with the 8p11-12 amplicon in which array CGH demonstrated genomic loss of the C-terminal region of the WHSC1L1 long isoform but with amplification of exons 1–10. We validated this finding in that particular breast cancer specimen by genomic PCR using primers specific for the short isoform exon 10 (S-10) and the long isoform exon 20 (L-20) as shown in figure 3C and supplemental figure S2. To further determine whether the WHSC1L1 short isoform protein, which only contains a PWWP domain, is also localized in the nucleus, we generated expression constructs containing the short isoform WHSC1L1 coding sequences fused to the EGFP epitope at the C-terminus. The constructs were transfected into MCF10A and HEK293 cells, and localization of the proteins was examined by fluorescence microscopy. The WHSC1L1 short isoform was localized to the nucleus as expected (Supplementary Figure S3). These results indicate that both WHSC1L1 protein isoforms are localized to the nucleus, and may act as transforming oncoproteins in breast cancer cells bearing the 8p11p12 amplicon.
Figure 3.
(A) Genomic copy number profiles of the WHSC1L1 region analyzed on the Agilent oligonucleotide array CGH in two SUM breast cancer cell lines (SUM-44 and SUM-52) and one primary breast cancer specimen (10173A). Array probes and genes are displayed horizontally by genome position. Log2 ratio in each sample is relative to normal female DNA. S represents WHSC1L1 short isoform and L represents long isoform. (B) WHSC1L1 protein levels were analyzed by western blot in two breast cancer cell lines, SUM-44, SUM-52 and control MCF10A line. (C) Genomic PCR using primers specific for the short isoform exon 10 (S-10) and the long isoform exon 20 (L-20) of WHSC1L1 were used to validate array-CGH data in breast cancer specimen 10173A.
Knock-down of WHSC1L1 inhibits cell proliferation in breast cancer cells
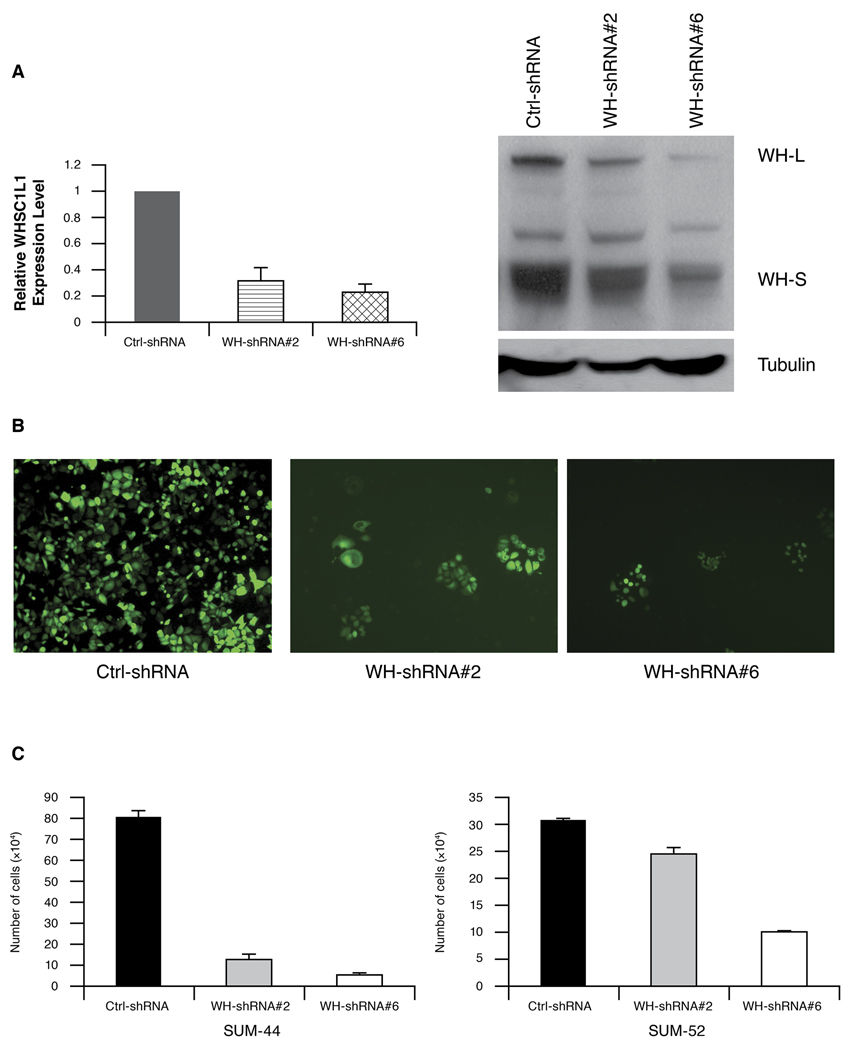
To directly assess the contribution of endogenous WHSC1L1 over expression on the transformation of HBC, we examined the effects of knock-down of WHSC1L1 in SUM-44 and SUM-52 cells where WHSC1L1 is amplified and over expressed, in SUM-149 cells that do not have the amplicon, and in the control cell line MCF10A. To perform RNAi knock-down experiments, we obtained eight pGIPZ-WHSC1L1 shRNA expression constructs from OpenBiosystems. (http://www.openbiosystems.com/). In this vector, TurboGFP and shRNA are part of a bicistronic transcript allowing the visual marking of the shRNA expressing cells. SUM-44, SUM-52 and control MCF10A cells were infected with these 8-shRNA lentivirus supernatants, pooled or individually, to determine which gave the best knock-down of WHSC1L1. Non-silencing shRNAmir lentiviral control, at the same titer as WHSC1L1 shRNA, was used in parallel as the negative control. First, the consequence of knock-down of WHSC1L1 on colony formation using all eight shRNAs was evaluated in all three cell lines. WHSC1L1 knock-down suppressed proliferation of SUM-44 and SUM-52 cells, while WHSC1L1 shRNAs had no effect on the growth of SUM-149 cells or MCF10A cells (Supplementary Figure S4). Next, we identified the two most efficient shRNAs with respect to knock-down of WHSC1L1 expression levels in SUM-44 and SUM-52 cells. Q-RT-PCR and western blot data revealed that the WHSC1L1-shRNAs #2 and #6 resulted in decreases in mRNA and protein levels to approximately 20–30% of the level seen in the non-silencing control-infected cells (Figure 4A). As shown in Figure 4B and C, WHSC1L1 knock-down with both shRNA constructs slowed cell growth of SUM-44 and SUM-52 cells. The results were most striking for SUM-44 cells in which WHSC1L1 knock-down inhibited cell proliferation by ~ 90% (Figure 4C). WHSC1L1 knock-down with these shRNA#2 and #6 had an undetectable effect on the cell growth of MCF10A cells (data not shown). Thus, knock-down of WHSC1L1 inhibits cell proliferation in breast cancer cells with WHSC1L1 gene amplification.
Figure 4.
(A) WHSC1L1 expression in SUM-44 cells was analyzed by semiquantitative RT-PCR and western blot after infection with non-silencing control shRNA or WHSC1L1 specific shRNA (shRNA#2 and #6). (B). The images showed the TurboGFP fluorescence of pGIPZ-WHSC1L1 shRNAs in SUM-44 cells after 3 weeks. (C) shRNA-mediated knock-down of WHSC1L1 inhibits cell growth in breast cancer cells SUM-44 and SUM-52 with WHSC1L1 amplification.
IRX3 is a novel target gene of WHSC1L1
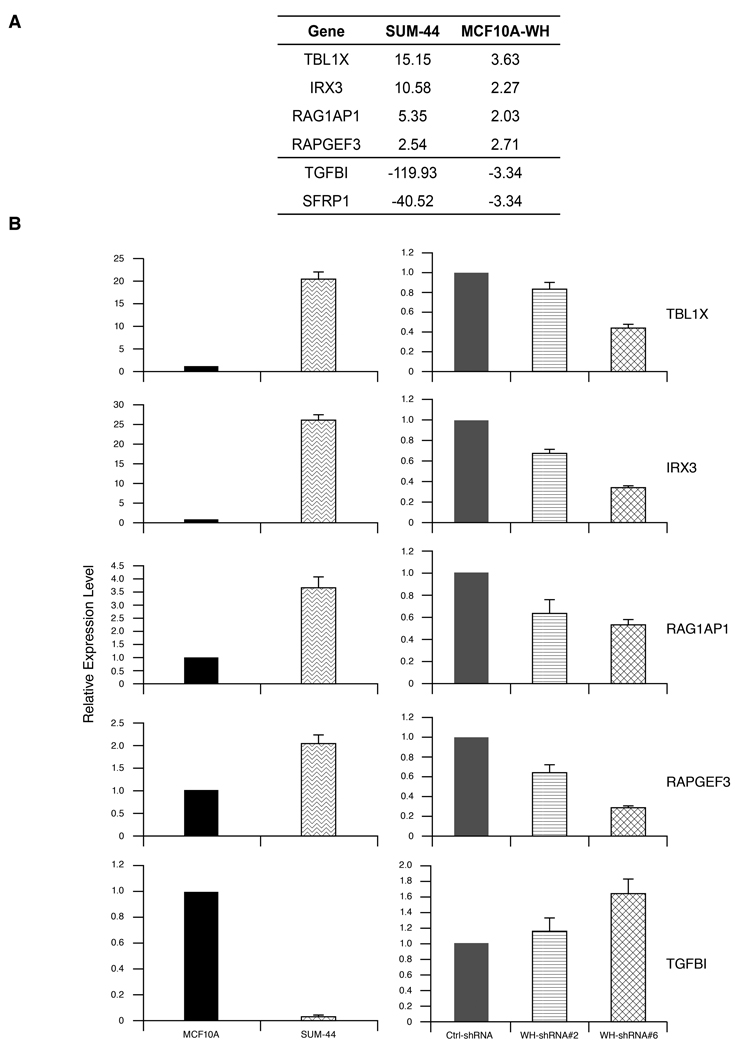
Because WHSC1L1 encodes a PWWP domain nuclear protein that has histone methyl transferase activity, it has been postulated that it can promote malignant transformation by altering the histone code and hence expression of specific target genes. To identify genes that may be altered in their expression by over expression of the short isoform of WHSC1L1, we performed expression profiling of MCF10A cells, MCF10A-WHSC1L1 cells, and SUM-44 cells. To identify genes most likely to be regulated by over expression of WHSC1L1 and relevant to human breast cancer, we determined which genes are differentially expressed in MCF10A-WHSC1L1 cells relative to parental MCF10A cells, and then determined which of those genes are also differentially expressed in SUM-44 cells compared to MCF10A cells. This orthogonal analysis resulted in the identification of 148 genes differentially expressed in both SUM-44 cells and MCF10A-WHSC1L1 cells, relative to MCF10A cells (Supplementary Table). Of the 148 differentially expressed genes, 36 are coordinately up-regulated in MCF10A-WHSC1L1 cells and SUM-44 cells. Figure 5A shows the four up-regulated genes (TBL1X, IRX3, RAG1AP1 and RAPGEF3), and two down-regulated genes (TFBI and SFRP1) in both SUM-44 cells and MCF10A-WHSC1L1 cells, relative to MCF10A cells. To validate these array-based observations, we examined expression of these genes by Q-RT-PCR in SUM-44 and MCF-10 cells, and in SUM-44 cells following knock-down of WHSC1L1. (Figure 5B). Figure 5 shows that IRX3, RAG1AP1, RAPGEF3, and TBL1X are significantly over expressed at the mRNA level in SUM-44 cells compared to MCF10A cells. Furthermore, knock-down of WHSC1L1 in SUM-44 cells using the shRNA constructs described previously resulted in significant down regulation of these four putative target genes. These results support the array-based analysis and indicate that WHSC1L1 regulates the expression of these target genes. Similarly, examination of one down-regulated gene, TGFBI also confirmed reduced expression in SUM-44 compared to MCF10A cells, and the expression of this gene was increased in SUM-44 cells bearing the WHSC1L1 shRNA constructs.
Figure 5.
(A) Six genes differentially expressed in both SUM-44 cells and MCF10A-WHSC1L1 cells, relative to MCF10A cells with the Illumina expression Beadarray. (B) TBL1X , IRX3, RAG1AP1, RAPGEF3 and TGFBI expression level was measured by semiquantitative RT-PCR in MCF10A, SUM-44 cell (left panels), and WHSC1L1 knock-down SUM-44 cells (right panels).
Since WHSC1L1 regulates the expression of IRX3 in SUM-44 cells and MCF10A-WHSC1L1 cells, we examined the genomic state of this potentially important target gene. Interestingly, we found that SUM-44 cells have an amplification at the IRX3 locus of chromosome 16q12. In addition, SUM-225 cells, which also have the 8p11-12 amplicon have an overlapping region of copy number increase in chromosome 16 (Supplementary Figure S5). We performed FISH analysis using an IRX3-specific probe prepared from BAC clone RP11-1061L23 and confirmed the presence of an independent IRX3 amplification in SUM-44 and SUM-225 cells (Supplementary Figure S6). Thus, these findings indicate that in SUM-44 cells, over expression of an amplified oncogene on chromosome 8p11-12 drives the expression of another amplified gene on a different chromosome. This genetic interaction explains the very high level of expression of IRX3 in SUM-44 cells compared to MCF10A-WHSC1L1 cells.
With respect to down regulated genes, the finding that over expression of WHSC1L1 resulted in down regulation of the negative regulator of WNT signaling, SFRP1 in MCF10A cells is intriguing. We have recently demonstrated that even though the SFRP1 gene is part of the 8p11-12 amplicon and is increased in copy number in SUM-44 cells, it is highly methylated and not expressed in these cells (28). Another down regulated gene is TGFBI, which encodes a secreted protein induced by transforming growth factor-β (Figure 5). Recent studies with TGFBI-null mice demonstrated that TGFBI loss promotes cell proliferation and predisposes mice to spontaneous tumor development (29). Thus, PWWP-protein WHSC1L1 may regulate a subset of genes involved in various functional pathways in breast cancer.
Discussion
The 8p11-12 amplicon has been the subject of a number of studies using high-resolution genomic analysis of copy number and gene expression in HBC (7–11, 30). Our first studies in this area demonstrated that the 8p11-12 amplicon has a complex genomic structure and the size of the amplicon is variable in three HBC lines, SUM-44, SUM-52 and SUM-225 (9, 31). In that work, we showed that FGFR1 was only one of several candidate oncogenes in the amplicon, and we provided evidence that FGFR1 is not the driving oncogene in every breast cancer with the 8p11 amplicon (31). In addition, our correlative evidence suggested that other genes in the region, including LSM-1, C8orf4 (TC-1), RAB11FIP1, WHSC1L1 and ERLIN2 were good candidate oncogenes based on their over expression associated with gene amplification (10). Our findings are consistent with those of other laboratories. Gelsi-Boyer et al. performed a comprehensive study combining genomic, expression, and chromosome break analyses of the 8p11-12 region in 37 HBC lines and 134 primary breast cancer specimens. (8). They identified four overlapping amplicon cores at 8p11-12 and 14 candidate oncogenes that are significantly over expressed in relation to amplification. In subsequent work, Bernard-Pierrot et al. carried out BAC-array CGH on 21 HBC lines and 152 ductal breast carcinomas and identified five genes (LSM1, BAG4, DDHD2, PPAPDC1B and WHSC1L1) within the 8p11-12 amplified region as consistently over expressed due to an increased gene copy number. Finally, Chin et al. published an analysis of the association of 8p11-12 gene amplification and disease-free survival and distant relapse in human breast cancer specimens and identified 23 genes from the 8p11-12 region as being correlated with progression, all of which have been named already (12). Thus, several groups have performed extensive analyses of the 8p11-12 genomic region in human breast cancer and there is substantial agreement on the candidate oncogenes present in this region. The candidate oncogenes consistently identified by all groups include, FGFR1, WHSC1L1, RAB11FIP1, LSM1, BAG4 and ERLIN2.
There are now several papers in the literature reporting experimental analysis of the transforming function of the candidate oncogenes from the 8p11-12 region. We reported that BAG4, LSM1, and C8orf4 (TC-1) are transforming when over expressed in MCF10A cells (10, 14, 15). In the present report, we provide evidence that three additional genes WHSC1L1, ERLIN2 and DDHD2 are transforming based on their ability to induce growth factor-independent proliferation, anchorage independent growth, and altered morphogenesis in Matrigel cultures. As reported in this paper, we find WHSC1L1 to be the most potently transforming of all the 8p11 oncogenes we have tested. Our results are consistent with those reported earlier by Bernard-Pierrot et al. who performed RNAi experiments to knock-down the expression of candidate genes in two cell lines (CAMA-1 and ZR-75-1) with 8p11-12 amplification (32). Their results suggest that PPAPDC1B and WHSC1L1 are two driving oncogenes from this amplicon. Knock-down of WHSC1L1 was found to inhibit the proliferation of ZR-75-1 and CAMA-1 cells, but had no effect on MCF-7 cells that lack the 8p11-12 amplicon. Further, inhibition of WHSC1L1 increased the number of apoptotic cells as assessed by TUNEL assay in ZR-75-1 and CAMA-1 cells (32). Recently, Zhang and colleagues applied a novel algorithm, termed TRIAGE (triangulating oncogenes through clinico-genomic intersects), to a collection of microarray expression profiles of primary human breast cancers in an effort to identify candidate genes in amplicons that could contribute to patient outcome (33). They identified RAP11FIP1 and also identified WHSC1L1 as being strongly associated with breast cancer subtype and outcome. They selected RAP11FIP1 for further transfection and knock-down studies and found that RAP11FIP1 is not sufficient to transform naive cells. However, over expression of RAP11FIP1 in breast cancer cell lines caused decreased growth factor dependence, increased survival under anoikis conditions, and increased motility and invasion. Furthermore, RAP11FIP1 over expression caused an epithelial-mesenchymal transition in vitro and increased tumor growth in vivo (33). In other studies, Luscher-Firzlaff et al. reported that ASH2L encodes the trithorax protein and cooperates with H-Ras to transform primary rat embryo fibroblasts (34).
Based on findings from several laboratories, WHSC1L1 is clearly emerging as an important transforming gene within the 8p11-12 amplicon in breast and other cancer types. WHSC1L1 is involved in a chromosomal translocation in acute myeloid leukemia, t(8;11)(p11.2;p15) (35). Recent published database of the Affymetrix 250K Sty array in a collection of 244 copy-number profiles of breast cancer samples demonstrated that WHSC1L1 amplification occurred in ~15% samples (Supplementary Figure S7) (36). With a CGH analysis program, GISTIC (Genomic Identification of significant Targets in Cancer), WHSC1L1 was identified at the peak of amplification in lung and esophageal squamous cell carcinoma (SCC) (37). siRNA-mediated knock-down of WHSC1L1 resulted in a 50% reduction in the number of soft agar colonies in a lung cancer cell line (H1703) with WHSC1L1 gene amplification and over expression (38). Furthermore, deep sequencing of a primary human breast cancer identified a deletion within the WHSC1L1 gene (39). In this study, we identified one primary breast cancer specimen with the 8p11-12 amplicon in which genomic analysis demonstrated a loss of the C-terminal region of the WHSC1L1 long isoform but amplification of exons 1–10 that coded for the short isoform. At the protein level, WHSC1L1 exists as two isoforms in breast cancer cells with 8p11-12 amplification. Alternative splicing of WHSC1L1 in breast cancer cells can be regulated at different steps of the spliceosome assembly by different splicing factors, and by many different mechanisms that rely on cis-acting elements (40). Future investigations are required to more precisely address the role and mechanism of action of WHSC1L1 isoforms in breast cancer,
A finding of particular interest from our study is that IRX3, a member of the homeobox gene family, and TBL1X are target genes of WHSC1L1. Interestingly, IRX3 is also amplified in SUM-44 cells and in SUM-225 cells. This is of interest because in embryonic stem cells IRX3 and TBL1X are linked in a gene expression network that regulates WNT signaling (41). In addition, we have previously shown that in breast cancers with the 8p11-12 amplicon, SFRP1, a negative regulator of WNT signaling, is silenced by promoter methylation, despite being present on the 8p11 amplicon and increased in copy number (28). These results suggest that over expression of WHSC1L1 and the silencing of SFRP1 result in potent activation of a transcriptional network linked to WNT signaling and expression of stem cell phenotypes.
FGFR1 has long been considered to be an important candidate breast cancer oncogene from the 8p11-12 region. However, we have consistently failed to find evidence for a direct role of FGFR1 in transformation in mammary epithelial cells. Recently, Turner et al. provided evidence for a functional role of FGFR1 in 8p11-12 amplified breast cancers (42). Many of the results reported by Turner et al. are consistent with our previously published negative results. However, they did demonstrate that over expression of FGFR1 increases the sensitivity and responsiveness of cells to FGF ligands, which influences the response of the cells to 4-OH Tamoxifen. These results suggest that FGFR1 over expression can play a role in endocrine therapy resistance, which may explain the consistent presence of FGFR1 in the amplicon‥
In the past, focal amplicons found in cancer specimens were considered to harbor a single driving oncogene that was responsible for the maintenance of the amplicon in the tumor, with the ERBB2 oncogene in the 17q12 amplicon being a prime example. In some cases, amplicons have been thought to harbor more than one driver oncogene that act independently such as the CCND1 and EMS1 genes present in the 11q12 amplicon. It is possible that the 8p11-p12 amplicon does not follow such a simple paradigm. Indeed, we have proposed that the 8p11-12 amplicon, rather than having a single driving oncogene, can act as an oncogenic unit consisting of multiple interacting transforming genes. This hypothesis is based on the consistent co-expression of several candidate oncogenes with transforming function when the amplicon is present in breast cancers. Within this oncogenic unit are two genes that can regulate the histone code (WHSC1L1, ASH2L), two genes that regulate RNA metabolism (LSM1, BRF2), a receptor tyrosine kinase (FGFR1), a gene that regulates the endoplasmic reticulum stress pathway (ERLIN2), and a gene that influences receptor trafficking (RAB11FIP1). While it remains possible that each of these genes act independently, and function as driver oncogenes in different tumors with the same amplicon, the possibility that the genes cooperate in mediating neoplastic transformation must now be considered.
Despite the significant and exciting progress in the understanding of the 8p11-12 genomic amplification in breast cancer, we are still in the early stages of functional studies for each 8p11-12 candidate oncogene and their role in breast cancer development. Understanding how the genes in this region influence fundamental cancer processes such as progression, metastasis, and drug resistance will provide potential new avenues for therapeutic development.
Supplementary Material
Achnowledgements
This work was supported by a grant from the Department of Defense Breast Cancer Program (BC083945) to Zeng-Quan Yang and a grant from the National Institutes of Health (RO1 CA100724) to Stephen P. Ethier. We thank Michele L. Dziubinski and Katie L. Streicher for technical assistance on the cell culture and three-dimensional morphogenesis assays. The array CGH work was facilitated by the Microarray and Bioinformatics Core Facility of the Wayne State University Environmental Health Sciences Center, NIEHS P30 ES06639.
Footnotes
Accession Numbers: The normalized data have been deposited in the GEO (GSE23734).
References
- 1.Albertson DG. Gene amplification in cancer. Trends Genet. 2006;22:447–455. doi: 10.1016/j.tig.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 2.Albertson DG, Collins C, McCormick F, Gray JW. Chromosome aberrations in solid tumors. Nature genetics. 2003;34:369–376. doi: 10.1038/ng1215. [DOI] [PubMed] [Google Scholar]
- 3.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nature medicine. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 4.Moasser MM. The oncogene HER2: its signaling and transforming functions and its role in human cancer pathogenesis. Oncogene. 2007 doi: 10.1038/sj.onc.1210477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arteaga CL, Moulder SL, Yakes FM. HER (erbB) tyrosine kinase inhibitors in the treatment of breast cancer. Seminars in oncology. 2002;29:4–10. doi: 10.1053/sonc.2002.34047. [DOI] [PubMed] [Google Scholar]
- 6.Badache A, Goncalves A. The ErbB2 signaling network as a target for breast cancer therapy. J Mammary Gland Biol Neoplasia. 2006;11:13–25. doi: 10.1007/s10911-006-9009-1. [DOI] [PubMed] [Google Scholar]
- 7.Garcia MJ, Pole JC, Chin SF, et al. A 1 Mb minimal amplicon at 8p11-12 in breast cancer identifies new candidate oncogenes. Oncogene. 2005;24:5235–5245. doi: 10.1038/sj.onc.1208741. [DOI] [PubMed] [Google Scholar]
- 8.Gelsi-Boyer V, Orsetti B, Cervera N, et al. Comprehensive profiling of 8p11-12 amplification in breast cancer. Mol Cancer Res. 2005;3:655–667. doi: 10.1158/1541-7786.MCR-05-0128. [DOI] [PubMed] [Google Scholar]
- 9.Yang ZQ, Albertson D, Ethier SP. Genomic organization of the 8p11-p12 amplicon in three breast cancer cell lines. Cancer genetics and cytogenetics. 2004;155:57–62. doi: 10.1016/j.cancergencyto.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 10.Yang ZQ, Streicher KL, Ray ME, Abrams J, Ethier SP. Multiple interacting oncogenes on the 8p11-p12 amplicon in human breast cancer. Cancer research. 2006;66:11632–11643. doi: 10.1158/0008-5472.CAN-06-2946. [DOI] [PubMed] [Google Scholar]
- 11.Pole JC, Courtay-Cahen C, Garcia MJ, et al. High-resolution analysis of chromosome rearrangements on 8p in breast, colon and pancreatic cancer reveals a complex pattern of loss, gain and translocation. Oncogene. 2006;25:5693–5706. doi: 10.1038/sj.onc.1209570. [DOI] [PubMed] [Google Scholar]
- 12.Chin K, DeVries S, Fridlyand J, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 13.Yang ZQ, Streicher KL, Ray ME, Abrams J, Ethier SP. Multiple interacting oncogenes on the 8p11-p12 amplicon in human breast cancer. Cancer Res. 2006;66:11632–11643. doi: 10.1158/0008-5472.CAN-06-2946. [DOI] [PubMed] [Google Scholar]
- 14.Yang ZQ, Moffa AB, Haddad R, Streicher KL, Ethier SP. Transforming properties of TC-1 in human breast cancer: interaction with FGFR2 and beta-catenin signaling pathways. International journal of cancer. 2007;121:1265–1273. doi: 10.1002/ijc.22831. [DOI] [PubMed] [Google Scholar]
- 15.Streicher KL, Yang ZQ, Draghici S, Ethier SP. Transforming function of the LSM1 oncogene in human breast cancers with the 8p11-12 amplicon. Oncogene. 2007;26:2104–2114. doi: 10.1038/sj.onc.1210002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stec I, van Ommen GJ, den Dunnen JT. WHSC1L1, on human chromosome 8p11.2, closely resembles WHSC1 and maps to a duplicated region shared with 4p16.3. Genomics. 2001;76:5–8. doi: 10.1006/geno.2001.6581. [DOI] [PubMed] [Google Scholar]
- 17.Angrand PO, Apiou F, Stewart AF, Dutrillaux B, Losson R, Chambon P. NSD3, a new SET domain-containing gene, maps to 8p12 and is amplified in human breast cancer cell lines. Genomics. 2001;74:79–88. doi: 10.1006/geno.2001.6524. [DOI] [PubMed] [Google Scholar]
- 18.Bergemann AD, Cole F, Hirschhorn K. The etiology of Wolf-Hirschhorn syndrome. Trends Genet. 2005;21:188–195. doi: 10.1016/j.tig.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 19.Rahman N. Mechanisms predisposing to childhood overgrowth and cancer. Curr Opin Genet Dev. 2005;15:227–233. doi: 10.1016/j.gde.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 20.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Molecular cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 21.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen T, Tsujimoto N, Li E. The PWWP domain of Dnmt3a and Dnmt3b is required for directing DNA methylation to the major satellite repeats at pericentric heterochromatin. Mol Cell Biol. 2004;24:9048–9058. doi: 10.1128/MCB.24.20.9048-9058.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Reddy B, Thompson J, et al. Regulation of Set9-mediated H4K20 methylation by a PWWP domain protein. Molecular cell. 2009;33:428–437. doi: 10.1016/j.molcel.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forozan F, Veldman R, Ammerman CA, et al. Molecular cytogenetic analysis of 11 new breast cancer cell lines. British journal of cancer. 1999;81:1328–1334. doi: 10.1038/sj.bjc.6695007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang ZQ, Imoto I, Fukuda Y, et al. Identification of a novel gene, GASC1, within an amplicon at 9p23-24 frequently detected in esophageal cancer cell lines. Cancer research. 2000;60:4735–4739. [PubMed] [Google Scholar]
- 26.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods (San Diego, Calif. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 27.Debnath J, Brugge JS. Modelling glandular epithelial cancers in three-dimensional cultures. Nature reviews. 2005;5:675–688. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- 28.Yang ZQ, Liu G, Bollig-Fischer A, Haddad R, Tarca AL, Ethier SP. Methylation-associated silencing of SFRP1 with an 8p11-12 amplification inhibits canonical and non-canonical WNT pathways in breast cancers. International journal of cancer. 2009;125:1613–1621. doi: 10.1002/ijc.24518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, Wen G, Shao G, et al. TGFBI deficiency predisposes mice to spontaneous tumor development. Cancer research. 2009;69:37–44. doi: 10.1158/0008-5472.CAN-08-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haverty PM, Fridlyand J, Li L, et al. High-resolution genomic and expression analyses of copy number alterations in breast tumors. Genes Chromosomes Cancer. 2008;47:530–542. doi: 10.1002/gcc.20558. [DOI] [PubMed] [Google Scholar]
- 31.Ray ME, Yang ZQ, Albertson D, et al. Genomic and expression analysis of the 8p11-12 amplicon in human breast cancer cell lines. Cancer research. 2004;64:40–47. doi: 10.1158/0008-5472.can-03-1022. [DOI] [PubMed] [Google Scholar]
- 32.Bernard-Pierrot I, Gruel N, Stransky N, et al. Characterization of the recurrent 8p11-12 amplicon identifies PPAPDC1B, a phosphatase protein, as a new therapeutic target in breast cancer. Cancer research. 2008;68:7165–7175. doi: 10.1158/0008-5472.CAN-08-1360. [DOI] [PubMed] [Google Scholar]
- 33.Zhang J, Liu X, Datta A, et al. RCP is a human breast cancer-promoting gene with Ras-activating function. The Journal of clinical investigation. 2009;119:2171–2183. doi: 10.1172/JCI37622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luscher-Firzlaff J, Gawlista I, Vervoorts J, et al. The human trithorax protein hASH2 functions as an oncoprotein. Cancer research. 2008;68:749–758. doi: 10.1158/0008-5472.CAN-07-3158. [DOI] [PubMed] [Google Scholar]
- 35.Rosati R, La Starza R, Veronese A, et al. NUP98 is fused to the NSD3 gene in acute myeloid leukemia associated with t(8;11)(p11.2;p15) Blood. 2002;99:3857–3860. doi: 10.1182/blood.v99.10.3857. [DOI] [PubMed] [Google Scholar]
- 36.Beroukhim R, Mermel CH, Porter D, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bass AJ, Watanabe H, Mermel CH, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nature genetics. 2009;41:1238–1242. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tonon G, Wong KK, Maulik G, et al. High-resolution genomic profiles of human lung cancer. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:9625–9630. doi: 10.1073/pnas.0504126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guffanti A, Iacono M, Pelucchi P, et al. A transcriptional sketch of a primary human breast cancer by 454 deep sequencing. BMC genomics. 2009;10:163. doi: 10.1186/1471-2164-10-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferrell CM, Dorsam ST, Ohta H, et al. Activation of stem-cell specific genes by HOXA9 and HOXA10 homeodomain proteins in CD34+ human cord blood cells. Stem cells (Dayton, Ohio) 2005;23:644–655. doi: 10.1634/stemcells.2004-0198. [DOI] [PubMed] [Google Scholar]
- 42.Turner N, Pearson A, Sharpe R, et al. FGFR1 Amplification Drives Endocrine Therapy Resistance and Is a Therapeutic Target in Breast Cancer. Cancer research. 70:2085–2094. doi: 10.1158/0008-5472.CAN-09-3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.