Abstract
Background
Previously, we identified 3,274 distinct differentially expressed genes in abdominal aortic aneurysm (AAA) tissue compared to non-aneurysmal controls. As transcriptional control is responsible for these expression changes, we sought to find common transcriptional elements in the promoter regions of the differentially expressed genes.
Methods and Results
We analyzed the up- and downregulated gene sets with Whole Genome rVISTA to determine the transcription factor binding sites (TFBSs) overrepresented in the 5 kb promoter regions of the 3,274 genes. The downregulated gene set yielded 144 TFBSs that were overrepresented in the subset when compared to the entire genome. In contrast, the upregulated gene set yielded only 13 distinct overrepresented TFBSs. Interestingly, as classified by TRANSFAC®, 8 of the 13 transcription factors (TFs) binding to these regions belong to the ETS family. Additionally, NFKB and its subunits p50 and p65 showed enrichment. Immunohistochemical analyses in 10 of the TFs from the upregulated analysis showed 9 to be present in AAA tissue. Based on Gene Ontology analysis of biological process categories of the upregulated target genes of enriched TFs, 10 TFs had enrichment in immune system process among their target genes.
Conclusions
Our genome-wide analysis provides further evidence of ETS and NFKB involvement in AAA. Additionally, our results provide novel insight for future studies aiming to dissect the pathogenesis of AAA and have uncovered potential therapeutic targets for AAA prevention.
Keywords: Aneurysm, Aorta, Genomics, Transcription Factors
Background
Abdominal aortic aneurysm (AAA) is a balloon-like dilatation that occurs in the infrarenal segment of the aorta.1 If left untreated, the aortic wall continues to weaken and becomes unable to withstand the forces of the luminal blood pressure resulting in progressive dilatation and eventual rupture.2, 3 With a mortality rate of ruptured AAA between 65 and 90%, AAA is among the leading causes of death in the United States.4 Currently, there are no pharmacological therapies approved for the treatment of AAA, leaving surgical intervention with high rates of morbidity and mortality as the only option for sufferers.
Recently, we generated genome-wide expression profiles comparing gene expression in AAA tissue to age, sex and ethnicity matched non-aneurysmal control aorta using two different microarray platforms yielding 3,274 distinct genes whose expression was significantly altered in AAA.5 These types of analyses offer critical insight into changes occurring at the mRNA level of a disease state. It is transcriptional control, however, that is fundamentally responsible for these expression changes. Therefore, integration of the expression data with genome sequence data to uncover transcriptional regulatory elements serves to provide more comprehensive information about complex diseases like AAA. An implicit hypothesis for this integrated approach is that sets of co-regulated genes share regulatory motifs that are similar and that regulatory motifs overrepresented in these sets are responsible for driving gene expression changes.6
Phylogenetic footprinting can be used to discover regulatory regions in human genomic DNA.7–10 This method relies on the concept that regions of non-coding DNA that are highly conserved across species have important biological functions and often contain binding sites for transcription factors (TFs).11 Comparative genomics, along with a computational approach to integrate gene expression data with sequence data has been used in a method coined “transcriptional genomics” and has uncovered novel transcriptional elements involved in human heart failure.10 A similar approach in the study of AAA holds great importance, as the pathobiology of the disease is still poorly understood.12 The present study focused on the regulatory elements contained in the 5 kb promoter regions of the 3,274 differentially expressed genes.
Methods
Gene Expression Studies
Our laboratory utilized two microarray platforms to generate global mRNA expression profiles for both aneurysmal (N = 7) and non-aneurysmal (N = 7) abdominal aorta. The details on these studies have been described previously,5 and the microarray data can be obtained at the Gene Expression Omnibus (GEO) database (Series# GSE7084; http://www.ncbi.nlm.nih.gov/geo/).
In Silico Analysis to Identify TFBSs Enriched in Differentially Expressed Gene Sets
We analyzed the up- and downregulated gene sets separately to elucidate the transcription factor binding sites (TFBSs) in the regulatory regions of the differentially expressed genes using a computational approach implemented in Whole Genome rVISTA13 (http://genome.lbl.gov/vista/index.shtml), a publicly available bioinformatics program. For a given set of genes, Whole Genome rVISTA generates a list of TFBSs that are over-represented in the set when compared to the entire genome. The rVISTA tool identifies TFBSs based on the TRANSFAC® Professional library14 in interspecies sequence alignments and then determines which of the TFBSs are conserved in order to maximize the identification of functional sites.15
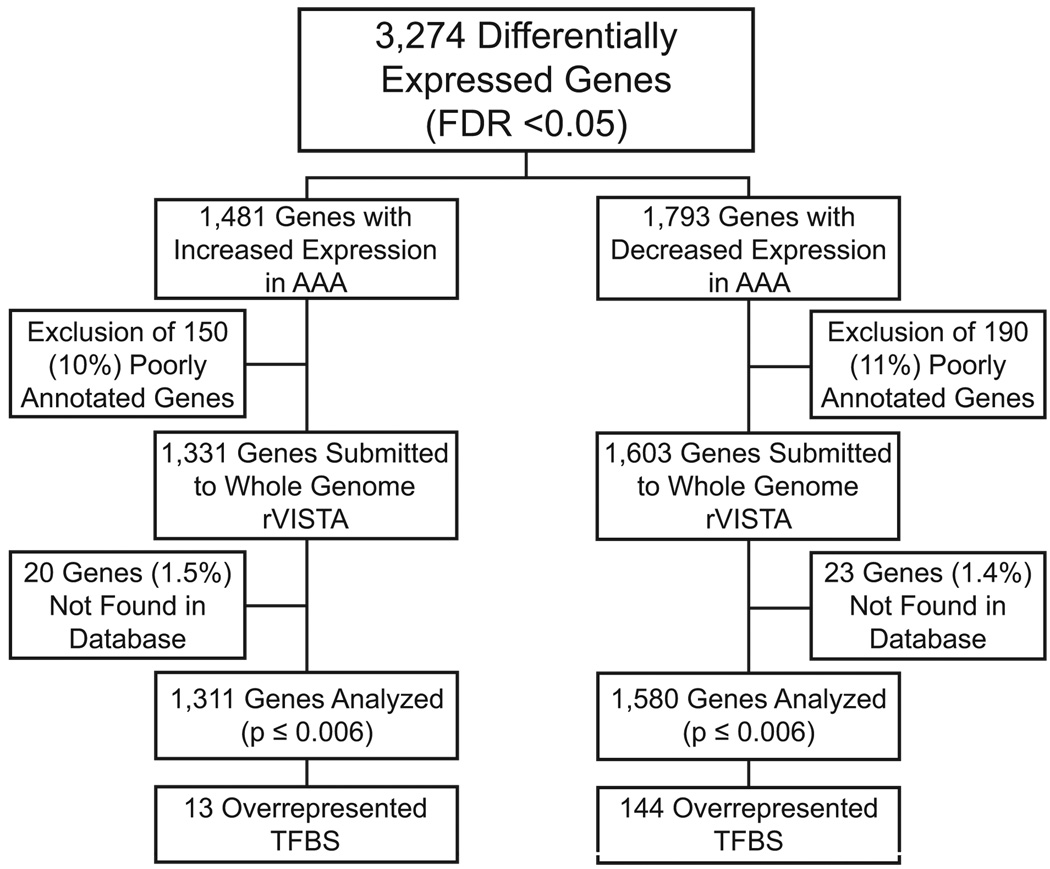
Uncharacterized and poorly annotated genes were removed from the 3,274 differentially expressed genes identified in our previous AAA microarray analysis.5 This resulted in the removal of 150 (10%) genes from the upregulated gene set and 190 (11%) genes from the downregulated gene set. Upregulated (1,331 genes) and downregulated (1,603 genes) gene sets were then analyzed separately in Whole Genome rVISTA to search for human sequences (UCSC version hg18, NCBI Build 36.1) conserved in alignment with mouse (UCSC version mm8, NCBI Build 36). The 5 kb region upstream from the transcription start site of each gene was examined. Binomial distribution was used for statistical analyses with a p-value cutoff of 0.006. It should be noted that 20 (1.5%) of the 1,331 upregulated and 23 (1.4%) of the 1,603 downregulated genes were not found in the database resulting in an analysis of 1,311 and 1,580 genes, respectively. An outline of the analyses is presented in Figure 1.
Figure 1.
Study design. Overall design of the enrichment analysis of transcription factor binding sites (TFBSs) in 5 kb promoter regions upstream from transcription start sites among differentially expressed genes in abdominal aortic aneurysm (AAA). The binomial distribution was used for statistical analyses with a p-value cutoff of 0.006. FDR, False-Discovery Rate.
The TRANSFAC® database version 2008.3 (BIOBASE GmbH, Wolfenbüttel, Germany) was used to classify the TFs, known to bind to the TFBSs, in our analysis into families.14
Immunohistochemical Analysis
Control aorta samples (n= 5; donor ages ranging from 58 to 87 years) were obtained at autopsy. Patient samples (6 AAA patients; ages ranging from 63 to 87 years) were tissues removed from the aneurysmal sac during surgical repair operations to trim the area for fitting the prosthesis (Table S1). Samples from other tissues were obtained during autopsies. The study was approved by The Institutional Review Board of Wayne State University.
Immunostaining was carried out with formalin-fixed paraffin-embedded tissue sections as described previously.16 The slides were incubated with a primary antibody against the protein of interest [Santa Cruz Biotechnology, Santa Cruz, CA: anti-PEA3, sc-113; anti-ELF1, sc-631; anti-ETS2, sc-22803; anti-ETS1, sc-350; anti-NFKB p65, sc-8008; anti-NFKB p50, sc-114; anti-GABP-alpha, sc-28311; anti-TEL2, sc-102131; anti-NERF, sc-6829; and anti-ISRE (anti-STAT1), sc-592. Genetex, San Antonio, TX: anti-RUNX1, GRX94183]. A secondary antibody with avidin-biotin peroxidase amplification (Dako, Glostrup, Denmark: LSAB2) was used and the signal was detected using diaminobenzidine as a chromogen. Antibodies for each protein were first tested on tissue known to contain the protein of interest as positive controls. Non-specific IgG antibody in lieu of primary antibody served as a negative control.
Biological Process Analysis of Target Genes
The down- and upregulated gene sets were used to identify enriched Gene Ontology (GO) biological process (BP) categories with a software package available using R and Bioconductor.17, 18 Then, Whole Genome rVISTA was used to sub-categorize genes in the upregulated gene set according to the overrepresented TFBSs they contain for ELF1, ETS2, ETS1, NFKB, NFKB P65, NFKB P50, GABP, TEL2, ISRE, AML1 and NERF. PEA3 and c-ETS 1–68 were excluded from the analysis, since no expression in aortic tissues was detected for PEA3, and no human TF binding to the TFBS c-ETS 1–68 was found. These gene lists were then used to identify enriched GO BP categories they have in common using WebGestalt (Web-based Gene SeT AnaLysis Toolkit; http://bioinfo.vanderbilt.edu/webgestalt).19, 20 GO analysis was performed using the hypergeometric test to determine overrepresentation of terms in the gene set compared to the 18,057 distinct genes represented on both Affymetrix and Illumina arrays based on our initial microarray studies.5 Directed acyclic graphs (DAGs) were generated to provide visual presentation of the hierarchical nature of the GO categories. They were re-drawn in Adobe Illustrator (version 12.0.1; Adobe Systems Inc., San Jose, CA) to provide high-resolution images.
Results
The 1,580 genes identified as downregulated in AAA in our previous microarray analyses5 contained 144 overrepresented TFBSs in their 5 kb promoter regions when compared to the entire genome (Table S2; Figure 1). Of these 1,580 genes, 1,550 (98%) contain at least one binding site for one of the 144 overrepresented TFBSs with a range of one (16 genes) to 128 (2 genes) enriched TFBSs and an average of 52 TFBSs per gene (not shown). The 144 TFs belong to 27 TF classes (Tables S2 and S3). A GO analysis, looking for enrichment in BP categories, showed that the group of 1,242 (77.5% of the total number of genes) downregulated genes with GO annotations was enriched among others for GO terms cell adhesion, muscle development, cell morphogenesis, and cell-matrix adhesion (Table S4).
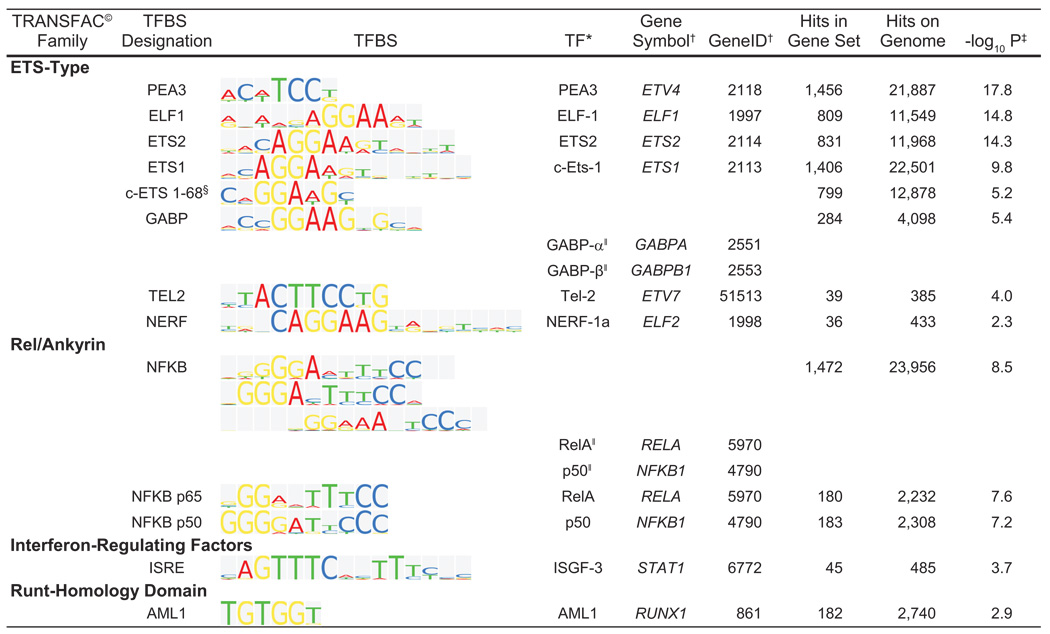
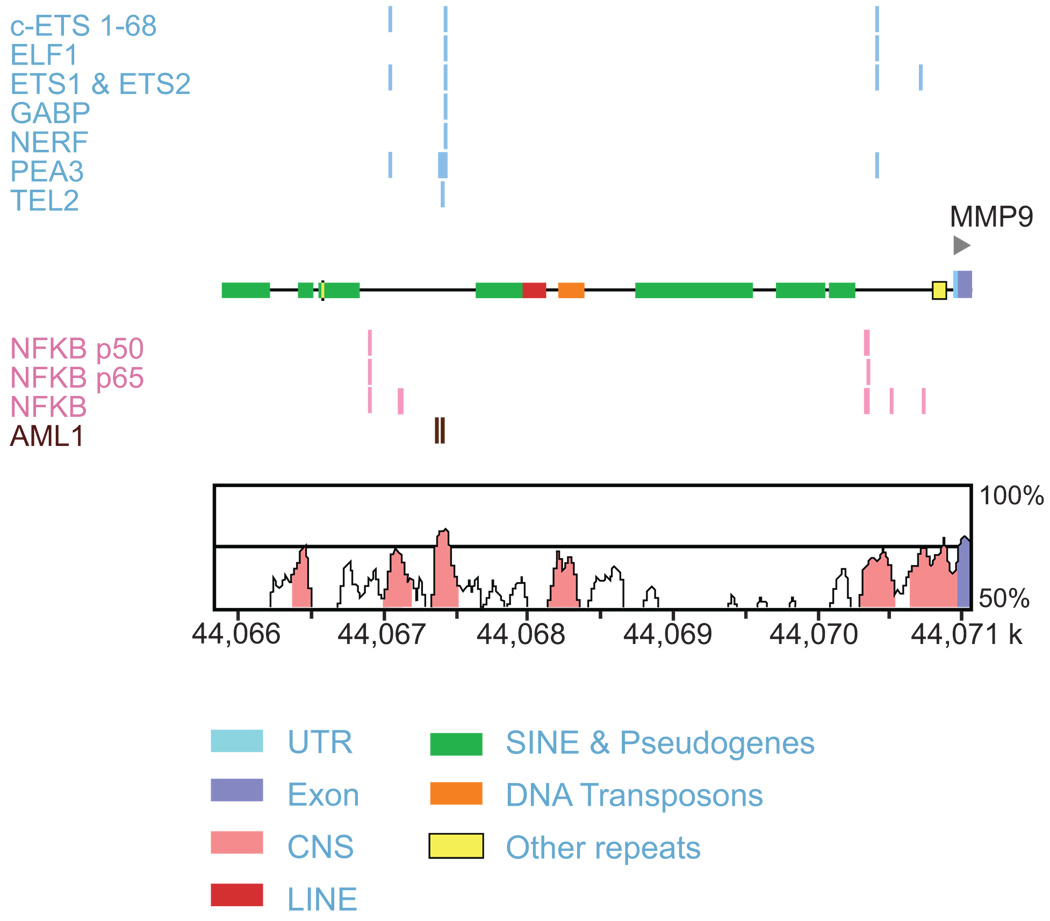
Analysis of the promoter regions of the 1,311 genes upregulated in AAA yielded only 13 overrepresented TFBSs when compared to the entire genome (Figure 1; Figure 2). Of these 1,311 genes, 981 (75%) genes contained binding sites for at least one of the 13 overrepresented TFs, with a range of one (154 genes) to 12 factors (MMP9; Figure 3 and an average of 4.2 TFs per gene (Table S5).
Figure 2.
Sequence logo representations for the transcription factor binding sites (TFBS) enriched in the upregulated gene set. Standard sequence logo representations of the binding site sequence matrices are shown in the column TFBS. Sites are identified, and grouped by family, according to the TRANSFAC© database. For each site the transcription factor is identified by protein name, gene symbol and gene ID. Also shown are the number of sites in the gene set, in the genome and the P value for the comparison. Symbols: *, transcription factors, also called binding proteins; †, gene symbol and gene identifier obtained from the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/); ‡, negative log base 10 of the P value from the binomial distribution; §, No transcription factor identified in human; and ‖, factors also bind as a heterodimer to the respective site. Whole Genome rVISTA reported monomeric binding sites for NFKB p65 and NFKB p50 in addition to the NFKB site.
Figure 3.
Schematic of the 5 kb promoter region in MMP9. Upper portion of the figure shows conserved, aligned binding sites in the 5 kb promoter region of MMP9 for the enriched TFBSs in the upregulated gene set analysis. In the lower portion of the figure the y-axis demonstrates the conservation (%) between mouse and human, and the x-axis represents the location on the human chromosome. The genomic sequence is in kilobase pairs (k). Conserved regions above the level of 70%/100 bp are highlighted under the curve, with pink indicating a conserved noncoding region; and blue, a conserved exon. 43 UTR, untranslated region; CNS, conserved non-coding sequence; LINE, long interspersed nuclear element; SINE, short interspersed nuclear element. Figure modified from Whole Genome rVISTA output.43
When classified by TRANSFAC®, 8 of the 13 enriched TFBSs in the upregulated gene set had binding proteins belonging to the ETS-type family of TFs (Figure 2. Additionally, NFKB and its heterodimeric subunits p65 and p50 showed enrichment in our analysis (Figure 2).
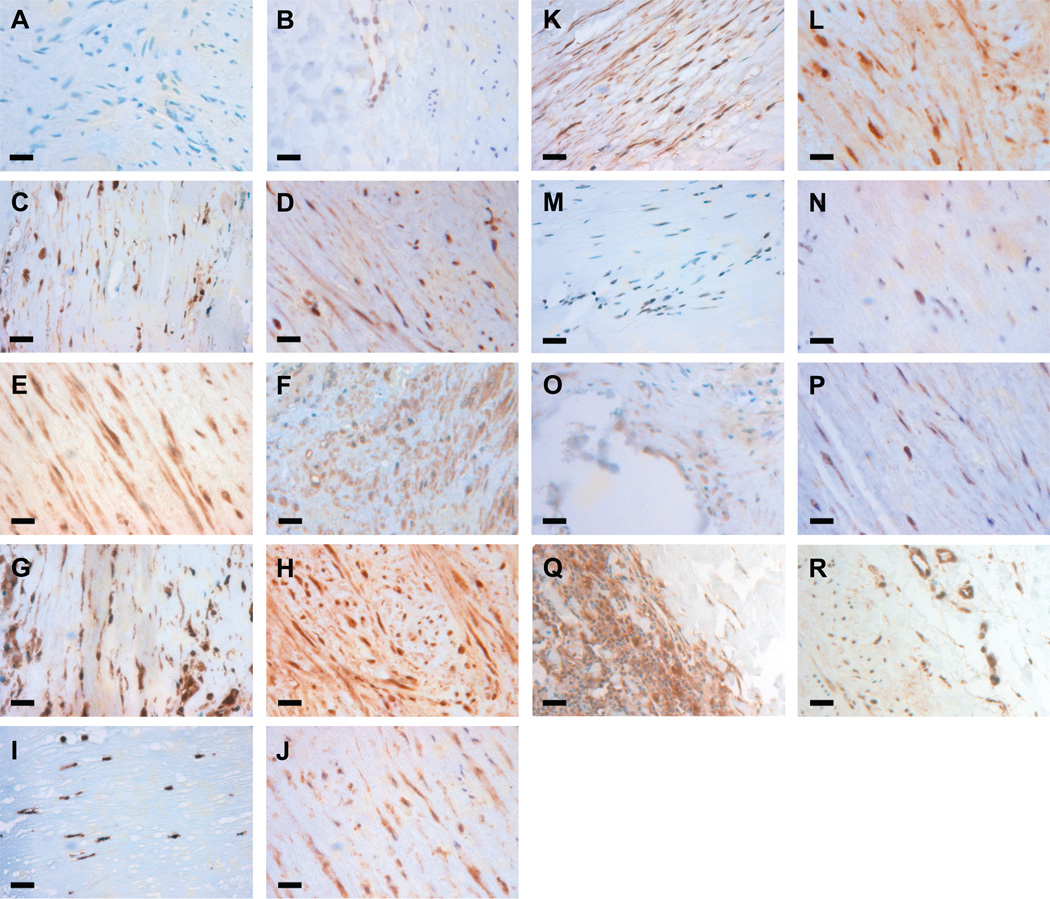
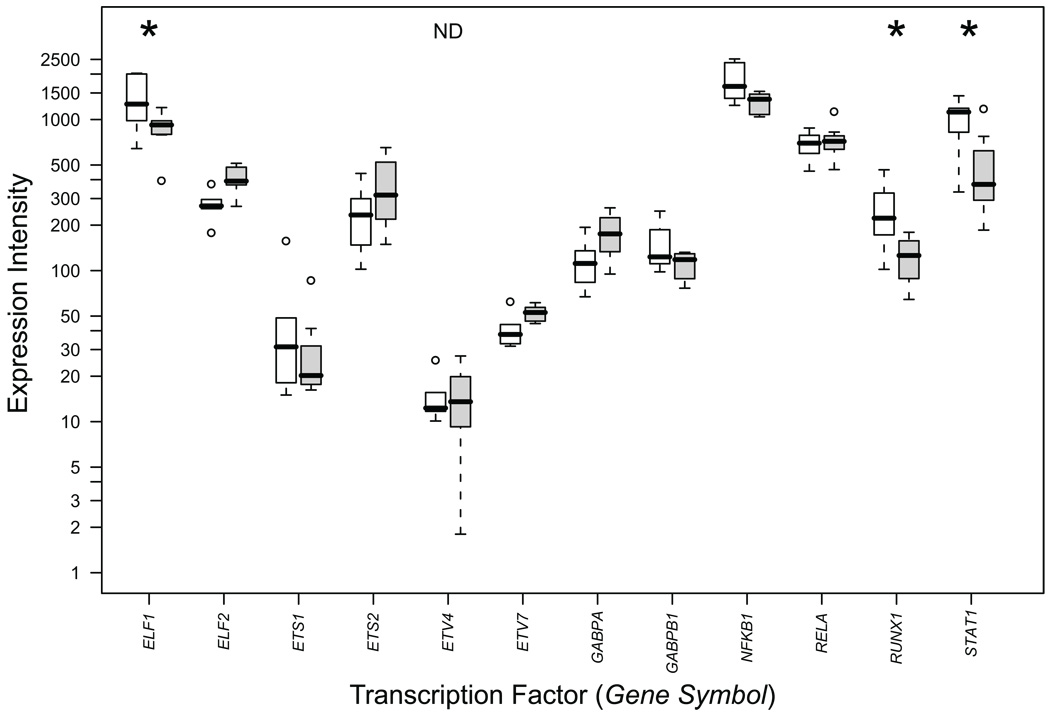
Immunohistochemical analysis was carried out for 10 of the 13 TFs enriched in the upregulated gene set to determine if the proteins were present both in non-aneurysmal aorta and AAA tissue. TF c-ETS 1–68 was not analyzed because no human protein binding to the TFBS has been identified, NERF was not analyzed, because the antibody used failed to show staining even in the positive control samples, and TEL2 was not analyzed due to high background in the antibody staining. Staining in aneurysmal tissue was positive for ELF1, ETS1, ETS2, NFKB p65, NFKB p50, GABPα, ISRE (STAT1) and AML1 in at least some component of the aortic media or adventitia (Figure 4) and negative for PEA3. The immunohistochemical results were in reasonable agreement with mRNA expression data from our microarray studies5 in that all of the proteins tested in our immunohistochemical analysis aside from PEA3 were detected (Figures 4 and 5; Table S6). PEA3 was not detectable on the mRNA level either (Table S6). TEL2 had a robust mRNA signal (Table S6), but due to technical problems, we did not succeed in the immunohistochemical analyses for TEL2. It is of interest that three of the TFs, ELF1, AML1 (RUNX1) and ISGF-3 (the gene symbol for ISGF-3 which binds to ISRE is STAT1) had significantly increased mRNA levels in AAA (Table S6; Figure 5).
Figure 4.
Immunohistochemical staining for transcription factors with enriched binding sites. Immunostaining was performed in both AAA tissue (first and third column) and non-aneurysmal control aorta (second and fourth column) for PEA3 (A,B), ELF1 (C,D), ETS2 (E,F), ETS1 (G,H), NFKB p65 (I,J), NFKB p50 (K,L), GABP alpha (M,N), AML1 (O,P), and STAT1 (Q,R). Positive staining is seen for all proteins in both AAA tissue and non-aneurysmal control aorta with the exception of PEA3. Bars=20 µm.
Figure 5.
Box and whisker plots of mRNA levels for transcription factors with statistically overrepresented binding sites in the promoters of the upregulated gene set. Results are based on microarray expression profiling which have been described previously.5 RNA expression levels in signal intensity units are indicated on the ordinate-axis in logarithmic scale. Open boxes represent AAA patients (n = 7) and shaded boxes represent abdominal aorta samples from non-aneurysmal autopsy samples (n = 7). Thick horizontal bars in the boxes indicate median values, boxes indicate interquartile range, whiskers indicate range of non-outlier values, circles indicate outliers less than 3 interquartile range units. Gene symbols available from the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/) were used. Asterisks indicate statistically significant results after FDR correction. ND, not detected, indicates that PEA3 (ETV4) was scored as not expressed since the signals were not statistically different from background noise at the 99% confidence level. For numerical values of the expression data, see Table S6.
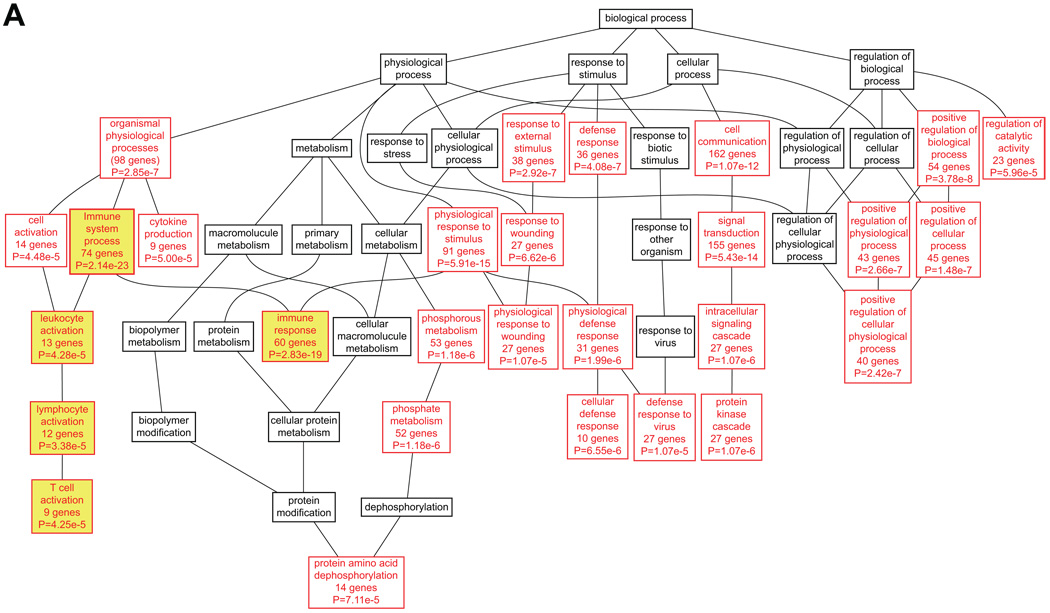
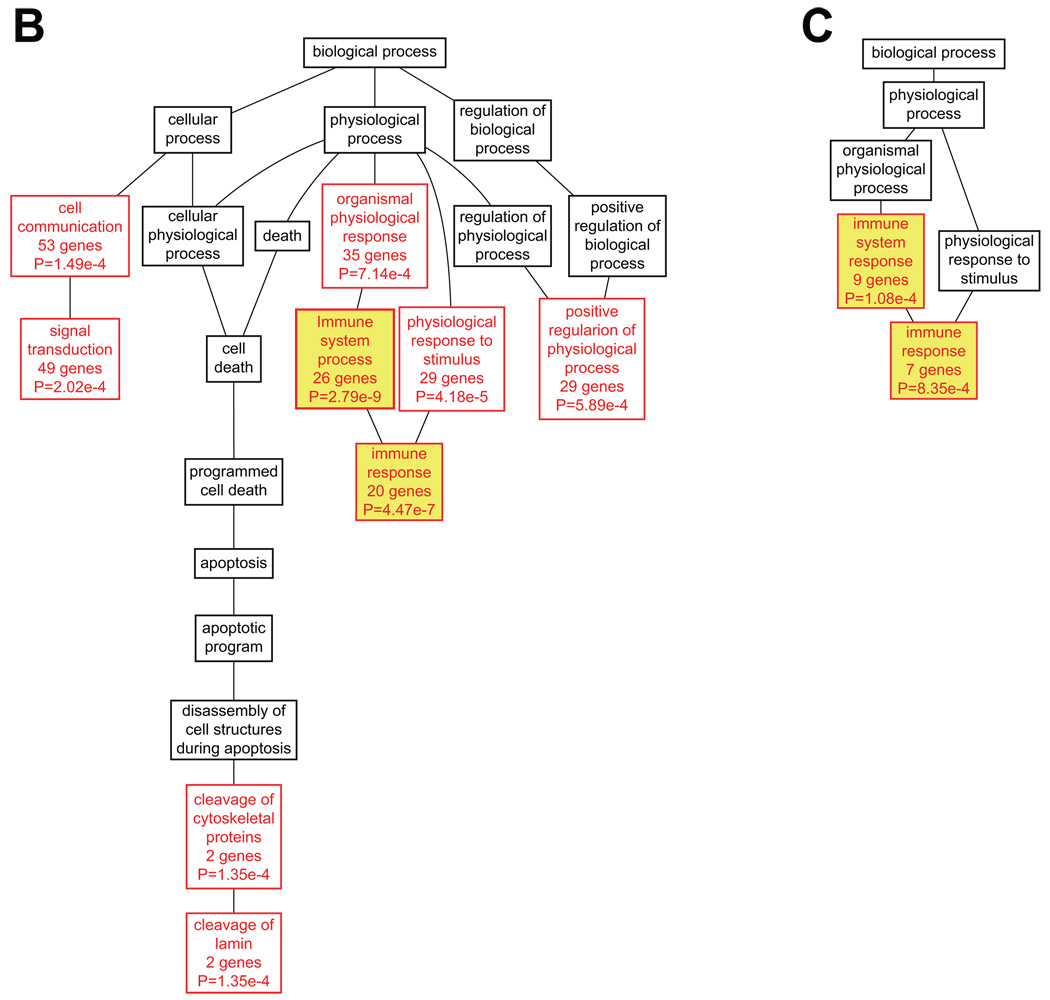
Next, a GO analysis was performed for the entire set of upregulated target genes. GO annotations were available for 91.8% of the genes. Many different GO categories related to immune function were among the most enriched GO categories (Table S7). This analysis was followed up by analyzing the GO categories for the different sub-categories of genes separately in order to determine if genes in each set defined by presence of a specific TF were enriched for, or participated in, the same BPs. The analysis presented a total of 65 enriched BP categories among the genes containing binding sites for the 11 TFs listed in Figure 2 (PEA3 and c-ETS 1–68 were excluded from this analysis; Tables S8 and S9). As of December 2008, 840 of the 955 (88%) target genes submitted for analysis had GO annotations. Among the most significant BP categories were immune system process and immune response, in that genes with 10 of the 11 TFBS showed enrichment for immune system process and 9 for immune response. Furthermore, one or both of these were either the first or second most significant category for 9 of the 11 gene lists (Table S9). Only the gene lists for ETS2 and NERF did not have these GO categories among the top most significantly enriched categories. The most enriched categories for ETS2 were signal transduction and cell communication, although immune system development, hemopoiesis, and hemopoietic or lymphoid organ development, which are immune-related, were also enriched categories. Genes containing NERF binding sites were also highly enriched for immune processes such as: hemopoiesis, hemopoietic or lymphoid organ development, immune system development, and immune system process. The DAGs for target genes of ELF1, AML1 (RUNX1) and ISGF-3 (STAT1) are shown in Figure 6.
Figure 6.
Functional classification of target genes for ELF1 (A), AML1 (RUNX1, B) and ISGF-3 (STAT1, C). DAGs were generated using WebGestalt19, 20 and re-drawn in Adobe Illustrator. BP categories with enrichment at P<0.001 using the hypergeometric test are shown in red, whereas parent nodes with P>=0.001 are shown in black. Immune-related categories are shown in yellow boxes. For a complete listing of these target genes and their GO annotations, see Tables S5, S8 and S9.
Discussion
Although the underlying pathobiology of AAA remains largely unknown, the aneurysmal aortic wall is characterized by inflammation, vascular smooth muscle cell (VSMC) apoptosis and degradation of the extracellular matrix (ECM) (reviewed in references 12, 21). Particular attention has focused on MMP9, which is an abundant proteinase secreted by human AAA organ cultures in vitro and is elevated in the plasma of AAA patients.22, 23 Additionally, targeted gene disruption of Mmp9 in mice prevented AAA formation in the elastase-perfusion model.23 Furthermore, inflammatory macrophage migration to the site of aneurysm requires activation of MMP9 by plasminogen in mice.24 Considering its proven involvement in AAA pathogenesis, it is interesting that MMP9 contains at least one binding site in its 5 kb promoter region for 12 of the 13 enriched TFBSs in the upregulated gene set (Figure 3).
ETS1 and NFKB, both of which have binding sites in the MMP9 promoter, have been previously investigated for their effect on MMP9 expression. In an organ culture of human aorta, transfection with ETS1 decoy oligodeoxynucleotides (ODNs) decreased MMP1 as well as MMP9 expression,25 and in human VSMCs, overexpression of NFKB inhibitor, IκBα, decreased expression of MMP9, MMP1 and MMP3.26
In addition to their effect on MMP gene expression, both NFKB and ETS1 also play a role in several other processes pertinent to AAA pathogenesis. AAAs exhibit VSMC apoptosis along with inflammation throughout the media and adventitia of the aortic wall (reviewed in 12). Immune infiltrates27–31 are present in AAA tissue and have been implicated in these processes. It is thought that loss of VSMC is a result of apoptosis and that macrophages and T-cells participate in this process through their production of cytotoxic mediators such as cytokines, perforin and Fas/FasL.29 FasL is a downstream target of ETS1 in VSMCs derived from atherosclerotic lesions and targeted gene disruption of ETS1 decreased production of natural killer cells and interleukin (IL) 2, as well as diminished Th1-mediated T-cell responses (reviewed in 32). NFKB is involved in T-cell activation and the induction of several inflammatory cytokines (reviewed in 33). In addition, ETS1 increased IL5 promoter activity in Jurkat T-cells.34 The release of pro-inflammatory cytokines such as IL5 and IL6 by T-cells is thought to promote vascular inflammation at the site of AAA.28
The above characteristics have made ETS1 and NFKB potential therapeutic targets in the AAA field. Inhibition of ETS1 by decoy ODNs in a rat AAA model prevented aneurysmal dilatation as well as the destruction of elastin fibers in the aorta.25 Simultaneous inhibition of ETS1 and NFKB using chimeric decoy ODNs prevented AAA formation in both rat and rabbit models of AAA35, 36 and demonstrated AAA regression in a rabbit model in which AAA was already established using elastase perfusion.37 Additionally, upregulation of NFKB and ETS coincided with accelerated AAA formation in a rat hypertensive AAA model.38 Surprisingly, we did not observe significant differences in the mRNA levels for the ETS1, ETS2, NFKBp50 or NFKBp65 between human AAA and control tissues.
Perhaps the most interesting discovery of the current study was the finding that in the upregulated AAA gene set, all of the genes containing binding sites for ETS2 also contain ETS1 binding sites in their promoters (Table S5). Based on the sequence similarity (AGGA; Figure 2) in their TFBS matrices (TRANSFAC®) and the fact that ETS2 sites are more restrictive based on the positional weight matrices model, ETS2 sites are actually a subset of ETS1 sites. One implication of the similarity in their recognition sequences, which has been mentioned previously,32 is that targeting ETS1 binding sites using the decoy ODN methods25, 35–37 mentioned above is unspecific and may lead to inhibition of ETS2 as well. It is, therefore, difficult to conclude whether the effects on MMP9 gene expression and AAA prevention resulted from ETS1 or ETS2 inhibition or some type of cooperative effect. This is further complicated by the fact that they both have conserved binding sites in the promoter region of MMP9 (Figure 3). A study examining the effect of ETS transcription factors on the MMP9 gene promoter found that both ETS1 and ETS2 were able to activate MMP9 in response to endothelial growth factor.39
The current genome-wide analysis reinforces the immune component of AAA as 10 of the 11 target gene lists among the upregulated genes showed enrichment in the BP category immune system process. Although immune-related categories were prominent for all of the target gene lists, a wide range of BPs were identified among the target genes, including: signal transduction, physiological response to stimulus and cell communication. The downregulated genes, however, showed enrichment in very different functional categories such as cell adhesion and cell morphogenesis, and no immune-related functions were identified among these genes.
Additionally, the study identified novel TFBSs enriched in the set of upregulated genes in AAA. AML1 (gene symbol: RUNX1; Runt-related transcription factor 1) had a particularly interesting protein expression pattern based on immunohistochemistry. AML1 stained positive in both the vasa vasorum and the endothelial layer of the aortic wall (Figure 4). The vasa vasorum have been implicated in AAA pathogenesis as both MMP2 and MMP9 as well as tissue inhibitors of MMPs (TIMPs) co-localize in the vasa vasorum in AAA tissue.40 Of additional importance is that many genes biologically relevant to AAA that were upregulated in our microarray studies contain binding sites for AML1 such as MMP9, several interleukins and chemokines, as well as PLAU (Table S5).41 Furthermore, the mRNA levels of RUNX1 were significantly increased in AAA compared to abdominal aortas obtained from age-, sex- and ethnicity-matched controls (Table S6; Figure 5).
Limitations
One of the limitations of the current study is that only the 5 kb promoter regions of the differentially expressed genes were examined for binding sites. Control elements can, and often do, exist outside of these regions.42 In addition only the conserved regions of these promoters were considered in the analysis. There is potential for regulatory binding sites in the human genome that are not shared with other species.42 Also, only conservation with mouse was considered in the analysis and it may be of benefit to extend the analysis with species phylogenetically closer and more distant to human. The lack of specificity in TFBSs is another limitation in the sense that it will artificially inflate the number of TFs assigned to each promoter of the target genes and thus creates the appearance that a family of TFs binds to a gene set. Most of the limitations can be addressed in future studies with experimental validation of the findings.
Conclusion
Our unbiased genome-wide analysis provides additional information on the role of the ETS family of TFs in the pathogenesis of AAA. Additionally, it identified other potential molecules for study. Future studies including chromatin immunoprecipitation and functional assays are needed to gain a better understanding of the roles the identified TFs play in AAA and the regulation of these gene sets. Taken together, these results might lead to the discovery of new therapeutic targets for the management of this deadly disease.
Abdominal aortic aneurysms (AAAs) are a chronic disease whose pathogenesis is poorly understood. The lack of knowledge about the underlying molecular mechanisms is hampering the development of treatment modalities. Currently surgical intervention is successful, but is used at a late stage of the disease: patients with small AAAs are managed conservatively until the AAA has grown large enough to justify surgical intervention (i.e, the risk of rupture and its attendant morbidity and mortality outweighs the risk of intervention). The development of methods to slow down the growth of small AAAs requires a better understanding of the molecular pathways involved in this process. We previously carried out a whole-genome-based expression analysis comparing aortic tissue samples obtained from patients with AAAs with those without the disease, and identified 3,274 genes whose expression was significantly decreased or increased in the AAA tissue. The current study used a genome-wide approach to analyze the control regions of the genes over-represented in AAA with the goal of finding transcriptional elements responsible for driving gene expression. We observed that the promoters were enriched for binding sites of a small number (N=13) of transcription factors, suggesting co-regulation and a likely significant role in the pathogenesis of AAAs. We confirmed protein expression of these transcription factors in control aorta and AAA tissue by immunohistochemical staining. These types of genome-wide analyses are critical for the identification of potential targets for new pharmacological therapies that may facilitate the prevention and management of AAA.
Supplementary Material
Acknowledgments
Sources of Funding: This project was funded in part by a grant from the National Heart, Lung, and Blood Institute (HL064310 to H.K.), as well as by the Office of the Vice President for Research and by the Department of Surgery of Wayne State University. G.M.L. was a recipient of an American Heart Association Predoctoral Fellowship (0510063Z and 0710099Z).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Journal Subject Codes: [146] Genomics; [142] Gene expression; [143] Gene regulation; [97] Other Vascular biology
Disclosures: None.
References
- 1.Sakalihasan N, Limet R, Defawe OD. Abdominal aortic aneurysm. Lancet. 2005;365:1577–1589. doi: 10.1016/S0140-6736(05)66459-8. [DOI] [PubMed] [Google Scholar]
- 2.Golledge J, Muller J, Daugherty A, Norman P. Abdominal aortic aneurysm: pathogenesis and implications for management. Arterioscler Thromb Vasc Biol. 2006;26:2605–2613. doi: 10.1161/01.ATV.0000245819.32762.cb. [DOI] [PubMed] [Google Scholar]
- 3.Vande Geest JP, Di Martino ES, Bohra A, Makaroun MS, Vorp DA. A biomechanics-based rupture potential index for abdominal aortic aneurysm risk assessment: demonstrative application. Ann N Y Acad Sci. 2006;1085:11–21. doi: 10.1196/annals.1383.046. [DOI] [PubMed] [Google Scholar]
- 4.Ernst CB. Abdominal aortic aneurysm. New England Journal of Medicine. 1993;328:1167–1172. doi: 10.1056/NEJM199304223281607. [DOI] [PubMed] [Google Scholar]
- 5.Lenk GM, Tromp G, Weinsheimer S, Gatalica Z, Berguer R, Kuivaniemi H. Whole genome expression profiling reveals a significant role for immune function in human abdominal aortic aneurysms. BMC Genomics. 2007;8:237. doi: 10.1186/1471-2164-8-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qiu P. Recent advances in computational promoter analysis in understanding the transcriptional regulatory network. Biochem Biophys Res Commun. 2003;309:495–501. doi: 10.1016/j.bbrc.2003.08.052. [DOI] [PubMed] [Google Scholar]
- 7.Gumucio DL, Heilstedt-Williamson H, Gray TA, Tarle SA, Shelton DA, Tagle DA, Slightom JL, Goodman M, Collins FS. Phylogenetic footprinting reveals a nuclear protein which binds to silencer sequences in the human gamma and epsilon globin genes. Mol Cell Biol. 1992;12:4919–4929. doi: 10.1128/mcb.12.11.4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boffelli D, McAuliffe J, Ovcharenko D, Lewis KD, Ovcharenko I, Pachter L, Rubin EM. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science. 2003;299:1391–1394. doi: 10.1126/science.1081331. [DOI] [PubMed] [Google Scholar]
- 9.Wasserman WW, Palumbo M, Thompson W, Fickett JW, Lawrence CE. Human-mouse genome comparisons to locate regulatory sites. Nat Genet. 2000;26:225–228. doi: 10.1038/79965. [DOI] [PubMed] [Google Scholar]
- 10.Hannenhalli S, Putt ME, Gilmore JM, Wang J, Parmacek MS, Epstein JA, Morrisey EE, Margulies KB, Cappola TP. Transcriptional genomics associates FOX transcription factors with human heart failure. Circulation. 2006;114:1269–1276. doi: 10.1161/CIRCULATIONAHA.106.632430. [DOI] [PubMed] [Google Scholar]
- 11.Forton JT, Kwiatkowski DP. Searching for the regulators of human gene expression. Bioessays. 2006;28:968–972. doi: 10.1002/bies.20466. [DOI] [PubMed] [Google Scholar]
- 12.Boddy AM, Lenk GM, Lillvis JH, Nischan J, Kyo Y, Kuivaniemi H. Basic research studies to understand aneurysm disease. Drug News Perspect. 2008;21:142–148. [PubMed] [Google Scholar]
- 13.Zambon AC, Zhang L, Minovitsky S, Kanter JR, Prabhakar S, Salomonis N, Vranizan K, Dubchak I, Conklin BR, Insel PA. Gene expression patterns define key transcriptional events in cell-cycle regulation by cAMP and protein kinase A. Proc Natl Acad Sci U S A. 2005;102:8561–8566. doi: 10.1073/pnas.0503363102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, Kloos DU, Land S, Lewicki-Potapov B, Michael H, Munch R, Reuter I, Rotert S, Saxel H, Scheer M, Thiele S, Wingender E. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loots GG, Ovcharenko I. rVISTA 2.0: evolutionary analysis of transcription factor binding sites. Nucleic Acids Res. 2004;32:W217–W221. doi: 10.1093/nar/gkh383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tromp G, Gatalica Z, Skunca M, Berguer R, Siegel T, Kline RA, Kuivaniemi H. Elevated expression of matrix metalloproteinase-13 in abdominal aortic aneurysms. Ann Vasc Surg. 2004;18:414–420. doi: 10.1007/s10016-004-0050-5. [DOI] [PubMed] [Google Scholar]
- 17.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.R: A Language and Environment for Statistical Computing [computer program]. Version 2.9.0. Austria: Vienna; 2009. [Google Scholar]
- 19.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33(Web Server issue):W741–W748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuivaniemi H, Platsoucas CD, Tilson MD., 3rd Aortic aneurysms: an immune disease with a strong genetic component. Circulation. 2008;117:242–252. doi: 10.1161/CIRCULATIONAHA.107.690982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aziz F, Kuivaniemi H. Role of matrix metalloproteinase inhibitors in preventing abdominal aortic aneurysm. Ann Vasc Surg. 2007;21:392–401. doi: 10.1016/j.avsg.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pyo R, Lee JK, Shipley JM, Curci JA, Mao D, Ziporin SJ, Ennis TL, Shapiro SD, Senior RM, Thompson RW. Targeted gene disruption of matrix metalloproteinase-9 (gelatinase B) suppresses development of experimental abdominal aortic aneurysms. J Clin Invest. 2000;105:1641–1649. doi: 10.1172/JCI8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gong Y, Hart E, Shchurin A, Hoover-Plow J. Inflammatory macrophage migration requires MMP-9 activation by plasminogen in mice. J Clin Invest. 2008;118:3012–3024. doi: 10.1172/JCI32750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miwa K, Nakashima H, Aoki M, Miyake T, Kawasaki T, Iwai M, Oishi M, Kataoka K, Ohgi S, Ogihara T, Kaneda Y, Morishita R. Inhibition of ets, an essential transcription factor for angiogenesis, to prevent the development of abdominal aortic aneurysm in a rat model. Gene Ther. 2005;12:1109–1118. doi: 10.1038/sj.gt.3302496. [DOI] [PubMed] [Google Scholar]
- 26.Bond M, Chase AJ, Baker AH, Newby AC. Inhibition of transcription factor NF-kappaB reduces matrix metalloproteinase-1, -3 and -9 production by vascular smooth muscle cells. Cardiovasc Res. 2001;50:556–565. doi: 10.1016/s0008-6363(01)00220-6. [DOI] [PubMed] [Google Scholar]
- 27.Ocana E, Bohórquez JC, Pérez-Requena J, Brieva JA, Rodríguez C. Characterisation of T and B lymphocytes infiltrating abdominal aortic aneurysms. Atherosclerosis. 2003;170:39–48. doi: 10.1016/s0021-9150(03)00282-x. [DOI] [PubMed] [Google Scholar]
- 28.Forester ND, Cruickshank SM, Scott DJ, Carding SR. Functional characterization of T cells in abdominal aortic aneurysms. Immunology. 2005;115:262–270. doi: 10.1111/j.1365-2567.2005.02157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henderson EL, Geng YJ, Sukhova GK, Whittemore AD, Knox J, Libby P. Death of smooth muscle cells and expression of mediators of apoptosis by T lymphocytes in human abdominal aortic aneurysms. Circulation. 1999;99:96–104. doi: 10.1161/01.cir.99.1.96. [DOI] [PubMed] [Google Scholar]
- 30.Forester ND, Cruickshank SM, Scott DJ, Carding SR. Increased natural killer cell activity in patients with an abdominal aortic aneurysm. Br J Surg. 2006;93:46–54. doi: 10.1002/bjs.5215. [DOI] [PubMed] [Google Scholar]
- 31.Bobryshev YV, Lord RS, Parsson H. Immunophenotypic analysis of the aortic aneurysm wall suggests that vascular dendritic cells are involved in immune responses. Cardiovascular surgery (London, England) 1998;6:240–249. doi: 10.1016/s0967-2109(97)00168-3. [DOI] [PubMed] [Google Scholar]
- 32.Oettgen P. Regulation of vascular inflammation and remodeling by ETS factors. Circ Res. 2006;99:1159–1166. doi: 10.1161/01.RES.0000251056.85990.db. [DOI] [PubMed] [Google Scholar]
- 33.Baeuerle PA, Henkel T. Function and activation of NF-kappa B in the immune system. Annu Rev Immunol. 1994;12:141–179. doi: 10.1146/annurev.iy.12.040194.001041. [DOI] [PubMed] [Google Scholar]
- 34.Blumenthal SG, Aichele G, Wirth T, Czernilofsky AP, Nordheim A, Dittmer J. Regulation of the human interleukin-5 promoter by Ets transcription factors. Ets1 and Ets2, but not Elf-1, cooperate with GATA3 and HTLV-I Tax1. J Biol Chem. 1999;274:12910–12916. doi: 10.1074/jbc.274.18.12910. [DOI] [PubMed] [Google Scholar]
- 35.Miyake T, Aoki M, Nakashima H, Kawasaki T, Oishi M, Kataoka K, Tanemoto K, Ogihara T, Kaneda Y, Morishita R. Prevention of abdominal aortic aneurysms by simultaneous inhibition of NFkappaB and ets using chimeric decoy oligonucleotides in a rabbit model. Gene Ther. 2006;13:695–704. doi: 10.1038/sj.gt.3302704. [DOI] [PubMed] [Google Scholar]
- 36.Nakashima H, Aoki M, Miyake T, Kawasaki T, Iwai M, Jo N, Oishi M, Kataoka K, Ohgi S, Ogihara T, Kaneda Y, Morishita R. Inhibition of experimental abdominal aortic aneurysm in the rat by use of decoy oligodeoxynucleotides suppressing activity of nuclear factor kappaB and ets transcription factors. Circulation. 2004;109:132–138. doi: 10.1161/01.CIR.0000105725.61763.A2. [DOI] [PubMed] [Google Scholar]
- 37.Miyake T, Aoki M, Masaki H, Kawasaki T, Oishi M, Kataoka K, Ogihara T, Kaneda Y, Morishita R. Regression of abdominal aortic aneurysms by simultaneous inhibition of nuclear factor kappaB and ets in a rabbit model. Circ Res. 2007;101:1175–1184. doi: 10.1161/CIRCRESAHA.107.148668. [DOI] [PubMed] [Google Scholar]
- 38.Shiraya S, Miwa K, Aoki M, Miyake T, Oishi M, Kataoka K, Ohgi S, Ogihara T, Kaneda Y, Morishita R. Hypertension accelerated experimental abdominal aortic aneurysm through upregulation of nuclear factor kappaB and Ets. Hypertension. 2006;48:628–636. doi: 10.1161/01.HYP.0000240266.26185.57. [DOI] [PubMed] [Google Scholar]
- 39.Watabe T, Yoshida K, Shindoh M, Kaya M, Fujikawa K, Sato H, Seiki M, Ishii S, Fujinaga K. The Ets-1 and Ets-2 transcription factors activate the promoters for invasion-associated urokinase and collagenase genes in response to epidermal growth factor. Int J Cancer. 1998;77:128–137. doi: 10.1002/(sici)1097-0215(19980703)77:1<128::aid-ijc20>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 40.Herron GS, Unemori E, Wong M, Rapp JH, Hibbs MH, Stoney RJ. Connective tissue proteinases and inhibitors in abdominal aortic aneurysms. Involvement of the vasa vasorum in the pathogenesis of aortic aneurysms. Arterioscler Thromb. 1991;11:1667–1677. doi: 10.1161/01.atv.11.6.1667. [DOI] [PubMed] [Google Scholar]
- 41.Carmeliet P, Moons L, Lijnen R, Baes M, Lemaitre V, Tipping P, Drew A, Eeckhout Y, Shapiro S, Lupu F, Collen D. Urokinase-generated plasmin activates matrix metalloproteinases during aneurysm formation. Nat Genet. 1997;17:439–444. doi: 10.1038/ng1297-439. [DOI] [PubMed] [Google Scholar]
- 42.Wilson MD, Barbosa-Morais NL, Schmidt D, Conboy CM, Vanes L, Tybulewicz VL, Fisher EM, Tavare S, Odom DT. Species-specific transcription in mice carrying human chromosome 21. Science. 2008;322:434–438. doi: 10.1126/science.1160930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brudno M, Poliakov A, Salamov A, Cooper GM, Sidow A, Rubin EM, Solovyev V, Batzoglou S, Dubchak I. Automated whole-genome multiple alignment of rat, mouse, and human. Genome Res. 2004;14:685–692. doi: 10.1101/gr.2067704. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.