Abstract
Dynamic regulation of diverse nuclear processes is intimately linked to covalent modifications of chromatin1,2. Much attention has focused on methylation at lysine 4 of histone H3 (H3K4), owing to its association with euchromatic genomic regions3,4. H3K4 can be mono-, di- or tri-methylated. Trimethylated H3K4 (H3K4me3) is preferentially detected at active genes, and is proposed to promote gene expression through recognition by transcription-activating effector molecules5. Here we identify a novel class of methylated H3K4 effector domains—the PHD domains of the ING (for inhibitor of growth) family of tumour suppressor proteins. The ING PHD domains are specific and highly robust binding modules for H3K4me3 and H3K4me2. ING2, a native subunit of a repressive mSin3a–HDAC1 histone deacetylase complex6, binds with high affinity to the trimethylated species. In response to DNA damage, recognition of H3K4me3 by the ING2 PHD domain stabilizes the mSin3a–HDAC1 complex at the promoters of proliferation genes. This pathway constitutes a new mechanism by which H3K4me3 functions in active gene repression. Furthermore, ING2 modulates cellular responses to genotoxic insults, and these functions are critically dependent on ING2 interaction with H3K4me3. Together, our findings establish a pivotal role for trimethylation of H3K4 in gene repression and, potentially, tumour suppressor mechanisms.
Binding of the ING2 PHD (ING2PHD) domain to the lipid signalling molecule phosphatidylinositol-5-phosphate (PtdIns(5)P) is critical for ING2 recruitment to chromatin during DNA damage responses7. Because some phosphatidylinositol phosphate (PtdInsP)-binding domains function through dual recognition of lipids and peptides8,9, we asked whether the ING2 PHD domain has protein, in addition to PtdInsP, ligands. In an in vitro pull-down screen for ING2PHD protein binding partners, we identified histone H3 (Supplementary Fig. S1a). ING2PHD-binding to H3 was independent of PtdIns(5)P-binding, and was confirmed in direct pull-down assays from purified bulk histones (Fig. 1a, c; Supplementary Fig. S1a).
Figure 1. The ING2 PHD domain specifically binds to H3K4me3 in vitro.
a, Coomassie-blue stain of glutathione S-transferase (GST)–ING2PHD and GST control pull downs from calf thymus histones. b, Gel shift showing ING2PHD-binding to purified native nucleosomes. Ethidium-bromide stain of nucleosomal DNA on a non-denaturing gel. c, Western blot analysis of GST pull-down assays as in a. d, The ING2 PHD domain preferentially binds H3K4me3 peptides. Western analysis of histone peptide pull downs with the indicated GST fusion proteins and biotinylated peptides. Peptide integrity was confirmed with known methyl-lysine-binding domains (Supplementary Fig. S1d, e). aa, amino acids. e, Methylation at H3K4 is required for ING2PHD-binding to chromatin in vitro. Western analysis of GST–ING2PHD pull-down assays of chromatin purified from wild-type or Set1-null S. cerevisiae strains. f, PHD domains of yeast and human ING family proteins preferentially bind to H3K4me2/3 in histone peptide binding assays.
The ING2 PHD domain bound robustly to native mononucleosomes purified from mammalian cells (Fig. 1b), but, in contrast to the ACF (ATP-dependent chromatin-assembly factor) and p300 (E1A binding protein p300) PHD domains, did not bind mononucleosomes reconstituted from recombinant histones (Supplementary Fig. S1b)10,11. Therefore, we postulated that this domain might recognize a post-translational H3 modification. Western blot analysis of ING2PHD-bound histones revealed significant methylation of H3 on lysine 4, but not lysine 9 (Fig. 1c). In in vitro binding assays of methylated histone peptides, the ING2 PHD domain bound most strongly to H3K4me3, with lower affinity to H3K4me2, and not at all to numerous other peptides (Fig. 1d; Supplementary Fig. S1c).
In the accompanying paper describing the crystal structure of the ING2PHD–H3K4me3 peptide complex, aspartate 230 (D230) of ING2 is implicated in H3K4me3 recognition12. Accordingly, substitution of D230 to alanine (ING2PHD-D230A) mostly abolished H3K4me3 peptide binding (Fig. 1d). The effect of this substitution is not due to unfolding of the PHD domain, as determined by NMR two-dimensional spectra12. Notably, this mutation has no effect on the lipid-binding activity of the ING2 PHD domain (Supplementary Fig. S2), and therefore allows selective abrogation of the ING2–H3K4me3 interaction for functional studies (see below). Finally, full-length ING2, but not a PHD domain deletion mutant (ING2ΔPHD), bound specifically to methylated H3K4 peptides (Fig. 1d). Thus, the ING2 PHD domain is necessary and sufficient for ING2 interaction with H3K4me3 in vitro.
The association of the ING2 PHD domain with H3 was increased by hypermethylation of bulk histones with the H3K4 methyltransferase SET7 (also known as SET9; ref. 13) and decreased by the H3K4 demethylase LSD1 (also known as AOF2; ref. 14) (Supplementary Fig. S3). Moreover, the ING2 PHD domain bound efficiently to H3 in chromatin purified from wild-type Saccharomyces cerevisiae strains, but not from strains lacking the H3K4 methyltransferase Set1 (Fig. 1e). Together, these data indicate that H3K4 methylation is critical for ING2PHD-binding to histone H3 at chromatin in vitro.
Like the ING2 PHD domain, the PHD domains of the S. cerevisiae and other human ING family members all bound preferentially to diand tri-methylated H3K4, whereas that of the Miα2 autoantigen bound to trimethylated H3K36 (Fig. 1f; Supplementary Fig. S4a, b). Thus, recognition of methylated histone peptides is a general feature of at least a subset of PHD domains, with the ING PHD family constituting a novel class of binding modules for methylated H3K4.
The functional role of ING2 in the context of mSin3a–HDAC1 complexes is unclear6. We postulated that ING2 bridges the mSin3a–HDAC1 complex with methylated H3K4 through its PHD domain, and thereby promotes deacetylation of nearby acetylated histone residues. To test this idea, we purified ING2–mSin3a–HDAC1 complexes containing ING2, ING2ΔPHD or ING2D230A. Neither mutation altered the composition of the ING2 complexes (Fig. 2a; Supplementary Fig. S5). In peptide pull-down assays, the wild-type, but not mutant, ING2 complexes bound preferentially to H3K4me3 (Fig. 2b). Furthermore, hypermethylation of H3K4 by SET7 enhanced the histone deacetylase activity of the wild-type, but not mutant, ING2 complexes (Figs 2c, d). Therefore, recognition of methylated H3K4 by the ING2 PHD domain promotes the histone deacetylase activity of mSin3a–HDAC1 complexes in vitro.
Figure 2. Methylated H3K4 recognition by the ING2 PHD domain enhances ING2-associated HDAC1 histone deacetylase activity in vitro.

a, Western analysis of affinity-purified wild-type and mutant Flag–ING2 complexes. Mock, empty vector control immunoprecipitation. b, Binding of ING2 complexes to methylated H3K4 peptides requires an intact PHD domain. Anti-ING2 western analysis of histone peptide pull downs from the indicated HDAC1–ING2 complexes. c, Histone deacetylation by HDAC1 in wild-type, but not mutant, ING2 complexes is increased by binding of the ING2 PHD domain to methylated H3K4. Western analysis of in vitro histone deacetylation reactions by ING2 complexes in the presence or absence of methylation by SET7. Histone deacetylase inhibition with trichostatin A (TSA) is shown as a control. d, Quantification of relative histone deacetylase activity of ING2 complexes as in c from three independent experiments. Error bars indicate the s.e.m.
We next studied the recognition of methylated H3K4 by the ING2 PHD domain in the context of physiological chromatin in cells. In protein–protein chromatin immunoprecipitation (ChIP) assays15, wild-type ING2 associated strongly with H3K4me3, but only weakly with H3K4me2 and not at all with other modified histones, suggesting that H3K4me3 is the preferred in vivo target of ING2 (Fig. 3a; data not shown). Moreover, the H3K4me3 interaction was mostly abolished by the D230A mutation and several additional substitutions that disrupt H3K4me3-binding in vitro12 (Fig. 3a, b; data not shown). We conclude that the PHD domain is required for the association of ING2 with H3K4me3 at chromatin in vivo.
Figure 3. The ING2 interaction with trimethylated H3K4 occurs in vivo and requires an intact PHD domain.
a, b, In vivo association of wild-type, but not PHD domain mutant, ING2 proteins with H3K4me3 at chromatin. Western analysis of proteins crosslinked in situ to wild-type or mutant Flag–ING2 proteins in protein–protein ChIPs. Note equal binding of mutant and wild-type ING2 to SAP30 (a) and HDAC1 (b). Input loaded represents 5% of total. c, Decreased H3K4me3 levels following WDR5 knockdown. Western analysis of HEK293T cells transfected with WDR5 or control RNA interference (RNAi) vectors. d, Flag–ING2 association with endogenous H3 requires H3K4me3. Western analysis of protein–protein ChIPs as in a with or without WDR5 knockdown. e, Endogenous association of H3 and ING2 requires H3K4me3. Western analysis of H3-bound proteins by protein–protein ChIP, with or without WDR5 knockdown. f, WDR5 knockdown decreases ING2 occupancy at the cyclin D1 promoter. Levels (per cent input = ChIP/input × 100) of cyclin D1 promoter and 3′ coding region sequences in the indicated ChIPs, with or without WDR5 knockdown. Error bars indicate the s.e.m. from three independent experiments.
To determine whether endogenous H3K4me3 is important for ING2 association with H3, we knocked down the expression of the methyltransferase component, the WD40-repeat protein WDR5, which leads to a specific reduction in H3K4me3 levels (Fig. 3c)16. This was accompanied by substantially reduced binding of both Flag-tagged and endogenous ING2 to H3 at chromatin (without affecting complex formation with HDAC1) (Fig. 3d, e). Thus, H3K4me3 is required for interaction of ING2 with H3, and endogenous ING2 specifically associates with trimethylated H3K4 at chromatin in vivo.
To investigate ING2 association with chromatin at a target gene, we focused on the cell-cycle regulator cyclin D1. This gene is transcriptionally inactivated by the mSin3a–HDAC1 complex17. In ChIP analyses, Flag–ING2 (and tri- and di-methylated H3K4, as expected3,4) are specifically detected at the cyclin D1 promoter but not at 3′ coding regions (Fig. 3f; Supplementary Fig. S6a). Furthermore, WDR5 knockdown decreased ING2 occupancy at the cyclin D1 promoter (Fig. 3f). Thus, trimethylation of H3K4 is important for ING2 association at the cyclin D1 promoter.
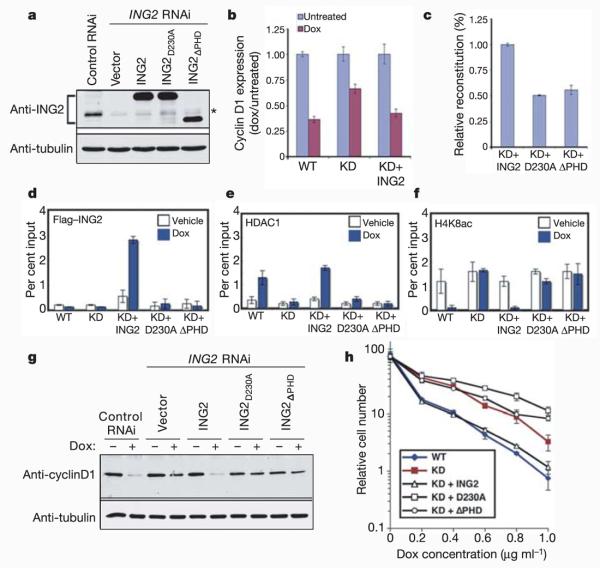
To investigate the biological significance of the ING2–H3K4me3 interaction, we used an ING2 complementation system7, whereby RNA interference (RNAi)-resistant wild-type or mutant (D230A or ΔPHD) ING2 constructs were stably expressed in ING2 knockdown cells. ING2 expression was substantially reduced in the knockdown cells, and the different ING2 constructs reconstituted expression at comparable levels (Fig. 4a). Notably, the ING2 knockdown cells showed impaired cyclin D1 mRNA repression in response to doxorubicin compared with control (wild-type) cells, and complementation of knockdown cells with wild-type ING2 mostly restored efficient cyclin D1 repression (Fig. 4b). In contrast, the ING2D230A and ING2ΔPHD proteins showed significantly reduced reconstitution activities (Fig. 4c). Thus, PHD domain recognition of H3K4me3 is important for ING2-mediated repression of cyclin D1 mRNA expression in response to DNA damage.
Figure 4. Recognition of H3K4me3 by ING2 PHD domain in vivo is critical for ING2 function.
a, Western analysis of ING2 expression in the indicated cell lines. *, endogenous ING2. b, ING2 is required for repression of cyclin D1 expression in response to doxorubicin (dox). Quantitative PCR of cyclin D1 mRNA, with or without doxorubicin (0.2μg ml−1, 24 h), in wild-type (“WT”), knockdown (“KD”) and knockdown reconstituted with ING2 (“KD+ING2”) cell lines. c, Impaired activity of mutant ING2 proteins in reconstitution of doxorubicin-dependent cyclin D1 repression, as in b, relative to wild-type ING2 (“KD+ING2”). In b and c, error bars indicate the s.e.m. of triplicate experiments; P-values < 0.01. d-f, DNA-damage-dependent increase of ING2-HDAC1 occupancy at the cyclin D1 promoter requires methylated H3K4-binding. ChIPs with antibodies to Flag (d), HDAC1 (e) and H4K8ac (f) at the cyclin D1 promoter in the indicated cell lines, with or without doxorubicin (2μM, 1 h). Error bars indicate the s.e.m. of at least three independent experiments. P-values < 0.05. g, Western analysis of cyclin D1 protein in the indicated cell lines with or without doxorubicin (0.2μg ml−1, 24 h). h, Impaired doxorubicin sensitivity following ING2 knockdown or reconstitution with H3K4me3-binding-defective ING2. Relative cell viability is normalized to untreated cells. Error bars indicate the s.e.m. of 3–6 independent experiments.
Next, we examined promoter occupancy of the ING2–HDAC1 complexes in response to DNA damage. Doxorubicin treatment increased promoter occupancy of wild-type, but not mutant, ING2 at the cyclin D1 promoter (Fig. 4d), as well as a second HDAC1-regulated DNA-damage response gene, c-Myc (Supplementary Fig. S7)18. Furthermore, HDAC1 occupancy and histone deacetylation at the cyclin D1 promoter both increased on DNA damage, and these responses, which were largely abrogated in the knockdown cells, were restored by reconstitution with wild-type, but not mutant, ING2 (Fig. 4e, f). H3K4me2, H3K4me3 and H3 levels were uniform across the different cell lines and not altered by doxorubicin treatment (Supplementary Fig. S6b). On western analysis, cyclin D1 protein levels were repressed following DNA damage in the presence of wild-type ING2 (in both control cells and knockdown cells reconstituted with ING2), but only slightly reduced in the knockdown cells or knockdown cells reconstituted with mutant ING2 proteins (Fig. 4g). We conclude that recognition of H3K4me3 by the ING2 PHD domain is critical for proper occupancy of the ING2–HDAC1 complex at target promoters during DNA damage responses and active transcriptional repression of the promoter's cognate gene.
Finally, H3K4me3 recognition by the PHD domain is critical for ING2 regulation of cellular functions. Specifically, ING2 knockdown significantly impairs cellular sensitivity to doxorubicin (Fig. 4h), and reconstitution of knockdown cells with wild-type, but not mutant, ING2 reverses this effect (Fig. 4h). Together, these data argue that the H3K4me3-binding activity of ING2 is critical for ING2 function in the cellular response to DNA damage.
Our findings and previous work demonstrate that the PHD domain of ING2 is a dual-specificity module that binds both PtdIns(5)P and H3K4me3. These activities are separable by specific mutations within the domain, and both seem critical for ING2 cellular functions. It is possible that PtdIns(5)P-binding could mediate trafficking of ING2 to target promoters, where binding to H3K4me3 then contributes to retention of ING2 at these promoters (see Supplementary Fig. S8). In this regard, the multi-functionality of the ING2 PHD domain may contribute to integrating complex nuclear signalling events that link DNA damage to chromatin responses.
Methylation of H3K4 is proposed to have a pivotal role in gene activation by serving as a binding platform for different transcription-promoting factors16,19-21. Our findings identify the ING2 PHD domain as the first effector domain for methylated H3K4 that links this modification to transcriptional repression. By focusing HDAC1 repressor complexes on actively transcribed genes, recognition of H3K4me3 by ING2 may be important for the efficiency of acute gene repression. Such a mechanism may be particularly important in the context of cellular responses to acute stress, such as DNA damage, in which rapid shut-off of proliferation genes is critical to prevent propagation of cells harbouring damaged DNA. The PHD domains of the other ING proteins also bind methylated H3K4, and thus may link this modification to additional cellular functions. Moreover, the PHD domain of the nucleosome remodelling factor (NURF) complex component BPTF (bromodomain and PHD domain transcription factor) also recognizes methylated H3K4 (ref. 22), suggesting a more general role for PHD domains as methyl-lysine effector domains. Together, our findings highlight the notion that the recognition of chromatin modifications by effector proteins, rather than the specific modification per se, determines biological function, and, as such, greatly expands the diversity of signalling at chromatin.
METHODS
Histone peptide binding assays
Biotinylated histone peptides were synthesized at Stanford Protein and Nucleic Acid facility or purchased from Upstate Biotechnology. Briefly, 0.5m g of peptides were incubated with 1 μg of protein in binding buffer (50 mM Tris-HCl, pH 7.5, 300 mM NaCl, 0.1% (v/v) NP-40, 1 mM phenylmethylsulphonyl fluoride (PMSF)) overnight at 4 °C. After 1 h of incubation, streptavidin beads (Amersham) were washed three times and subjected to western analysis.
Protein–protein ChIP assays for detection of in situ ING2–histone interactions was performed as described15. Cell viability assays were performed as described23. Information about antibodies, recombinant vectors, RNAi sequences and primers are available in Supplementary Information and on request. Detailed methods can be found in Supplementary Information.
Supplementary Material
Acknowledgements
We thank Y. Zhang for the SET7 expression vector, C. Harris for ING3, ING4 and ING5 complementary DNAs, J. Wysocka and W. Herr for a WDR5 antibody, J. Wysocka and C. D. Allis for communicating unpublished results, J. Yuan for ING2 antibodies and A. Sanchez for peptide synthesis. This work was supported by NIH grants to O.G., Y.S. and B.R.C. O.G. is a recipient of a Burroughs Wellcome Career Development Award in Biomedical Sciences.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
The authors declare no competing financial interests.
References
- 1.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 2.Kurdistani SK, Grunstein M. Histone acetylation and deacetylation in yeast. Nature Rev. Mol. Cell Biol. 2003;4:276–284. doi: 10.1038/nrm1075. [DOI] [PubMed] [Google Scholar]
- 3.Schneider R, et al. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nature Cell Biol. 2004;6:73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein BE, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 5.Bannister AJ, Kouzarides T. Histone methylation: recognizing the methyl mark. Methods Enzymol. 2004;376:269–288. doi: 10.1016/S0076-6879(03)76018-2. [DOI] [PubMed] [Google Scholar]
- 6.Doyon Y, et al. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol. Cell. 2006;21:51–64. doi: 10.1016/j.molcel.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 7.Gozani O, et al. The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell. 2003;114:99–111. doi: 10.1016/s0092-8674(03)00480-x. [DOI] [PubMed] [Google Scholar]
- 8.Zimmermann P, et al. PIP2–PDZ domain binding controls the association of syntenin with the plasma membrane. Mol. Cell. 2002;9:1215–1225. doi: 10.1016/s1097-2765(02)00549-x. [DOI] [PubMed] [Google Scholar]
- 9.Stolt PC, et al. Origins of peptide selectivity and phosphoinositide binding revealed by structures of disabled-1 PTB domain complexes. Structure (Camb.) 2003;11:569–579. doi: 10.1016/s0969-2126(03)00068-6. [DOI] [PubMed] [Google Scholar]
- 10.Ragvin A, et al. Nucleosome binding by the bromodomain and PHD finger of the transcriptional cofactor p300. J. Mol. Biol. 2004;337:773–788. doi: 10.1016/j.jmb.2004.01.051. [DOI] [PubMed] [Google Scholar]
- 11.Eberharter A, Vetter I, Ferreira R, Becker PB. ACF1 improves the effectiveness of nucleosome mobilization by ISWI through PHD–histone contacts. EMBO J. 2004;23:4029–4039. doi: 10.1038/sj.emboj.7600382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Penña PV, et al. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006 May 21; doi: 10.1038/nature04814. advance online publication, doi:10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang H, et al. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol. Cell. 2001;8:1207–1217. doi: 10.1016/s1097-2765(01)00405-1. [DOI] [PubMed] [Google Scholar]
- 14.Shi Y, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 15.Ricke RM, Bielinsky AK. Easy detection of chromatin binding proteins by the Histone Association. Assay. Biol. Proced. Online. 2005;7:60–69. doi: 10.1251/bpo106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wysocka J, et al. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 17.Dannenberg JH, et al. mSin3A corepressor regulates diverse transcriptional networks governing normal and neoplastic growth and survival. Genes Dev. 2005;19:1581–1595. doi: 10.1101/gad.1286905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho JS, Ma W, Mao DY, Benchimol S. p53-dependent transcriptional repression of c-myc is required for G1 cell cycle arrest. Mol. Cell. Biol. 2005;25:7423–7431. doi: 10.1128/MCB.25.17.7423-7431.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santos-Rosa H, et al. Methylation of histone H3 K4 mediates association of the Isw1p ATPase with chromatin. Mol. Cell. 2003;12:1325–1332. doi: 10.1016/s1097-2765(03)00438-6. [DOI] [PubMed] [Google Scholar]
- 20.Pray-Grant MG, Daniel JA, Schieltz D, Yates JR, III, Grant PA. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature. 2005;433:434–438. doi: 10.1038/nature03242. [DOI] [PubMed] [Google Scholar]
- 21.Sims RJ, III, et al. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J. Biol. Chem. 2005;280:41789–41792. doi: 10.1074/jbc.C500395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wysocka J, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006 May 21; doi: 10.1038/nature04815. advance online publication, doi:10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 23.Mostoslavsky R, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.