Abstract
Specific genotypes of hepatitis B virus (HBV) are increasingly recognized for their clinical significance and association with particular viral mutations. Although many HBV genotyping methods exist, there has been no standardized or commercially available method for direct molecular typing of the HBV genome. A newly available line probe assay (INNO-LiPA HBV Genotyping assay; Innogenetics N.V., Ghent, Belgium) that allows the identification of HBV genotypes A to G was assessed by comparison with pre-S1/pre-S2 sequence analysis of the isolates in 188 serum specimens. All seven genotypes were detected by the line probe assay (LiPA), and complete concordance between LiPA and sequence analysis was observed for 152 specimens (81%). LiPA was able to detect 19 mixed genotype infections not detected by amplicon sequencing, which for the most part were confirmed by cloning and sequencing of the pre-S1/pre-S2 amplicon. Four specimens had discrepant results between the two methods, and five specimens had indeterminate results by LiPA. The HBV DNA in four specimens was unable to be amplified by the nested INNO-LiPA HBV DR amplification primers; however, the HBV DNA in six specimens unable to be genotyped by sequencing was clearly genotyped by LiPA. The INNO-LiPA HBV Genotyping assay appears to be useful for the rapid genotyping of HBV, particularly for the sensitive detection of mixed genotype infections.
It is estimated that more than 350 million individuals worldwide are chronically infected with hepatitis B virus (HBV), of which approximately 20 to 30% risk death from HBV-related liver failure or hepatocellular carcinoma (7, 18). In the past several years, the clinical significance of different HBV genotypes has become increasingly recognized in patients with both acute and chronic HBV infections. For example, genotype C has been more closely associated with a poor prognosis and a more aggressive clinical phenotype (8, 26), while genotype B has been associated with earlier HBeAg seroconversion (2), possibly leading to a lower prevalence of HBV-related cirrhosis. Core promoter and lamivudine resistance mutations were found to be more common in genotypes C and A, while precore stop mutations have been observed more frequently in genotypes B and D (6, 9, 30, 34, 35). Therefore, knowledge of the genotype infecting a patient may assist a physician in making clinical and therapeutic decisions.
Although many HBV genotyping methods exist, there has been no standardized or commercially available method for direct molecular typing of the HBV genome. There are seven genotypes of HBV, based on nucleotide differences of 8% or greater along the entire length of the HBV genome or 4% or greater within the small S gene (HBsAg) (23). Recently, an assay based on the line probe assay (LiPA; INNO-LiPA HBV Genotyping assay, Innogenetics N.V., Ghent, Belgium) was released for research purposes. This method is based on the reverse hybridization principle, such that biotinylated amplicons hybridize to specific oligonucleotide probes that are immobilized as parallel lines on membrane-based strips. The amplified region analyzed overlaps the sequence encoding the major hydrophilic region of HBsAg, which is often investigated as a reliable measure of HBV genetic comparison (11, 19). The present study evaluated the INNO-LiPA HBV Genotyping assay by testing 188 serum specimens positive for HBV DNA that had been genotyped by sequencing and phylogenetic analysis of the pre-S1/pre-S2 region of the HBV genome.
MATERIALS AND METHODS
Serum specimens.
Serum specimens were selected from among routine diagnostic specimens sent from laboratories across Canada for detection of HBV serological markers and DNA. Serum specimens were sent from January 1999 to August 2002. Several samples were also selected from directed studies undertaken from 1990 to 1995. A total of 188 specimens were chosen on the basis of the sequence-determined genotypes in the specimens. Representatives of genotypes A (n = 49), B (n = 35), C (n = 48), and D (n = 43) were selected; genotypes not routinely detected in Canada (genotype E, n = 3; genotype F, n = 1) or not often detected as the dominant strain (genotype G, n = 3) were less well represented. Also, several specimens for which the HBV DNA was not typeable by sequencing due to an inability to PCR amplify a pre-S region amplicon (n = 1) or due to poor sequence data (n = 5) were included in the assessment of the LiPA kit.
DNA extraction.
DNA was extracted from 150 μl of serum by the proteinase K-sodium dodecyl sulfate lysis and phenol-chloroform extraction methods as described previously (24) and was resuspended in a final volume of 30 μl of sterile, nuclease-free water. The extracted DNA was used for amplification in both the sequencing and the LiPA procedures. Sequencing and LiPA analysis were performed within approximately 5 days following DNA extraction. If DNA extracts were not used immediately, they were stored at −20°C.
Sequencing and phylogenetic analysis.
Extracted DNA was amplified by using primers specific for the pre-S1/pre-S2 region of HBV (nucleotides 3025 to 80 from the theoretical EcoRI site of the 3,221-nucleotide HBV sequence). The sense primer was preS1F (5′-AGGTRGGAGYGGGAGCATTCGG-3′), and the antisense primer was preS1R 5′-CCTGAACTGGAGCCACCAGCAGG-3′ (R is A and G and Y is C and T). Thermal cycling parameters involved 40 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s. The expected product of 277 bp was gel purified prior to cycle sequencing with an ABI Prism 377 DNA sequencer by standard dye terminator chemistry.
If the 277-bp product was not initially detected, the extracted DNA was amplified by nested PCR with outer primers that produce a 479-bp product (sense primer, 5′-TCACCATATTCTTGGGAACAAGA-3′; antisense primer, 5′-TTCCTGAACTGGAGCCACCA-3′) (15), followed by second-stage amplification with the pre-S primers described above. Reaction tubes for PCR contained 5 μl of DNA extract or the first-stage PCR product, AmpliTaq Gold reaction buffer (Applied Biosystems, Foster City, Calif.), 0.2 mM deoxynucleoside triphosphates (Invitrogen Life Technologies, Burlington, Ontario, Canada), 2.5 mM MgCl2, 25 pmol of each primer, and 2.5 U of AmpliTaq Gold polymerase. All oligonucleotides used for amplification were custom synthesized by the DNA Core Facility at the National Microbiology Laboratory.
In order to prevent PCR carryover contamination during nested PCR, each step of the procedure was performed in a separate room with dedicated equipment, with a directional flow from the beginning of the procedure to the end. Negative serum and water controls were also included in each extraction run, and an extra water control was included at each stage of the PCR.
Sequence data for the amplified region of HBV DNA in each specimen were aligned with pre-S1/pre-S2 sequences from among 102 GenBank sequences of known genotype representing the seven HBV genotypes. Alignment of the sequences was performed with ClustalX software (version 1.81) (31). Genetic distances were estimated by Kimura two-parameter analysis, and phylogenetic trees were constructed by the neighbor-joining method with 500 bootstrap replicates with MEGA (version 2.1) software (http://www.megasoftware.net/).
LiPA amplification and detection.
The extracted DNA was amplified by nested PCR according to the instructions of the manufacturer (Innogenetics) for amplification of the HBsAg region to provide a biotinylated product. The HBV genomic region amplified extends from nucleotides 415 to 824 for the outer primers and nucleotides 456 to 798 for the nested inner primers (the numbering is based on the sequence with GenBank accession number AY128092). These procedures have been described previously (17). Each stage of amplification was monitored by visualization of the PCR products on a 2% agarose gel. If the first-stage PCR products were visible, they were used for hybridization to the test strips; otherwise, the second-stage products were used.
Genotype detection was performed according to the instructions of the manufacturer. In brief, the biotinylated PCR products were first denatured and were then incubated with a test strip for hybridization of the denatured amplicon to genotype-specific probes immobilized as parallel lines on each strip. Following hybridization, the strips were stringently washed and incubated with a streptavidin conjugate to allow color development from the biotinylated DNA bound to the strip.
LiPA amplicons showing a single genotype that was different from the genotype determined by sequencing of the pre-S1/pre-S2 region were sequenced and analyzed by using an HBsAg phylogenetic tree to confirm the LiPA genotype designation. The nested products were sequenced with primers HBPr75 (5′-CAAGGTATGTTGCCCGTTTGTCC-3′) and HBPr94 (5′-GG(T/C)A(A/T)AAAGGGACTCA(A/C)GATG-3′) (28).
Clonal analysis of PCR products.
Specimens showing discrepant genotypes between sequencing and LiPA were further analyzed by cloning of the pre-S region amplicon used for sequencing and phylogenetic analysis. Of the 23 specimens with discrepant results, 4 were unable to be further analyzed, as there was insufficient serum for extraction or the pre-S region could not be amplified. Specific amplicons were gel purified and cloned into a pCR2.1-TOPO plasmid vector (Invitrogen Life Technologies) according to the instructions of the manufacturer. Ligated products were transformed into Escherichia coli TOP10F (Invitrogen Life Technologies), individual colonies were picked, and the plasmid DNA inserts were sequenced. Insert sequence data were aligned with GenBank pre-S1/pre-S2 sequences, as described above, to determine the genotype of each individual clone.
RESULTS
LiPA amplification and detection.
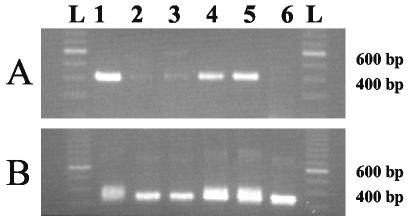
The HBV DNA in all except 4 of 188 specimens were amplified by the first- or second-stage nested primers provided with the LiPA kit. The HBV DNA in 53 specimens required amplification by second-stage primers in order to observe a product, while a PCR product was evident for the HBV DNA in 131 specimens following first-stage amplification. PCR products were visualized on a 2% agarose gel stained with ethidium bromide. A representative gel showing the PCR products obtained with both sets of primers is shown in Fig. 1. The outer primer set generates an amplicon of 409 bp, while the nested primer set generates an amplicon of 342 bp. As can be seen in Fig. 1, nested products from HBV DNA showing obvious first-round products often electrophoresed as doublet bands (lanes 1, 4, and 5), while nested products from weak or nonvisible first-round products electrophoresed as sharp, discrete bands (lanes 2, 3, and 6). As described in the kit instructions, visible products generated by the outer primers were used preferentially for LiPA analysis; otherwise, the nested products were used.
FIG. 1.
Agarose gel electrophoresis of each stage of nested PCR for the LiPA. (A) First-stage PCR; (B) second-stage PCR. A 100-bp ladder (lanes L) is shown on each gel. The genotypes of the representative specimens determined by LiPA are as follows: C (lanes 1), A and G (lanes 2), A and B (lanes 3), B (lanes 4), C (lanes 5), and A and C (lanes 6).
Genotype detection by hybridization of the PCR products to the kit membrane strips was performed as described above and required approximately 3.5 to 4 h to complete. A representation of the membrane strip with all the immobilized control and genotype-specific oligonucleotide bands is shown in Fig. 2. For all genotypes except genotype G, several reactive bands can indicate a specific genotype. Interpretation of the test strips was relatively straightforward; however, in certain cases faint bands appeared, and these made interpretation of the genotype unclear.
FIG. 2.
Graphical representation of the INNO-LiPA HBV Genotyping assay strip. Bands representing oligonucleotide probes specific for controls and each HBV genotype are shown as they are arranged on the test strip.
The HBV DNA levels in the four specimens for which HBV could not be genotyped were measured (Cobas AMPLICOR HBV Monitor test; Roche Molecular Systems, Inc., Branchburg, N.J.) to ensure that sufficient DNA was available for genotyping by LiPA. The lower limit of detection of the Cobas AMPLICOR Monitor test is 100 copies/ml, while the lower limit of detection of LiPA is 700 copies/ml (Erwin Sablon, personal communication). The four specimens had HBV DNA levels below the limit of detection by the Cobas AMPLICOR Monitor test and, thus, had HBV DNA levels below the limit of detection of LiPA.
Comparison of HBV genotypes determined by sequencing and LiPA.
Genotyping of the HBV DNA in 188 specimens was performed by sequencing and phylogenetic analysis of the pre-S1/pre-S2 region, and these genotypes were compared to the genotypes determined by LiPA for the same specimens. Phylogenetic analysis of the pre-S region by using more than 100 sequences from GenBank for comparison provided clear branching of the seven different genotypes (data not shown), with bootstrap values of 99 to 100 for all genotypes other than genotype A (for which the bootstrap value was 84).
Genotypes determined by sequencing of the HBV DNA in all specimens resulted in a single genotype determination, while LiPA detected 19 mixed genotypes. Table 1 shows the number of samples positive for each single or mixed genotype as determined by sequencing and LiPA. The results of the two methods for single genotypes were concordant for 152 of 188 (81%) specimens (Table 1). The results of the methods for the single genotype were discrepant for four of the specimens (genotype A by sequencing and genotype C by LiPA, n = 2; genotype D by sequencing and genotype A by LiPA, n = 1; genotype D by sequencing and genotype C by LiPA, n = 1). The genotypes in five specimens were also indeterminate by LiPA, and the HBV DNA in four other specimens did not yield a PCR product following LiPA nested PCR (Table 1). The HBV DNA in six specimens that could not be genotyped by sequencing due to poor sequence data or a lack of a pre-S1/pre-S2 amplicon was tested by LiPA, and the HBV DNA in all six specimens could be clearly typed by this method. If a discrepancy was observed between the results of LiPA and sequencing, the HBV DNA was reextracted from the serum sample and retested by both methods to ensure that an actual discrepancy existed prior to further clonal analysis.
TABLE 1.
Comparison of HBV genotype as determined by sequencing and LiPA
| Genotype by LiPA | No. of specimens with the following genotype(s) by sequencing:
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | A/B | A and C | A and G | A, C, and G | A and F | INDa | NAb | |
| A | 38 | —c | — | 1 | — | — | — | — | — | — | — | — | — | — |
| B | — | 29 | — | — | — | — | — | — | — | — | — | — | — | — |
| C | 2 | — | 42 | 1 | — | — | — | — | — | — | — | — | 3 | — |
| D | — | — | — | 39 | — | — | — | — | — | — | — | — | 1 | — |
| E | — | — | — | — | 3 | — | — | — | — | — | — | — | — | — |
| F | — | — | — | — | — | 1 | — | — | — | — | — | — | — | — |
| G | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| A and B | 1 | 3 | — | — | — | — | — | — | — | — | — | — | — | — |
| A and C | 3 | — | 3 | — | — | — | — | — | — | — | — | — | 1 | 1 |
| A and G | 2 | — | — | — | — | — | 3 | — | — | — | — | — | — | — |
| A, C, and G | — | — | 1 | — | — | — | — | — | — | — | — | — | — | — |
| A and F | 1 | — | — | — | — | — | — | — | — | — | — | — | — | — |
| IND | — | 3 | 1 | 1 | — | — | — | — | — | — | — | — | — | — |
| NA | 2 | — | 1 | 1 | — | — | — | — | — | — | — | — | — | — |
IND, indeterminate.
NA, no amplification.
—, zero samples.
Single genotype discrepancies.
For all four specimens with discrepant results for a single genotype, the genotype determined by LiPA was confirmed following sequencing and phylogenetic analysis of the LiPA-derived amplicon. Therefore, the discrepancy was further investigated by clonal analysis of the pre-S amplicon. Among the four specimens with discrepant results, clonal analysis identified the same genotypes in both specimens determined by either method for two of the specimens, thus showing the presence of mixed infections in these samples. Two specimens were not further analyzed due to a lack of serum.
Mixed genotype discrepancies.
Clonal analysis of the pre-S region amplified from specimens demonstrating a mixed genotype by LiPA (n = 19) was performed. Two specimens were not further analyzed due to a lack of serum or an inability to amplify the pre-S region. Up to 12 clones from each transformation were sequenced to analyze the mixture of genotypes present. For all except six of the analyses, a mixture of genotype clones equivalent to the genotypes determined by LiPA was observed. Only four of the specimens provided one genotype following sequencing of all clones (the same genotype determined by sequencing of the pre-S region). Interestingly, all specimens with genotype G clones, as determined by sequencing, were confirmed to contain multiple infections with genotype A and G clones, as has been reported previously (11). The specimen with genotype C showing a mixed genotype A, C, and G infection by LiPA (Table 1) had a mixture of genotype A and C clones upon clonal analysis, but no genotype G clones were observed among the clones tested. One specimen with genotype A clones showing a mixed genotype A and G infection by LiPA also had a mixture of genotype A and C clones upon clonal analysis. Upon closer inspection of the original membrane strip, band 8 (Fig. 2, genotype C) was also observed to be reactive; however, according to the interpretation instructions provided with the LiPA kit, this single band should be ignored in case of the presence of multiple genotypes.
DISCUSSION
A greater demand for genotyping of patient strains of HBV is likely to occur as specific clinical associations with each genotype becomes increasingly apparent (3, 6, 16). To date, a number of HBV genotyping methods have been published. These methods consist of PCR with genotype-specific primers (12, 22, 25) or PCR amplification of the pre-S region (15) or S region (19) followed by restriction fragment length polymorphism analysis. These methods offer a simple and rapid means of determining the HBV genotype; however, these methods may fail to type all isolates (27) and interpretation of results may be difficult, particularly in the case of mixed genotype infections (13). Several HBV genotyping methods are now commercially available, including the LiPA kit and an enzyme immunoassay (EIA) with monoclonal antibodies raised against genotype-specific epitopes in the pre-S2 region (HBV Genotype EIA; Institute of Immunology, Tokyo, Japan) (32, 33). The EIA kit offers a convenient, serologically based assay, although it, too, may fail to type the HBV DNA in clinical samples due to the presence of mixtures of genotypes (32) or low levels of HBsAg in the sample (14, 20). Therefore, a distinct advantage of the LiPA kit is the sensitive detection of mixed genotype infections, as observed in the present study. The LiPA detected 19 of 188 (10%) mixed genotype infections that were not detected by sequencing of the pre-S region.
LiPA detected all seven HBV genotypes in HBV-infected clinical specimens submitted to laboratory reference services in Canada. An ongoing Canadian laboratory surveillance program that monitors specimens from patients with acute cases of HBV infection has verified that all seven genotypes are present in Canada. The isolates in surveillance specimens have been genotyped by sequencing and phylogenetic analysis of the pre-S1/pre-S2 region or by a restriction digest algorithm (E. Giles and C. Osiowy, unpublished data). The most predominant genotype observed since our surveillance of HBV began in 1999-2000 has been genotype D, followed by genotypes A, C, and B. Genotypes E, F, and G have been infrequently detected by these methods. In the present study, the number of genotype G clones detected increased approximately twofold by LiPA, with all genotype G infections detected as mixed genotype G and A infections. This finding confirms previous observations that patients infected with genotype G are frequently coinfected with genotype A (10, 11). We also observed multiple genotypes in many of the specimens tested by clonal analysis. A high frequency of genotypic coinfection in patients with chronic HBV infection has been observed previously (5, 21). However, one genotype will often predominate, and so sequence analysis provides information only on the majority strain, as was observed in the present study. LiPA appears to overcome this limitation by its sensitive detection of mixed genotypes.
LiPA is based on S-gene analysis, which has been shown to provide genotypic determinations similar to those provided by full genome analysis (19). The pre-S region has also been used for genotyping purposes (15), and it occasionally distinguishes genotypes not able to be typed by analysis of the S gene alone (1, 15, 29). Even so, our results demonstrated a high degree of correlation between the genotypes determined by genotyping of the pre-S region and genotyping of the S region, as has been observed previously (15).
Discrepancies between the two methods appear to be due to the presence of mixed genotype infections. We detected 19 samples with genotypic coinfections by LiPA, and of the 17 samples that could be further analyzed, genotypic coinfections were confirmed in 11 by clonal analysis. Coinfecting genotype clones may have been missed in six of the specimens because an insufficient number of clones were sequenced, and therefore, this suggests that LiPA has a very high sensitivity for the detection of multiple genotypes. Alternatively, band reactivity may have been incorrectly interpreted due to the presence of faint bands that are nonspecific. A different possibility is the occurrence of recombination in these specimens. Recombination between coinfecting HBV isolates may be a relatively frequent event (4, 21). Within the region of the HBsAg gene analyzed by LiPA, recombination crossover points that produce a mosaic genotype A and D genome have been reported previously (21). In addition, the possibility exists that PCR artifacts were produced during PCR amplification of a mixture of closely related target sequences. This may lead to the observation of an apparent hybrid between two genotypes. Further sequencing and analysis of the entire genome of these samples would be required to determine if actual recombination had occurred. We also observed what appeared to be the lack of specific hybridization to the test strip of certain amplicons from mixed infections, as was observed with the LiPA result indicating the presence of genotypes A and G (genotypes A and C by clonal analysis) by showing a single band for genotype C on the LiPA test strip, which is interpreted as genotypes A and G. It is possible that in rare cases certain genotype C strains may not react specifically with the oligonucleotide probes for genotype C on the membrane strip (bands 8 and 9 in Fig. 2).
In summary, the INNO-LiPA HBV genotyping assay is a very convenient and rapid method for the specific detection of HBV genotypes. The method correlates very well with existing genotyping methods and has the distinct advantage of sensitively detecting mixed genotype infections. This method should provide a sensitive and reliable means of genotyping for clinical diagnostic laboratories.
Acknowledgments
We gratefully acknowledge Jingxin Cao and Erwin Sablon for helpful discussions and editorial comments.
REFERENCES
- 1.Alestig, E., C. Hannoun, P. Horal, and M. Lindh. 2001. Hepatitis B virus genotypes in Mongols and Australian aborigines. Arch. Virol. 146:2321-2329. [DOI] [PubMed] [Google Scholar]
- 2.Chu, C., M. Hussain, and A. S. F. Lok. 2002. Hepatitis B virus genotype B is associated with earlier HBeAg seroconversion compared with hepatitis B virus genotype C. Gastroenterology 122:1756-1762. [DOI] [PubMed] [Google Scholar]
- 3.Chu, C., and A. S. F. Lok. 2002. Clinical significance of hepatitis B virus genotypes. Hepatology 35:1274-1276. [DOI] [PubMed] [Google Scholar]
- 4.Cui, C., J. Shi, L. Hui, H. Xi, Zhuoma, Quni, Tsedan, and G. Hu. 2002. The dominant hepatitis B virus genotype identified in Tibet is a C/D hybrid. J. Gen. Virol. 83:2773-2777. [DOI] [PubMed] [Google Scholar]
- 5.Hannoun, C., K. Krogsgaard, P. Horal, M. Lindh, and INTERPRED Trial Group. 2002. Genotype mixtures of hepatitis B virus in patients treated with interferon. J. Infect. Dis. 186:752-759. [DOI] [PubMed] [Google Scholar]
- 6.Kao, J. 2002. Hepatitis B viral genotypes: clinical relevance and molecular characteristics. J. Gastroenterol. Hepatol. 17:643-650. [DOI] [PubMed] [Google Scholar]
- 7.Kao, J., and D. Chen. 2002. Global control of hepatitis B virus infection. Lancet Infect. Dis. 2:395-403. [DOI] [PubMed] [Google Scholar]
- 8.Kao, J., P. Chen, M. Lai, and D. Chen. 2002. Genotypes and clinical phenotypes of hepatitis B virus in patients with chronic hepatitis B virus infection. J. Clin. Microbiol. 40:1207-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kao, J., C. Liu, and D. Chen. 2002. Hepatitis B viral genotypes and lamivudine resistance. J. Hepatol. 36:303-304. [DOI] [PubMed] [Google Scholar]
- 10.Kato, H., E. Orito, R. G. Gish, N. Bzowej, M. Newsom, F. Sugauchi, S. Suzuki, R. Ueda, Y. Miyakawa, and M. Mizokami. 2002. Hepatitis Be antigen in sera from individuals infected with hepatitis B virus of genotype G. Hepatology 35:922-929. [DOI] [PubMed] [Google Scholar]
- 11.Kato, H., E. Orito, R. G. Gish, F. Sugauchi, S. Suzuki, R. Ueda, Y. Miyakawa, and M. Mizokami. 2002. Characteristics of hepatitis B virus isolates of genotype G and their phylogenetic differences from the other six genotypes (A through F). J. Virol. 76:6131-6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kato, H., E. Orito, F. Sugauchi, R. Ueda, R. G. Gish, S. Usuda, Y. Miyakawa, and M. Mizokami. 2001. Determination of hepatitis B virus genotype G by polymerase chain reaction with hemi-nested primers. J. Virol. Methods 98:153-159. [DOI] [PubMed] [Google Scholar]
- 13.Kato, H., R. Ruzibakiev, N. Yuldasheva, T. Hegay, F. Kurbanov, B. Achundjanov, L. Tuichiev, S. Usuda, R. Ueda, and M. Mizokami. 2002. Hepatitis B virus genotypes in Uzbekistan and validity of two different systems for genotyping. J. Med. Virol. 67:477-483. [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi, M., Y. Arase, K. Ikeda, A. Tsubota, Y. Suzuki, S. Saitoh, M. Kobayashi, F. Suzuki, N. Akuta, T. Someya, M. Matsuda, J. Sato, K. Takagi, Y. Miyakawa, and H. Kumada. 2002. Viral genotypes and response to interferon in patients with acute prolonged hepatitis B virus infection of adulthood in Japan. J. Med. Virol. 68:522-528. [DOI] [PubMed] [Google Scholar]
- 15.Lindh, M., J. E. Gonzalez, G. Norkrans, and P. Horal. 1998. Genotyping of hepatitis B virus by restriction pattern analysis of a pre-S amplicon. J. Virol. Methods 72:163-174. [DOI] [PubMed] [Google Scholar]
- 16.Locarnini, S. 2002. Clinical relevance of viral dynamics and genotypes in hepatitis B virus. J. Gastroenterol. Hepatol. 17:S322-S328. [DOI] [PubMed] [Google Scholar]
- 17.Lok, A. S. F., F. Zoulim, S. Locarnini, A. Mangia, G. Niro, H. Decraemer, G. Maertens, F. Hulstaert, K. DeVreese, and E. Sablon. 2002. Monitoring drug resistance in chronic hepatitis B virus (HBV)-infected patients during lamivudine therapy: evaluation of performance of INNO-LiPA HBV DR assay. J. Clin. Microbiol. 40:3729-3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Minuk, G. Y. 2002. Hepatitis B viral mutants and their relevance to the Canadian health care system. Can. J. Gastroenterol. 16:45-54. [DOI] [PubMed] [Google Scholar]
- 19.Mizokami, M., T. Nakano, E. Orito, Y. Tanaka, H. Sakugawa, M. Mukaide, and B. H. Robertson. 1999. Hepatitis B virus genotype assignment using restriction fragment length polymorphism patterns. FEBS Lett. 450:66-71. [DOI] [PubMed] [Google Scholar]
- 20.Moriya, T., I. K. Kuramoto, H. Yoshizawa, and P. V. Holland. 2002. Distribution of hepatitis B virus genotypes among American blood donors determined with a preS2 epitope enzyme-linked immunosorbent assay kit. J. Clin. Microbiol. 40:877-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morozov, V., M. Pisareva, and M. Groudinin. 2000. Homologous recombination between different genotypes of hepatitis B virus. Gene 260:55-65. [DOI] [PubMed] [Google Scholar]
- 22.Naito, H., S. Hayashi, and K. Abe. 2001. Rapid and specific genotyping system for hepatitis B virus corresponding to six major genotypes by PCR using type-specific primers. J. Clin. Microbiol. 39:362-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Norder, H., A. Courouce, and L. O. Magnius. 1994. Complete genomes, phylogenetic relatedness, and structural proteins of six strains of the hepatitis B virus, four of which represent two new genotypes. Virology 198:489-503. [DOI] [PubMed] [Google Scholar]
- 24.Osiowy, C. 2002. Sensitive detection of HBsAg mutants by a gap ligase chain reaction assay. J. Clin. Microbiol. 40:2566-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Repp, R., S. Rhiel, K. H. Heermann, S. Schaefer, C. Keller, P. Ndumbe, F. Lampert, and W. H. Gerlich. 1993. Genotyping by multiplex polymerase chain reaction for detection of endemic hepatitis B virus transmission. J. Clin. Microbiol. 31:1095-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakugawa, H., H. Nakasone, T. Nakayoshi, E. Orito, M. Mizokami, T. Yamashiro, T. Maeshiro, F. Kinjo, A. Saito, and Y. Miyagi. 2002. Preponderance of hepatitis B virus genotype B contributes to a better prognosis of chronic HBV infection in Okinawa, Japan. J. Med. Virol. 67:484-489. [DOI] [PubMed] [Google Scholar]
- 27.Sanchez-Tapias, J. M., J. Costa, A. Mas, M. Bruguera, and J. Rodes. 2002. Influence of hepatitis B virus genotype on the long-term outcome of chronic hepatitis B in western patients. Gastroenterology 123:1848-1856. [DOI] [PubMed] [Google Scholar]
- 28.Stuyver, L., C. Van Geyt, S. De Gendt, G. Van Reybroeck, F. Zoulim, G. Leroux-Roels, and R. Rossau. 2000. Line probe assay for monitoring drug resistance in hepatitis B virus-infected patients during antiviral therapy. J. Clin. Microbiol. 38:702-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sugauchi, F., E. Orito, H. Kato, S. Suzuki, S. Kawakita, Y. Sakamoto, K. Fukushima, T. Akiba, N. Yoshihara, R. Ueda, and M. Mizokami. 2003. Genotype, serotype, and phylogenetic characterization of the complete genome sequence of hepatitis B virus isolates from Malawian chronic carriers of the virus. J. Med. Virol. 69:33-40. [DOI] [PubMed] [Google Scholar]
- 30.Sumi, H., O. Yokosuka, N. Seki, M. Arai, F. Imazeki, T. Kurihara, T. Kanda, K. Fukai, M. Kato, and H. Saisho. 2003. Influence of hepatitis B virus genotypes on the progression of chronic type B liver disease. Hepatology 37:19-26. [DOI] [PubMed] [Google Scholar]
- 31.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Usuda, S., H. Okamoto, H. Iwanari, K. Baba, F. Tsuda, Y. Miyakawa, and M. Mayumi. 1999. Serological detection of hepatitis B virus genotypes by ELISA with monoclonal antibodies to type-specific epitopes in the preS2-region product. J. Virol. Methods 80:97-112. [DOI] [PubMed] [Google Scholar]
- 33.Usuda, S., H. Okamoto, T. Tanaka, K. Kidd-Ljunggren, P. V. Holland, Y. Miyakawa, and M. Mayumi. 2000. Differentiation of hepatitis B virus genotypes D and E by ELISA using monoclonal antibodies to epitopes on the preS2-region product. J. Virol. Methods 87:81-89. [DOI] [PubMed] [Google Scholar]
- 34.Yuen, M., E. Sablon, H. Yuan, D. Wong, C. Hui, B. Wong, A. Chan, and C. Lai. 2003. Significance of hepatitis B genotype in acute exacerbation, HBeAg seroconversion, cirrhosis-related complications, and hepatocellular carcinoma. Hepatology 37:562-567. [DOI] [PubMed] [Google Scholar]
- 35.Zollner, B., J. Petersen, P. Schafer, M. Schroter, R. Laufs, M. Sterneck, and H. Feucht. 2002. Subtype-dependent response of hepatitis B virus during the early phase of lamivudine treatment. Clin. Infect. Dis. 34:1273-1277. [DOI] [PubMed] [Google Scholar]