Abstract
Radiation metabolomics has aided in the identification of a number of biomarkers in cells and mice by ultra-performance liquid chromatography-coupled time-of-flight mass spectrometry (UPLC-ESI-QTOFMS) and in rats by gas chromatography-coupled mass spectrometry (GCMS). These markers have been shown to be both dose- and time-dependent. Here UPLC-ESI-QTOFMS was used to analyze rat urine samples taken from 12 rats over 7 days; they were either sham-irradiated or γ-irradiated with 3 Gy after 4 days of metabolic cage acclimatization. Using multivariate data analysis, nine urinary biomarkers of γ radiation in rats were identified, including a novel mammalian metabolite, N-acetyltaurine. These upregulated urinary biomarkers were confirmed through tandem mass spectrometry and comparisons with authentic standards. They include thymidine, 2′-deoxyuridine, 2′deoxyxanthosine, N1-acetylspermidine, N-acetylglucosamine/galactosamine-6-sulfate, N-acetyltaurine, N-hexanoylglycine, taurine and, tentatively, isethionic acid. Of these metabolites, 2′-deoxyuridine and thymidine were previously identified in the rat by GCMS (observed as uridine and thymine) and in the mouse by UPLC-ESI-QTOFMS. 2′Deoxyxanthosine, taurine and N-hexanoylglycine were also seen in the mouse by UPLC-ESI-QTOFMS. These are now unequivocal cross-species biomarkers for ionizing radiation exposure. Downregulated biomarkers were shown to be related to food deprivation and starvation mechanisms. The UPLC-ESI-QTOFMS approach has aided in the advance for finding common biomarkers of ionizing radiation exposure.
INTRODUCTION
Identifying noninvasive biomarkers of ionizing radiation would permit rapid assessment and triage of exposed individuals for palliative and follow-up care. It may also facilitate the elucidation of novel molecular mechanisms associated with the ionizing radiation DNA damage and repair response. Historically biomarkers of radiation have been those related to DNA damage [e.g., γ-H2AX (1)] and gene expression changes [e.g., XPC (2, 3), CDKN1A (4), GADD45 (4), MDM2 (5)], but small molecule indicators have not been explored or developed as extensively. A benefit of using small molecule screening (i.e., metabolomics) compared to other “omics” technologies such as proteomics, genomics or transcriptomics is that the data produced from the latter may not accurately reflect the current physiological status. Metabolites, as the end products of cellular processes, are likely to reflect any changes occurring at the epigenomic, genomic, proteomic or transcriptomic level. Furthermore, first-pass assessment of exposed individuals with urinary biomarkers measured by miniaturized devices using differential mobility spectrometry can be done in the field quickly and accurately. Exposed individuals can then be assessed later using more time-intensive “gold standard” biodosimetry approaches such as dicentric analysis.
Radiation metabolomics has been used successfully to identify numerous biomarkers in cells (6) and in mouse urine using ultra-performance liquid chromatography (UPLC) coupled with electrospray ionization time-of-flight mass spectrometry (ESI-QTOFMS) (7, 8) and in rat urine using gas chromatography-coupled mass spectrometry (GCMS) (9). The numerous analytical methods employed in these studies have provided broad coverage of the many chemical constituents in urine, both polar and nonpolar, and reassuringly have all pointed to similar dose-responsive urinary constituents.
Here we analyzed urine samples from γ-irradiated rats by UPLC-ESI-QTOFMS. This study used UPLC-ESI-QTOFMS-based metabolomics to analyze 24-h urine samples collected from rats housed in metabolism cages for 4 days before irradiation and 3 days after sham/γ irradiation. The aim was to uncover novel biomarkers of radiation exposure and identify already known biomarkers seen in rats and other species, which could aid in the development of rapid screening protocols for radiation-exposed individuals. Using multiple platforms for analysis of the irradiated rat urine samples has provided an effective method for biomarker discovery and revealed the optimal number of metabolites and the limitations of each platform.
MATERIALS AND METHODS
Compounds
Isethionic acid, N8-acetylspermidine dihydrochloride, spermidine, N-acetyl-D-galactosamine 6-sulfate sodium salt, N-acetyl-D-glucosamine 6-sulfate sodium salt, 2′-deoxyuridine (dU), 2′-deoxycytidine (dC), thymidine (dT), pimelic acid, taurine, azelaic acid, dodecanedioic acid, chlorpropamide, N-hexanoylglycine, xanthine and xanthosine were all obtained from Sigma Aldrich (St. Louis, MO). N-isovalerylglycine was obtained from TCI America (Portland, OR). 5-Methyl-2′-deoxycytidine (5-MedC) was purchased from MP Biomedicals (Solon, OH). 2′-Deoxyxanthosine (dX) was a kind gift from Dr. Peter Dedon, Department of Biological Engineering, MIT. All other chemicals were of the highest purity grade.
Synthesis
To verify the identities of N-acetyltaurine and N1-acetylspermidine, they were synthesized in-house. For N-acetyltaurine, taurine (0.5 mg) was stirred under low heat in water (7 ml). Pyridine (2.5 ml) and acetic anhydride (2 ml) were added, stirred at 4°C for 2 h, and then left overnight at 4°C. The solvent was then removed under a steady stream of nitrogen gas and a solid mass formed. Ethyl alcohol and dichloromethane were added to the solid and boiled in a fume hood. The sample was frozen and resuspended in ice-cold ethyl alcohol. The solution was filtered with additional ice-cold ethanol and the filter paper was left to dry on a watch glass. The crystals were scraped off and dried under vacuum. The yield was 429 mg (32%). The sample was sent for 1H NMR spectroscopic analysis for confirmation. N1-Acetylspermidine was synthesized using a method similar to that proposed by Tabor et al. (10). A solution of spermidine in water and pyridine was treated with two equivalent volumes of acetic anhydride and stirred for 3 days at ambient temperature. The mixture was evaporated and water was added; the solution was then acidified to pH 1–2 with 2 M HCl. The sample was concentrated under low pressure and formed a white solid. This was extracted with 2-propanol and filtered to remove spermidine trihydrochloride. The filtrate was cooled to −20°C to precipitate N1-acetylspermidine as a white solid, which was collected on a sintered glass funnel. 1H-NMR of N1-acetylspermidine 2HCl in D2O showed 1H-NMR (400MHz, D2O) δ 3.19 (t, 2H, J = 6.7 Hz, CH3CONHCH2), 2.97 (m, 6H, CH2NHCH2 and CH2NH2), 1.91 (s, 3H, CH3CO), 1.80 (m, 2H, CH3CONHCH2CH2), 1.67 (m, 4H, CH2CH2CH2NH2).
Radiation Exposure and Urine Collection
The radiation exposure and urine control methods are described in Lanz et al. (9). In brief, 12 male Wistar rats were age-matched and fed with no. 3430 mouse and rat diet (Provimi Kliba AG, Kaiseraugst, Switzerland) ad libitum. The rats were housed individually in metabolic cages for 7 days and urine was collected every 24 h; days were numbered −4, −3, −2, −1, +1, +2, +3, where − and + refer to preirradiation and postirradiation, respectively. The urine was collected and diluted to a volume of 14 ml with purified water after rinsing the metabolic cages, except for day +1, when the final volume was 20 ml. The urine samples were then frozen at −20°C. After day −1 the rats were removed from the metabolic cages and half were subjected to sham irradiation and half were exposed to 3.0 Gy of γ radiation. Use of direct-reading dosimeters revealed the dose received by the rats was 2.80–2.85 Gy. After radiation exposure the rats were placed back in their respective metabolic cages and urine was collected for the next 3 days (days +1 to +3).
Sample Preparation for UPLC-ESI-QTOFMS
Urine samples were thawed and 50 μl was added to a microcentrifuge tube containing 50 μl 50:50 acetonitrile:water and 5 μM chlorpropamide kept at 4°C. The samples were vortexed for 1 min each and centrifuged at 13,000g for 20 min at 4°C to remove proteins and particulates. The supernatant was transferred to a UPLC vial. A pooled sample containing 5 μl of each sample was also made for quality control.
UPLC-ESI-QTOFMS Analysis
The samples were randomized and analyzed by UPLC-ESI-QTOFMS as described previously (7). In brief, the following mobile-phase linear gradient consisting of 0.1% formic acid (A) and acetonitrile containing 0.1% formic acid (B) was used with a flow rate of 0.5 ml/min: 98% A for 0.5 min to 20% B at 4.0 min to 95% B at 8 min. The column was washed with 100% B for 1 min and then equilibrated with 100% A before subsequent injections. The pooled sample was injected after every seven samples. In brief, samples were injected onto a reverse-phase 50 × 2.1-mm ACQUITY® 1.7 μl C18 column (Waters Corp, Milford, MA) using an ACQUITY® UPLC system (Waters) with a gradient mobile phase of 0.1% formic acid (solution A) and acetonitrile containing 0.1% formic acid (solution B). Mass spectrometry was performed on a Waters SYNAPT-MS operating in negative and positive electrospray ionization (ESI) mode.
Multivariate Data Analysis and Biomarker Identification
The mass spectral data were centroided, integrated and deconvoluted to generate a multivariate data matrix using MarkerLynx® (Waters). Peak picking, alignment, deisotoping and integration were performed automatically by the software with the following parameters: mass tolerance = 0.05 u, peak width at 5% height = 1 s, peak-to-peak baseline noise 10, intensity threshold = 100 counts, mass window = 0.05 Da, retention time window = 0.20 min, and noise elimination level = 10. The raw data were transformed into a multivariate matrix containing aligned peak areas with matched mass-to-charge ratios and retention times. The data were normalized to the peak area of the internal standard chlorpropamide, which appeared at a retention time of 5.3 min, 275.024 [M-H]− and 277.041 [M+H]+and exported in SIMCA-P+ software (Umetrics, Kinnelon, NJ). The ESI+ and ESI− data were Pareto-scaled to increase the importance of low-abundance ions without significant amplification of noise and analyzed by principal components analysis (PCA) and orthogonal projection to latent structures discriminate analysis (OPLS-DA). For identification of biomarkers specific to γ radiation, OPLS-DA models were constructed comparing control on day −1 and day +1 sham (y = 0), to those γ-irradiated on day +1 (y = 1), and was repeated for day +2 and day +3 where day −1 and sham were maintained as controls. Ions with a pcorr value above 0.8 and peak area above 100 were subjected to tandem MS. Further confirmation of identity was then carried out by repeating the tandem MS fragmentation using authentic standards at 100 μM in water and in urine.
Quantification
Biomarkers were quantified by multiple reaction monitoring (MRM) on an Acquity UPLC coupled to a XEVO triple-quadrupole tandem MS (Waters). This was to obtain the actual concentration of each metabolite normalized to the endogenous creatinine levels. Standard calibration curves were made for creatinine, taurine, dU, dX, dT, dC, N-acetyltaurine, N-acetylgalactosamine-6-sulfate, N1-acetylspermidine, N-hexanoylglycine, pimelic acid, azelaic acid, dodecanedioic acid and N-isovalerylglycine using authentic standards. Urine samples were deproteinized in 50% acetonitrile and diluted 1:2, except for pimelic acid (1:64), dU (1:64) and azelaic acid (1:512). An internal standard of chlorpropamide was added to each sample at a final concentration 1 μM. The samples were quantified using TargetLynx (Waters) software.
Statistics
All concentrations were expressed as means ± SEM after one-way ANOVA using GraphPad Prism 4 software. For comparisons between preirradiation and postirradiation samples, Bonferroni’s correction for multiple comparisons was performed. Days −3, −2 and −1 were compared to days +1, +2, +3 sham and irradiated; a comparison with P < 0.05 was considered to be statistically significant. Day −4 was not used for comparison due to increases in metabolites related to acclimatization in the metabolic cages.
RESULTS
Multivariate Data Analysis
Unsupervised PCA analyses were initially carried out and samples were seen to cluster by day and dose (2.8–2.85 Gy or sham irradiation). The aims of these analyses were to identify any systematic variation in the data and to find patterns or groupings. All the observations from the ESI+ MS data fitted within the 95% confidence interval of the modeled variation, while for the ESI− MS data there were three outliers in one class of samples (day −4). The PCA score scatter plots constructed using the ESI+ and ESI− data from all the rat urine samples are shown in Fig. 1A and B, respectively. The samples collected before irradiation, days −4 to −1, visibly cluster by day in Fig. 1A. On day −4 the rats were removed from group housing and placed in metabolic cages. The urine collected from those rats contained metabolites that caused a clear separation from the rest of the observations. As the rats became acclimatized from days −3 to −1, the urine samples are seen to cluster within their day classes, but they also overlap between those days; the variation between those samples was small. For the ESI− data, day −4 is separated from the rest of the observations, but days −3 to −1 cluster together and are not clearly separated by day class as seen with the ESI+ mode data. After the rats were taken from their cages to be sham irradiated or γ-irradiated, the PCA scores plots separate again. The samples from the γ-irradiated rats on days +1 to +3 are clustered together and are completely separated from the other observations.
FIG. 1.
PCA scores plots from 24-h rat urine samples analyzed by UPLC-ESI-QTOFMS ESI+ (panel A) UPLC-ESI-QTOFMS ESI− (panel B). Black squares, day −4; black triangles, day −3; black circles, day −2; black diamonds day −1; blue squares, day 1 sham-irradiated; red squares, day +1 irradiated with 3 Gy; inverted blue triangles, day +2 sham-irradiated; inverted red triangles, day +2 irradiated with 3 Gy; blue diamonds, day +3 sham-irradiated; red diamonds, day +3 irradiated with 3 Gy.
Variables Contributing to Satellite Day −4 Grouping by PCA
The variables contributing to the separation of the day −4 urine samples from those collected on days −3 to −1 were identified from the PCA loadings plot in both ESI− and ESI+ mode data and PCA loadings plot from the ESI− data can be seen in Fig. 2. These ions were subjected to tandem MS fragmentation and compared to authentic standards. They were identified as hippurate, phenylacetylglycine (PAG), 2,8-quinolinediol and 2,8-quinolinediol glucuronide.
FIG. 2.
PCA loadings plot of ESI− urine data run by UPLC-ESI-QTOFMS. Metabolites shown are upregulated at day −4.
Metabolomic Analysis to Identify Biomarkers of γ-Radiation Exposure
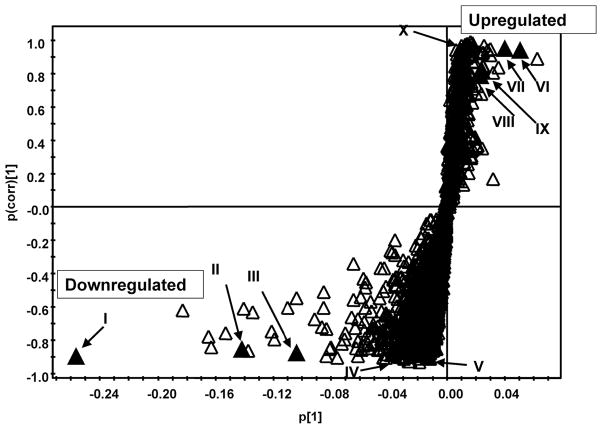
To identify upregulated and downregulated biomarkers of radiation exposure, day −1 samples were compared to day +1, +2 and +3 samples individually in both ESI+ and ESI− modes through construction of OPLS-DA models. By day −1 the rats were acclimatized to the metabolic cages as confirmed by the lack of variation seen between the day −3 to −1 samples in Fig. 1. An example of an OPLS-DA S-plot is shown in Fig. 3, which compares the data from day −1 samples to day +1 γ-irradiated samples. The ions in the bottom lefthand quadrant are those that decreased 1 day after γ irradiation and in the top righthand quadrant increased after 1 day postirradiation. The ions with a correlation coefficient (pcorr value) greater than 0.8 were investigated further by database searching, tandem MS fragmentation, and confirmation using authentic standards. The tandem MS fragmentation patterns of samples and standards can be seen in Fig. 4, and the confirmed ions are listed in Tables 1 (upregulated biomarkers) and 2 (downregulated biomarkers). The tables also show the pcorr values of the metabolites taken from OPLS-DA modeling from comparisons of day −1 samples to days +1, +2 and +3. There were a number of ions observed by OPLS-DA that were highly correlated to radiation exposure but could not be identified after fragmentation or from searching against databases. The confirmed upregulated and downregulated ions were quantified; in addition, taurine, xanthine, xanthosine and N-hexanoylglycine were quantified because these were observed to be upregulated in mouse urine after irradiation but were not seen to be correlated here after OPLS-DA modeling (7, 8). The ions that were successfully quantified are shown in Figs. 5 and 6. Some ions were of too low concentration to be quantified; these included N-acetyl-D-glucosamine sulfate, xanthine, xanthosine and dC.
FIG. 3.
OPLS-DA loadings S-plot from control and irradiated rats 1 day irradiation and 1 day postirradiation analyzed by UPLC-ESI-QTOFMS ESI-. Ions marked with filled black triangles are identified in Tables 1 and 2.
FIG. 4.
Extracted mass spectra from tandem MS of upregulated markers of γ radiation in rat urine (upper panels) compared against standards (lower panels). Panel A: N-Acetyltaurine, panel B: N-acetylglucosamine-6-sulfate, panel C: N1-acetylspermidine, panel D: 2′-deoxyuridine, panel E: 2′-deoxyxanthosine, panel F: thymidine.
TABLE 1.
Upregulated Urinary Biomarkers of γ Radiation in the Rat
| Symbol on Fig. 3 | Retention time (min) | Experimental m/z | Calculated m/z | Mass error (ppm) | Identity | pCorr value
|
||
|---|---|---|---|---|---|---|---|---|
| Predose vs day 1 | Predose vs day 2 | Predose vs day 3 | ||||||
| VI | 0.33 | 166.0171 [M-H]− | 166.0174 | 1.8 | N-Acetyltaurine | 0.953 | 0.955 | 0.610 |
| VII | 0.33 | 124.9911 [M-H]− | 124.9909 | 1.6 | Isethionic acida | 0.945 | 0.972 | 0.647 |
| VIII | 0.32 | 300.0394 [M-H]− | 300.0389 | 1.7 | N-Acetyl-D-glucosamine/galactosamine-6-sulphate | 0.791 | 0.952 | 0.678 |
| IX | 0.55 | 227.0686 [M-H]− | 227.0668 | 7.9 | 2′-Deoxyuridine | 0.826 | 0.000 | 0.000 |
| X | 0.78 | 267.0727 [M-H]− | 267.0729 | 0.7 | 2′-Deoxyxanthosine | 0.953 | 0.334 | 0.000 |
| N/A | 1.10 | 265.0810 [M+Na]+ | 265.0800 | 3.7 | Thymidine sodium adduct | 0.849 | 0.000 | 0.000 |
| N/A | 0.27 | 188.1760 [M+H]+ | 188.1763 | 1.6 | N′-Acetylspermidine | 0.852 | 0.871 | 0.844 |
| N/A | 0.33 | 124.0067 [M-H]− | 124.0068 | 0.8 | Taurine | 0.174 | 0.096 | 0.000 |
| N/A | 3.20 | 172.0983 [M-H]− | 172.0974 | 5.2 | N-Hexanoylglycine | 0.000 | 0.398 | 0.472 |
Tentative.
FIG. 5.
Mean concentrations of upregulated quantified biomarkers. Panel A: Thymidine, panel B: 2′-deoxyuridine, panel C: 2′-deoxyxanthosine, panel D: taurine, panel E: N-acetyltaurine, panel F: N1-acetylspermidine, panel G: N-hexanoylglycine. Error bars are SEM. Significance as determined by one-way ANOVA between °1 day and +1, +2 and +3 days is ***P < 0.001, **P < 0.01 and *P < 0.05.
FIG. 6.
Mean concentrations of downregulated quantified biomarkers. Panel A: Azelaic acid, panel B: N-isovalerylglycine, panel C: pimelic acid, panel D: dodecanedioic acid. Error bars show ±SEM. Significance as determined by one-way ANOVA between −1 day and +1, +2 and +3 days is ***P < 0.001, **P < 0.01, *P < 0.05.
Upregulated Biomarkers
Table 1 shows a list of urinary metabolites upregulated after 3 Gy γ irradiation as identified by multivariate data analysis. It includes taurine, N-acetyltaurine, isethionic acid, N-acetyl-D-glucosamine/galactosamine-6-sulfate, N1-acetylspermidine, N-hexanoylglycine, dU, dX and dT.
The deoxynucleosides dU, dX and dT were transiently upregulated for 24 h postirradiation. The concentration of dU increased 4.3-fold from 18 ± 4 to 77 ± 15 nmol/mmol creatinine (P < 0.001). dX increased 2.9-fold from 150 ± 10 to 440 ± 30 pmol/mmol creatinine (P < 0.001), and dT 6.7-fold from 0.9 ± 0.2 to 6.0 ± 0.6 nmol/mmol creatinine (P < 0.001). Through targeted detection of N-hexanoylglycine and taurine, these ions were seen to be increased after irradiation. N-hexanoylglycine was significantly elevated from day +2 onward, from 0.6 ± 0.1 before irradiation to 1.4 ± 0.2 nmol/mmol creatinine (P < 0.001) on day +2 (2.2-fold), and 1.7 ± 0.2 nmol/mmol creatinine (P < 0.001) on day +3 (3.1-fold). This ion, however, had pcorr values of 0.40 and 0.47 on days +2 and +3 after modeling by OPLS-DA, as shown in Table 1. Taurine increased 2.5-fold in concentration from 0.56 ± 0.03 μmol/mmol creatinine to 1.39 ± 0.20 (P < 0.001) and 2.1-fold to 1.20 ± 0.12 μmol/mmol creatinine (P < 0.001) on days +1 and +2, respectively. N-Acetyl-D-glucosamine-6-sulfate was also seen to increase in OPLS-DA modeling but could not be quantified due to the small peak size.
The metabolites N-acetyltaurine and isethionic acid had pcorr values ranging from 0.95 upward for 48 h postirradiation by OPLS-DA. However, after quantification, the upregulation of N-acetyltaurine was found to be significant for no more than 24 h after irradiation. The ion 124.9911 [M-H]− had a fragmentation pattern matching that of isethionic acid, with a mass error of 1.6 ppm, but it also co-eluted with taurine, which was elevated after irradiation; consequently it was not possible to distinguish between them. Urine samples were compared against authentic standards for isethionic acid and taurine prepared in control urine by tandem MS (125 [M-H]−). Both the standards and the samples had the same fragmentation patterns; this was also true for when the tandem MS mode was set to 124 [M-H]−. Therefore, the ion 124.9911 [M-H]− could not be positively identified. N-Acetyltaurine was identified after fragmentation studies, which revealed a taurine-related metabolite with an additional 42.01 mass units: an acetyl conjugation (+COCH3). N-Acetyltaurine was synthesized in-house and subjected to fragmentation, which gave an identical mass spectrum as seen in Fig. 4. The metabolite had a relative increase of 1.6, from 11.2 ± 1.3 predose to 18.1 ± 1.7 nmol/mmol creatinine (P < 0.05) for day +1.
In ESI+ mode a significant metabolite was observed with a m/z of 188.176 [M+H]+, suggesting N8-acetylspermidine. Similar fragmentation patterns were seen in the sample and the N8-acetylspermidine standard, but two fragments in the irradiated urine were not seen in the standard, 100.0783 [M+H]+ and 117.1064 [M+H]+, and two fragments in the standard were not present in the sample, 131.1154 [M+H]+ and 114.0919 [M+H]+. It was determined that these fragments could arise from the N1 isomer. This standard was synthesized in-house as described in the Materials and Methods section. The fragmentation patterns of the urine sample, N1-acetylspermidine standard and N8-acetylspermidine standard are shown in Fig. 7A–C, respectively, and the metabolite 188.176 [M+H]+ can be positively identified as N1-acetylspermidine. This metabolite was increased 3.5-fold from 0.40 ± 0.04 nmol/mmol creatinine predose to 1.40 ± 0.06 nmol/mmol creatinine (P < 0.001) 24 h after irradiation and 2.6-fold to 1.1 ± 0.1 nmol/mmol creatinine (P < 0.001) on day +2.
FIG. 7.
Extracted mass spectra from tandem MS (188.1 [M+H]+) of (panel A) rat urine 3 days after irradiation (panel B) N1-acetylspermidine spiked in urine and (panel C) N8-acetylspermidine spiked in urine.
The other upregulated N-acetylated biomarker was N-acetyl-D-glucosamine/galactosamine-6-sulfate, an amino sugar, which increased after irradiation but could not be quantified. This metabolite had an m/z ratio of 300.0394 [M-H]−, but both N-acetyl-D-glucosamine-6-sulfate and N-acetyl-D-galactosamine-6-sulfate had the same retention time and fragmentation pattern, so the identity could not be confirmed.
Downregulated Biomarkers of γ-Radiation Exposure
Table 2 shows the biomarkers that were downregulated after radiation exposure. Three of them, pimelic, dodecanedioic and azelaic acids, are dicarboxylic acids and were decreased for all 3 days postirradiation. N-isovalerylglycine, an acyl glycine, was significantly decreased for all 3 days, with P values of <0.05 on day +1 and <0.001 on days +2 and +3. A decrease in dicarboxylic acids may indicate an increase in β-oxidation for gluconeogenesis during food deprivation, and indeed the γ-irradiated rats decreased food consumption by approximately 80% [Fig. 3B (9)]. A further experiment was carried out to assess the effect of food consumption on the metabolome. Ten rats were split into two groups; one group was allowed food and water ad libitum, while the other group had access to water only, and 24-h urine samples were collected. These data showed quite clearly that the dicarboxylic acids and N-isovalerylglycine were decreased during food deprivation and were not down-regulated biomarkers specific to radiation. An OPLS-DA S-plot in Fig. 8 shows that dodecanedioic acid, azelaic acid, pimelic acid and N-isovalerylglycine were downregulated after 24 h of food deprivation and had pcorr values of 0.81, 0.89, 0.87 and 0.73, respectively.
TABLE 2.
Downregulated Urinary Biomarkers of γ Radiation in the Rat after Food Deprivation
| Symbol on Fig. 3 | Retention time (min) | Experimental m/z | Calculated m/z | Mass error (ppm) | Identity | pCorr value
|
||
|---|---|---|---|---|---|---|---|---|
| Predose vs day 1 | Predose vs day 2 | Predose vs day 3 | ||||||
| I | 4.20 | 187.0964 [M-H]− | 187.0970 | 3.2 | Azelaic acid | 0.898 | 0.966 | 0.590 |
| II | 4.20 | 125.0950 [M-H]− | 125.0966 | 12.8 | In-source fragment of I | 0.853 | 0.948 | 0.615 |
| III | 2.35 | 159.0637 [M-H]− | 159.0657 | 12.6 | Pimelic acid | 0.873 | 0.898 | 0.451 |
| IV | 4.76 | 229.1440 [M-H]− | 229.1440 | 1.3 | Dodecanedioic acid | 0.883 | 0.935 | 0.645 |
| V | 1.40 | 158.0809 [M-H]− | 158.0817 | 5.1 | N-Isovalerylglycine | 0.888 | 0.923 | 0.417 |
FIG. 8.
OPLS-DA loadings S-plot from control (fed) and unfed rats at 24 h analyzed by UPLC-ESI-QTOFSMS. Ions marked with filled black triangles are identified in Table 2.
DISCUSSION
UPLC-ESI-QTOFMS metabolomics was used to identify novel metabolites of γ-radiation exposure in rats and to confirm established biomarkers previously identified in mice by UPLC- ESI-QTOFMS (7, 8) and rats by GCMS (9). Upregulated biomarkers included dT, dU and dX, N1-acetylspermidine, N-acetylglucosamine-6-sulfate, N-acetyltaurine, taurine and N-hexanoylglycine.
dT, dU and dX were previously reported as biomarkers of radiation exposure in mice (8, 9), and dU and dT were seen as their corresponding uracil and thymine bases in rat urine after γ irradiation by GCMS-based metabolomics analysis (a result of the derivatization process for GCMS) (9). In mice, the excretion of dT increased 7-fold and was similarly upregulated here in rats by 6.7-fold; a 7-fold increase in rats has also been observed by Zharkov et al. (12). The metabolites dU and dX were increased 4.3-fold and 2.9-fold, respectively, and increased to the same extent in mice (8). dU, dX and dT are markers of DNA damage and can be formed by hydrolytic or enzymatic deamination of dC, 2′-deoxyguanosine (dG) and 5′-MedC, respectively. They can also be formed through interactions with ROS generated during exposure to ionizing radiation and redox reactions (11). A previous publication reported an increase in dU in mice subjected to 1–3 Gy γ radiation (8) and a decrease in dC. Both dC and 5′-MedC were mined for and were decreased after irradiation with pcorr values from 0.65–0.7 by OPLS-DA; however, they could not be quantified. dG was below the limit of detection by UPLC-ESI-QTOFMS. dX had the lowest concentration and relative change compared to dT and dU; therefore, it is not surprising that dG could be observed. dG is also known to form 2′-deoxyoxanosine (dO) (13). This metabolite was mined for in the UPLC-ESI-QTOFMS data (267.0729 [M-H]−); it had the same m/z as dX but a different structure and would thus be expected to form different fragments when subjected to tandem MS fragmentation. 267.0729 [M-H]− was identified and fragmented to reveal dX (confirmed against an authentic standard) and not dO. Pyrimidine bases in DNA are known to be more susceptible to hydrolytic deamination than purine bases, but purines are more prone to deamination by nitrous acid; therefore, the results here indicated that spontaneous deamination was a more prevalent route of deamination than ROS (11, 14). Further work must be performed to determine whether the increases in dU and dT resulted from radiation-induced enzymatic activity (cytidine deaminase and/or dC deaminase and thymidylate synthase) or were due to the actions of ROS. The repair of DNA bases is important in cells exposed to ROS, and increases in other biomarkers past day +1 are of interest because of their relationship to this process.
The excretion of taurine was seen to increase for 2 days after radiation exposure by 2.5- and 2.1-fold. Taurine has previously been observed to be elevated in mouse urine after 8 Gy irradiation by 1.2-fold and was not seen at sublethal doses (7). Taurine is metabolized from methionine and cysteine and is involved in many physiological functions such as bile acid conjugation, osmoregulation, antioxidation and detoxication. It is generally seen as a marker of hepatotoxicity. Because exposure to radiation can increase ROS, the role of taurine here may have been protective, inhibiting ROS, or an enhancement in cysteine or glutathione turnover could have occurred (7). Another mechanism was proposed by Dilley (15) in which an increase in taurine urinary excretion in radiation-exposed dogs and humans was a result of radiation-induced destruction of circulating lymphocytes.
Taurine is known to regulate ion channels. The urine collected from this study was previously analyzed by GCMS as described by Lanz et al. (9). Water and food consumption and ion levels were documented. A large increase in phosphate with a decrease in calcium, sodium, potassium and chloride ions was seen after radiation exposure; the latter three could be attributed to a decrease in food intake, but regulation of calcium ion channels could also play a role in this here. Another ion that was highly correlated to radiation exposure was 124.9911 [M-H]−, which was tentatively assigned as isethionic acid but could not be resolved from taurine. Isethionic acid is the hydroxyl analogue of taurine and is formed from taurine via a sulfoacetaldehyde intermediate. It has been proposed that mammals cannot directly metabolize taurine to isethionate; instead gut microflora perform this role (16). Therefore, it is hypothesized that isethionic acid could be a biomarker produced as a response of the gut microflora to radiation. p-Cresol, another gut microfloral metabolite, was previously identified by GCMS analyses of urine from radiation-exposed rats (9).
N-Acetyltaurine was also identified as an upregulated biomarker of ionizing radiation exposure, and it is a novel mammalian metabolite. It is a derivative of taurine and has only been observed previously in orb spiders (17). It had a high correlation to radiation exposure, with a pcorr value of 0.95 by OPLS-DA for 1–2 days postirradiation. After in-house synthesis of the N-acetyltaurine pyridine salt, quantification was possible. N-acetyltaurine does not derive directly from taurine because a concomitant decrease of that metabolite was not observed.
N1-Acetylspermidine was increased after radiation exposure and forms through reversible N-acetylation of spermidine by spermidine/spermine N1-acetyltransferase (SSAT). The transcription of SSAT is induced in the presence of ROS, increased polyamine levels, interleukin-1 and hepatocyte growth factor. Polyamine oxidase (PAO) can convert the acetylated compound back to putrescine, producing H2O2 and aldehydes, which in turn can cause apoptosis and cell damage (18). Spermidine and N1-acetylspermidine form from arginine via putrescine; they play vital roles in cellular processes involving nucleic acids. Other polyamines in the putrescine pathway were also mined for in the data generated by MarkerLynx, but correlation to radiation was not observed. However, a previous study with TK6 cells showed an attenuation of spermine after exposure to 1 Gy γ radiation (6). Spermine is formed from spermidine; it is possible that the spermidine precursor in both these cases was N-acetylated instead of undergoing conversion to spermine by spermine synthase, producing the observed decrease in spermine. A similar effect was observed in rat spleen after 3 Gy γ irradiation; decreases in putrescine, spermine and spermidine were seen for 5 days after irradiation (19). Increases in N1-acetylspermidine have been observed previously in rat and human colon cancers as well as in many other types of cancer (20–23). It has been observed that SSAT is enhanced at the beginning of growth arrest and cell death in HeLa S3 and myeloid cell lines after X and γ irradiation (24, 25). The induction of SSAT could potentially regulate growth arrest that results in cell death after toxic stress and play a protective role in response to ionizing radiation.
Another N-acetylated biomarker seen to be upregulated after exposure to ionizing radiation was N-acetyl-D-glucosamine/galactosamine-6-sulfate, an amino sugar. N-Acetyl-D-glucosamine-6-sulfate and N-acetyl-D-galactosamine-6-sulfate are key components of the proteoglycans dermatan, keratan and chondroitin sulfates. Exposure to X radiation in a rabbit chondrocyte culture system revealed a change in proteoglycan synthesis and a stimulation of proteoglycan degradation (26); in this study, an increase in N-acetylated amino sugars could result from ionizing radiation-induced degradation. However, it was not possible here to distinguish between N-acetyl-D-glucosamine-6-sulfate and N-acetyl-D-galactosamine-6-sulfate; they had the same retention time and fragmentation patterns.
The downregulated metabolites included a number of dicarboxylic acids and an acyl glycine. A reduced urinary excretion of dicarboxylic acids was observed previously by GCMS in the rat urine; it was proposed that this was a result of changes in renal tubular cells (9). These dicarboxylic acids can be completely oxidized through β-oxidation to produce succinyl-CoA, which can be used in the gluconeogenesis pathway during starvation (27); therefore, the decrease in these dicarboxylic acids was proposed to be a result of decreased food intake after irradiation. A further study was carried out in which rats were deprived of food for 24 h and urine was collected. Metabolomic analysis of this urine showed a decrease in the metabolites downregulated after irradiation. The ions that were increased after food deprivation were also mined for the upregulated radiation markers, but they were not found.
The samples taken at day −4 showed a number of metabolites that were present only at this time: hippurate, PAG, 2,8-quinolinediol and its glucuronide. These metabolites were produced as the rats became acclimatized to the metabolic cages. Hippurate is known to be formed from the microbial breakdown of larger dietary phenolics and benzoic acids, which are then excreted as hippurate after glycine conjugation in the liver (28). PAG is derived from the metabolism of phenylalanine also aided by microbial metabolism and subsequent conjugation of phenylacetic acid with glycine (29). Therefore, the movement of the rats from their standard group-housed cages to individual metabolic cages resulted in a disruption of the gut microfloral metabolized compounds that started to stabilize after 24 h.
Evaluation of GCMS and UPLC-ESI-QTOFMS for Metabolic Profiling
As mentioned originally, this metabolomic study was implemented previously using GCMS (9) to analyze the urine samples and was repeated here using UPLC-ESI-QTOFMS. This was to determine how effective the two technologies are for metabolomics analysis of rat urine after radiation exposure and to maximize metabolite recovery. Initial PCA models were constructed from the UPLC-ESI-QTOFMS data and had identical groupings and patterns as seen by GCMS. A PCA scores plot showing the GCMS data taken from Lanz et al. (9) (Fig. 9) is similar to the UPLC-ESI-QTOFMS PCA scores plots in Fig. 1. There are some differences, however; the UPLC-ESI-QTOFMS variables cluster together within their group classifications, while the GCMS variables are spread out across PC1 and PC2. The more distinctive grouping within class by UPLC-ESI-QTOFMS could be due to the acquisition of the data or to pre-processing methods such as peak alignment and detection; different software was used to process the UPLC-ESI-QTOFMS and GCMS data.
FIG. 9.
PCA scores plots from 24 h rat urine samples analyzed by GCMS. Black squares, day −4; black triangles, day−3; black triangles, day −2; black diamonds, day −1; blue squares, day 1 sham-irradiated; red squares, day +1 irradiated with 3.5 Gy; inverted blue triangles, day +2 sham-irradiated; inverted red triangles, day +2 irradiated 3.5 Gy; blue diamonds, day +3 sham-irradiated; red diamonds, day +3 irradiated with 3.5 Gy.
For biomarker identification, both methods revealed upregulated nucleosides, but they were seen as their respective free bases by GCMS due to the sample processing step of derivitization. These nucleosides (dU and dT) could be confirmed in both systems; however, dX could be seen only by UPLC-ESI-QTOFMS due its low abundance. Therefore, UPLC-ESI-QTOFMS is more effective for identifying low-abundance metabolites than GCMS. There were a number of other biomarkers that were observable only by UPLC-ESI-QTOFMS compared to GCMS and vice versa. Of these were polar compounds that would not derivatize well and thus were not identified by GCMS, for example, taurine and the N-acetylated metabolites. Three metabolites, glyoxylate, threonate and p-cresol, were observed by GCMS and not by UPLC-ESI-QTOFMS. The sample processing methods for UPLC-ESI-QTOFMS may have resulted in extraction of these metabolites into the pellet, and thus they could have been lost before loading onto the column. Alternatively, derivatization for GCMS may have improved the ability to observe these metabolites.
Conclusions
UPLC-ESI-QTOFMS-based metabolomics allowed effective identification of nine urinary biomarkers of γ radiation in rats, including a novel mammalian metabolite. These upregulated urinary biomarkers included dT, dU and dX, N1-acetylspermidine, N-acetylglucosamine/galactosamine-6-sulfate, N-acetyltaurine, N-hexanoylglycine, taurine and tentatively isethionic acid. Some of these metabolites were also identified in the mouse (dT, dU, dX, taurine, N-hexanoylglycine) and helped to reveal cross-species biomarkers for radiation. It is now of interest to look at other mammals, in particular non-human primates or humans, to ascertain whether these metabolites are shared outside of mice and rats. Further studies include analysis of the rat tissues to look for changes in gene transcription and regulation of enzymes to relate the metabolites discovered to molecular mechanisms.
Acknowledgments
This work was supported by the National Cancer Institute Intramural Research Program and also performed as part of the Columbia University Center for Medical Countermeasures against Radiation (P.I. David Brenner) funded by NIH (NIAID) grant U19 AI067773-05/06.
References
- 1.Mah LJ, El-Osta A, Karagiannis TC. γH2AX: a sensitive molecular marker of DNA damage and repair. Leukemia. 2010;24:679–686. doi: 10.1038/leu.2010.6. [DOI] [PubMed] [Google Scholar]
- 2.Amundson S, Do K, Shahab S, Bittner M, Meltzer P, Trent J, Fornace AJ. Identification of potential mRNA biomarkers in peripheral blood lymphocytes for human exposure to ionizing radiation. Radiat Res. 2000;154:342–346. doi: 10.1667/0033-7587(2000)154[0342:iopmbi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 3.Wiebalk K, Schmezer P, Kropp S, Chang-Claude J, Celebi O, Debus J, Bartsch H, Popanda O. In vitro radiation-induced expression of XPC mRNA as a possible biomarker for developing adverse reactions during radiotherapy. Int J Cancer. 2007;121:2340–2345. doi: 10.1002/ijc.22981. [DOI] [PubMed] [Google Scholar]
- 4.Amundson S, Do K, Fornace AJ. Induction of stress genes by low doses of gamma rays. Radiat Res. 1999;152:225–231. [PubMed] [Google Scholar]
- 5.Amundson S, Lee R, Koch-Paiz C, Bittner M, Meltzer P, Trent J, Fornace AJ. Differential responses of stress genes to low dose-rate gamma irradiation. Mol Cancer Res. 2003;1:445–452. [PubMed] [Google Scholar]
- 6.Patterson A, Li H, Eichler G, Krausz K, Weinstein J, Fornace AJ, Gonzalez F, Idle J. UPLC-ESI-TOFMS-based metabolomics and gene expression dynamics inspector self-organizing metabolomic maps as tools for understanding the cellular response to ionizing radiation. Anal Chem. 2008;80:665–674. doi: 10.1021/ac701807v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tyburski JB, Patterson AD, Krausz KW, Slavik J, Fornace AJ, Jr, Gonzalez FJ, Idle JR. Radiation metabolomics. 1. Identification of minimally invasive urine biomarkers for gamma-radiation exposure in mice. Radiat Res. 2008;170:1–14. doi: 10.1667/RR1265.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tyburski J, Patterson A, Krausz K, Slavík J, Fornace AJ, Gonzalez F, Idle J. Radiation metabolomics. 2. Dose- and time-dependent urinary excretion of deaminated purines and pyrimidines after sublethal gamma-radiation exposure in mice. Radiat Res. 2009;172:42–57. doi: 10.1667/RR1703.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lanz C, Patterson A, Slavík J, Krausz K, Ledermann M, Gonzalez F, Idle J. Radiation metabolomics. 3. Biomarker discovery in the urine of gamma-irradiated rats using a simplified metabolomics protocol of gas chromatography-mass spectrometry combined with random forests machine learning algorithm. Radiat Res. 2009;172:198–212. doi: 10.1667/RR1796.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tabor H, Tabor CW, de Meis L. Chemical synthesis of N-acetyl-1,4-diamihobutane, N1-acetylpermidine, and N8-acetylspermidine. In: Tabor TH, White C, editors. Methods in Enzymology. Academic Press; New York: 1971. pp. 829–833. [Google Scholar]
- 11.Vongchampa V, Dong M, Gingipalli L, Dedon P. Stability of 2′-deoxyxanthosine in DNA. Nucleic Acids Res. 2003;31:1045–1051. doi: 10.1093/nar/gkg177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zharkov I, Fedorova T, Mikhailova L. The excretion of thymidine in the urine of rats after total body x-ray irradiation in various doses. Radiobiologiia. 1965;5:675–680. [PubMed] [Google Scholar]
- 13.Suzuki T, Yamaoka R, Nishi M, Ide H, Makino K. Isolation and characterization of a novel product, 2′-deoxyoxanosine, from 2′-deoxyguanosine, oligodeoxynucleotide, and calf thymus DNA treated by nitrous acid and nitric oxide. J Am Chem Soc. 1996;118:2515–2516. [Google Scholar]
- 14.Caulfield J, Wishnok J, Tannenbaum S. Nitric oxide-induced deamination of cytosine and guanine in deoxynucleosides and oligonucleotides. J Biol Chem. 1998;273:12689–12695. doi: 10.1074/jbc.273.21.12689. [DOI] [PubMed] [Google Scholar]
- 15.Dilley J. The origin of urinary taurine excretion during chronic radiation injury. Radiat Res. 1972;50:191–196. [PubMed] [Google Scholar]
- 16.Fellman J, Roth E, Avedovech N, McCarthy K. The metabolism of taurine to isethionate. Arch Biochem Biophys. 1980;204:560–567. doi: 10.1016/0003-9861(80)90068-5. [DOI] [PubMed] [Google Scholar]
- 17.Mayer J, Denger K, Smits T, Hollemeyer K, Groth U, Cook A. N-acetyltaurine dissimilated via taurine by Delftia acidovorans NAT. Arch Microbiol. 2006;186:61–67. doi: 10.1007/s00203-006-0123-7. [DOI] [PubMed] [Google Scholar]
- 18.Zahedi K, Wang Z, Barone S, Prada A, Kelly C, Casero R, Yokota N, Porter C, Rabb H, Soleimani M. Expression of SSAT, a novel biomarker of tubular cell damage, increases in kidney ischemia-reperfusion injury. Am J Physiol Renal Physiol. 2003;284:F1046–F1055. doi: 10.1152/ajprenal.00318.2002. [DOI] [PubMed] [Google Scholar]
- 19.Becciolini A, Porciani S, Lanini A, Balzi M. Polyamine content as a marker of radiation injury in the rat spleen. Int J Radiat Biol Relat Stud Phys Chem Med. 1987;52:767–774. doi: 10.1080/09553008714552281. [DOI] [PubMed] [Google Scholar]
- 20.O’Brien B, Hankewych M, McCormick D, Jacoby R, Brasitus T, Halline A. Urinary N1-acetylspermidine and N8-acetylspermidine excretion in normal humans and in patients with colorectal cancer. Dig Dis Sci. 1995;40:1269–1274. doi: 10.1007/BF02065536. [DOI] [PubMed] [Google Scholar]
- 21.Löser C, Fölsch U, Paprotny C, Creutzfeldt W. Polyamine concentrations in pancreatic tissue, serum, and urine of patients with pancreatic cancer. Pancreas. 1990;5:119–127. doi: 10.1097/00006676-199003000-00001. [DOI] [PubMed] [Google Scholar]
- 22.Halline A, Dudeja P, Lashner B, Brasitus T. Urinary excretion of N1-acetylspermidine and other acetylated and free polyamines in the 1,2-dimethylhydrazine model of experimental rat colon cancer. Cancer Res. 1989;49:4721–4723. [PubMed] [Google Scholar]
- 23.Persson L, Rosengren E. Increased formation of N1-acetylspermidine in human breast cancer. Cancer Lett. 1989;45:83–86. doi: 10.1016/0304-3835(89)90140-7. [DOI] [PubMed] [Google Scholar]
- 24.Mita K, Fukuchi K, Hamana K, Ichimura S, Nenoi M. Accumulation of spermidine/spermine N1-acetyltransferase and alternatively spliced mRNAs as a delayed response of HeLa S3 cells following X-ray irradiation. Int J Radiat Biol. 2004;80:369–375. doi: 10.1080/09553000410001695886. [DOI] [PubMed] [Google Scholar]
- 25.Amundson S, Bittner M, Chen Y, Trent J, Meltzer P, Fornace AJ. Fluorescent cDNA microarray hybridization reveals complexity and heterogeneity of cellular genotoxic stress responses. Oncogene. 1999;18:3666–3672. doi: 10.1038/sj.onc.1202676. [DOI] [PubMed] [Google Scholar]
- 26.Jikko A, Hiranuma H, Iwamoto M, Kato Y, Okada Y, Fuchihata H. Effects of X irradiation on metabolism of proteoglycans. Radiat Res. 1996;146:93–99. [PubMed] [Google Scholar]
- 27.Salinari S, Bertuzzi A, Gandolfi A, Greco AV, Scarfone A, Manco M, Mingrone G. Dodecanedioic acid overcomes metabolic inflexibility in type 2 diabetic subjects. Am J Physiol Endocrinol Metab. 2006;291:E1051–E1058. doi: 10.1152/ajpendo.00631.2005. [DOI] [PubMed] [Google Scholar]
- 28.Phipps A, Stewart J, Wright B, Wilson I. Effect of diet on the urinary excretion of hippuric acid and other dietary-derived aromatics in rat. A complex interaction between diet, gut microflora and substrate specificity. Xenobiotica. 1998;28:527–537. doi: 10.1080/004982598239443. [DOI] [PubMed] [Google Scholar]
- 29.Haley C, Harper A. The importance of transamination and decarboxylation in phenylalanine metabolism in vivo in the rat. Arch Biochem Biophys. 1978;189:524–530. doi: 10.1016/0003-9861(78)90242-4. [DOI] [PubMed] [Google Scholar]