Abstract
Background
2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) is an environmental contaminant that elicits a broad spectrum of toxic effects in a species-specific manner. Current risk assessment practices routinely extrapolate results from in vivo and in vitro rodent models to assess human risk. In order to further investigate the species-specific responses elicited by TCDD, temporal gene expression responses in human HepG2, mouse Hepa1c1c7 and rat H4IIE cells were compared.
Results
Microarray analysis identified a core set of conserved gene expression responses across species consistent with the role of AhR in mediating adaptive metabolic responses. However, significant species-specific as well as species-divergent responses were identified. Computational analysis of the regulatory regions of species-specific and -divergent responses suggests that dioxin response elements (DREs) are involved. These results are consistent with in vivo rat vs. mouse species-specific differential gene expression, and more comprehensive comparative DRE searches.
Conclusions
Comparative analysis of human HepG2, mouse Hepa1c1c7 and rat H4IIE TCDD-elicited gene expression responses is consistent with in vivo rat-mouse comparative gene expression studies, and more comprehensive comparative DRE searches, suggesting that AhR-mediated gene expression is species-specific.
Background
2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) is a ubiquitous environmental contaminant that elicits a broad spectrum of biochemical and physiological effects in a species-specific manner [1]. These effects include lethality, cancer, developmental abnormalities, immunotoxicity, skin lesions, hepatotoxicity, and xenobiotic enzyme metabolism induction. They result from altered gene expression mediated by the aryl hydrocarbon receptor (AhR), a ligand activated transcription factor [1,2]. Briefly, TCDD binds to the cytoplasmic AhR causing nuclear translocation and heterodimerization with the AhR nuclear translocator (ARNT). The heterodimer then binds to specific DNA elements, termed dioxin response elements (DREs), within the regulatory regions of targeted genes to modulate expression, resulting in downstream physiological responses [3]. Although the structure and function of the AhR are highly conserved [4], the sensitivity to and the responses elicited by TCDD vary widely across species, suggesting TCDD and related compounds may activate species-specific AhR-mediated gene expression networks.
Risk assessment assumes that there is a conserved mode of action and comparable toxic responses between species. However, there are inherent differences between species that compromise the extrapolation of rodent data to estimate potential human risks. Moreover, there is a discord between preclinical animal testing compared to human clinical trials regarding toxicity [5]. Similarly, species differ widely in response to TCDD exposure. For example, LD50 values range from 1 μg/kg in the guinea pig [6] to >1000 μg/kg for the hamster, and several responses exhibit species-specific sensitivities and toxicities. These differential effects are not attributed to differences in binding affinity or AhR complex stability [7-9]. Collectively, these data suggest that although the AhR is well conserved, subsequent differential gene expression responses are species-specific.
To further investigate differences in TCDD elicited differential gene expression, global gene expression was assessed in human HepG2, mouse Hepa1c1c7 and rat H4IIE hepatoma cells following treatment with TCDD. Comparative analysis indicates there are significant differences in gene expression between species, suggesting AhR-mediated gene expression may not be conserved.
Results
Temporally Conserved Gene Expression Responses Elicited by TCDD
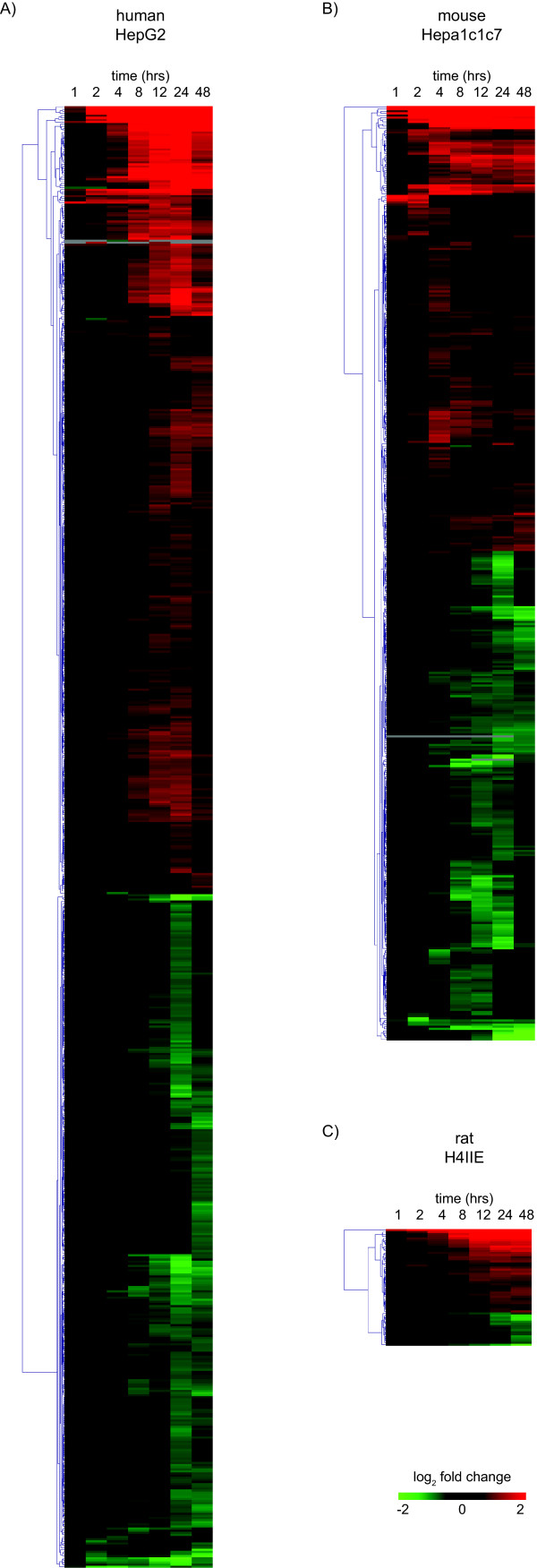
Species-specific, cDNA microarrays were used to profile the temporal gene expression elicited by TCDD in human HepG2, mouse Hepa1c1c7 and rat H4IIE cells. The microarrays queried 6995, 8478 and 5169 unique human, mouse and rat genes, respectively (Table 1). Empirical Bayes analysis identified 691, 439 and 57 differentially expressed genes (P1(t) > 0.999 and |fold change| > 1.4) in HepG2, Hepa1c1c7 and H4IIE cells, respectively. Complete cDNA microarray data sets are provided in Additional files 1, 2, 3. HepG2 cells were the most responsive as indicated by both the overall number of differentially expressed genes as well as the number of responsive genes at each time point (Figure 1). H4IIE cells exhibited significantly less differentially expressed genes, partially explained by the smaller microarray and the immaturity of rat genome annotation compared to the human and mouse. Differentially expressed genes were hierarchically clustered based on Euclidian distance and distinct clusters of temporal gene expression patterns were identified (Figure 2).
Table 1.
Gene coverage of species-specific cDNA microarray platforms and number of differentially regulated genes
| Human HepG2 | Mouse Hepa1c1c7 | Rat H4IIE | ||||
|---|---|---|---|---|---|---|
| Total | Differentially Regulateda | Total | Differentially Regulateda | Total | Differentially Regulateda | |
| Unique Genesb | 6,995 | 624 | 8,478 | 438 | 5,169 | 56 |
| Orthologsc | 6,825 | 616 | 8,233 | 432 | 4,871 | 52 |
a|fold change| > 1.4 and P1(t) > 0.999
bbased on Entrez GeneID
cbased on HomoloGeneID
Figure 1.
Number of TCDD-elicited differentially expressed genes in human HepG2, mouse Hepa1c1c7 and rat H4IIE. Global gene expression changes were detected using cDNA microarray analysis of cells treated with 10 nM TCDD for 1, 2, 4, 8, 12, 24 and 48 hrs. Differentially expressed genes are defined as having P1(t) > 0.999 and |fold change| > 1.4-fold at one or more time points.
Figure 2.
Hierarchical clustering of differentially expressed genes (P1(t) > 0.999 and |fold change| > 1.4) from cDNA microarray time course studies in HepG2, Hepa1c1c7 and H4IIE hepatoma cells.
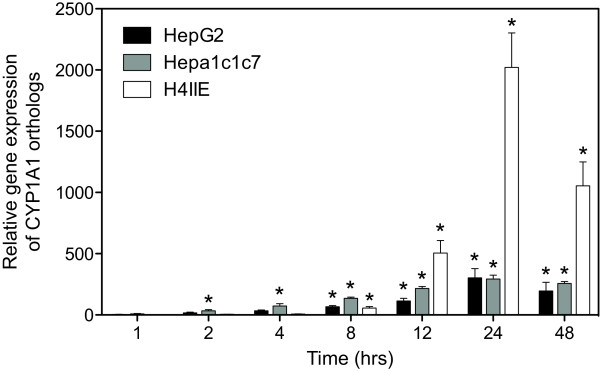
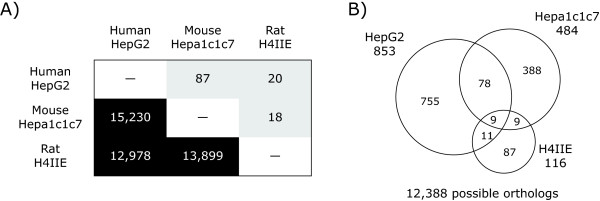
Pair-wise comparisons of differentially expressed genes were conducted using HomoloGene (build 35) defined orthologs (Figure 3A). Human and mouse cDNA microarrays shared 4546 orthologous genes, however only 0.9% (41 orthologs; Additional file 4) were differentially regulated by TCDD in HepG2 and Hepa1c1c7 cells. Comparison of the rodent platforms identified 3850 orthologs with only 0.2% (8 orthologs; Additional file 5) responding in both Hepa1c1c7 and H4IIE cells. The lack of conserved ortholog differential expression in Hepa1c1c7 and H4IIE cells is consistent with the reported differences in differential expression observed in vivo between mice and rats [10-12]. Time dependent profiling of HepG2 and H4IIE gene expression identified only 5 conserved responses out of 2625 possible orthologs, representing only 0.2% (Additional file 6). Comparative analysis across all three species with 2252 shared orthologous probes identified only one ortholog that was differentially regulated in all three cell lines (immediate early response 3, IER3; HomoloGene ID 2894). Note that other members of the AhR gene battery, namely CYP1A1, ALDH3A1 and NQO1, were not present across all of the cDNA microarray platforms. However, their responses were also conserved across all three cell lines when measured using QRTPCR. The results for CYP1A1 are shown in Figure 4.
Figure 3.
Cross-species comparison of TCDD-elicited temporal gene expression responses using cDNA microarrays. (A) The lower half (dark boxes) of the matrix show the number of orthologs represented between pairs of species-specific cDNA microarray platforms. The upper half (grey boxes) of the matrix provides the number of TCDD elicited differentially expressed orthologs between pairs of cell lines. (B) The Venn diagram shows the overlap of all differentially expressed genes (P1(t) > 0.999 and |fold change| > 1.4) from 2,252 possible orthologs represent across all three microarray platforms. Additional files 4, 5, 6 lists the shared responses between pairs of species.
Figure 4.
QRTPCR verification of the conserved induction of CYP1A1 across the human HepG2, mouse Hepa1c1c7 and rat H4IIE cell lines. The same RNA used for microarray analysis was examined by QRTPCR. Fold changes were calculated relative to time-matched vehicle controls. Error bars represent the standard error of measurement for the average fold change and the asterisk (*) indicates p < 0.05.
Identification of Putative Primary Gene Expression Responses
In order to further investigate AhR-mediated responses, TCDD-elicited differential gene expression was examined in the presence of cycloheximide (CHX), a protein translation inhibitor. Putative primary responses were defined in this study as those where CHX co-treatment either maintained or enhanced the response elicited by TCDD, while responses that were attenuated or blocked by CHX co-treatment were classified as secondary responses based on their assumed dependence on additional protein translation. cDNA microarray analysis confirmed the superinduction of CYP1A1 mRNA in CHX co-treated Hepa1c1c7 cells [13,14], consistent with the superinduction in human MCF10A cells treated with TCDD [15]. Additionally, Hepa1c1c7 ARNT-deficient c4 mutants treated with TCDD did not exhibit induction of the prototypical AhR battery genes including CYP1A1 [16]. Collectively, these results indicate that CYP1A1 is a primary gene expression response consistent with the direct interaction of the AhR with DREs within the promoter region.
For each species, differentially expressed orthologs were classified as primary or secondary responses based on CHX co-treatment studies at both 4 and 12 hrs. Overall 61, 38 and 2 human, mouse and rat orthologs, respectively, were considered primary responses (Additional files 7, 8, 9). Furthermore, 45, 12 and 10 orthologous genes in the HepG2, Hepa1c1c7 and H4IIE cells were classified as secondary AhR responses (Additional files 10, 11, 12). Comparative examination of the CHX co-treatment data suggested that each cell line had its own unique set of primary responsive orthologs.
Whole-Genome Analysis of Conserved TCDD-Elicited Gene Expression Responses
The lack of whole genome coverage on the human, mouse and rat cDNA microarrays limited the number of orthologs that could be investigated. Therefore, whole genome expression analysis was performed at 24 hrs, one of the most active time points in terms of the number of differentially expressed genes (Figure 1), using 4 × 44 k Agilent oligonucleotide microarrays. Each microarray contained more than 41,000 individual probes, representing more than 18,000 unique genes (Table 2). Despite the increased coverage, the number of TCDD elicited differentially expressed genes were surprisingly small relative to the cDNA microarray results. For example, the human Agilent microarray consisted of 19,406 known genes, representing a 2.8-fold increase in coverage compared to the human cDNA microarray. However, only 899 unique genes were differentially expressed, a modest increase from the 691 genes identified using cDNA microarrays. Similarly, only 519 and 121 genes were responsive in the Hepa1c1c7 and H4IIE cells, respectively. Complete Agilent microarray data sets are provided in Additional files 13, 14, 15.
Table 2.
Gene coverage of species-specific Agilent microarray platforms and number of differentially regulated genes
| Human HepG2 | Mouse Hepa1c1c7 | Rat H4IIE | ||||
|---|---|---|---|---|---|---|
| Total | Differentially Regulateda | Total | Differentially Regulateda | Total | Differentially Regulateda | |
| Unique Genesb | 18,499 | 865 | 20,929 | 508 | 18,244 | 129 |
| Orthologsc | 16,781 | 828 | 17,543 | 477 | 15,705 | 116 |
a|fold change| > 1.4 and P1(t) > 0.999
bbased on Entrez GeneID
cbased on HomoloGeneID
The use of whole genome microarrays also increased the number of orthologs that could be examined (Figure 5A). As seen with the cDNA microarray dataset pair-wise comparisons, HepG2 and Hepa1c1c7 cells shared the greatest number of TCDD responsive orthologs (Additional files 16, 17, 18). Ortholog coverage between all three species increased from 2252 on the cDNA microarrays to 12,388 across the Agilent platforms. Comparative analysis (P1(t) > 0.999 and |fold change| > 1.4) identified only 10 orthologs that were differentially expressed by TCDD at 24 hrs (Figure 5B; Table 3). Whole genome expression profiling identified the species-conserved induction of CYP1A1, TIPARP and UGT1A6 in all three cell lines. Despite this increased coverage, the number of differentially expressed orthologs across all three cell lines remained small, consistent with our cDNA microarray results.
Figure 5.
Cross-species comparison of TCDD-elicited gene expression responses at 24 hrs using 4 × 44 k Agilent microarrays. (A) The lower half (dark boxes) of the matrix show the number or orthologous genes represented between pairs of species-specific microarrays. The upper half (grey boxes) provides the number of TCDD-elicited differentially expressed orthologs between pairs of cell lines. (B) The 3-way Venn diagram shows the overlap of all differentially expressed genes (P1(t) > 0.999 and |fold change| > 1.4) from 12,388 possible orthologous genes represented across all three platforms. The 9 genes differentially expressed in all three cell lines are listed in Table 3. Additional files 16, 17, 18 lists the shared responses between pairs of species.
Table 3.
List of common genes identified as differentially expressed by TCDD treatment from whole genome Agilent microarray analysis.
| Gene Symbol | Gene Name | Homologene ID | Fold changea | |||||
|---|---|---|---|---|---|---|---|---|
| Human | Mouse | Rat | ||||||
| NQO1 | NAD(P)H dehydrogenase, quinone 1 | 695 | 3.22 | ▲ | 4.88 | ▲ | 4.82 | ▲ |
| CCND1 | cyclin D1 | 1334 | 1.57 | ▲ | -1.56 | ▼ | -1.47 | ▼ |
| ID3 | inhibitor of DNA binding 3, dominant negative helix-loop-helix protein | 1633 | 1.74 | ▲ | -1.83 | ▼ | -1.68 | ▼ |
| TIPARP | TCDD-inducible poly(ADP-ribose) polymerase | 9167 | 1.47 | ▲ | 7.05 | ▲ | 2.91 | ▲ |
| POC1A | POC1 centriolar protein homolog A | 51460 | -1.64 | ▼ | -1.67 | ▼ | -1.57 | ▼ |
| CYP1A1 | cytochrome P450, family 1, subfamily A, polypeptide 1 | 68062 | 13.64 | ▲ | 153.52 | ▲ | 134.16 | ▲ |
| GSTA5 | glutathione S-transferase A5 | 74378 | -2.47 | ▼ | 1.47 | ▲ | 5.85 | ▲ |
| UGT1A6 | UDP glucuronosyltransferase 1 family, polypeptide A6 | 85959 | 2.09 | ▲ | 2.24 | ▲ | 2.06 | ▲ |
| MT1E | metallothionein 1E | 108228 | 4.89 | ▲ | -1.66 | ▼ | 2.65 | ▲ |
aMaximum absolute foldchange determined by microarray analysis
bDifferentially regulated genes with P1(t) > 0.999 and |foldchange| > 1.4
Species-Specific & Species-Divergent Gene Expression Responses
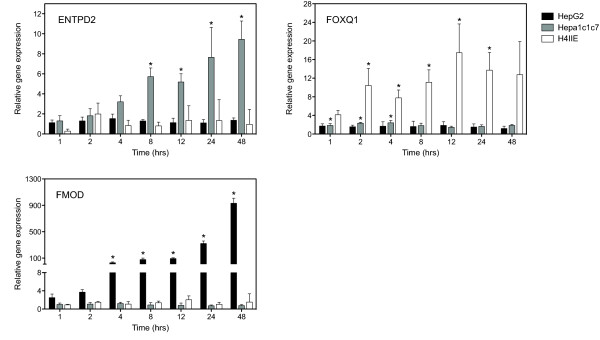
Whole genome comparative analysis of HepG2, Hepa1c1c7 and H4IIE responses identified genes that were species-specific, i.e. differentially expressed in only a single species. For example, microarray analysis at 24 hrs found that fibromodulin (FMOD, HomoloGene ID 1530) was significantly up-regulated 17.2-fold in the HepG2 cells while no significant change in expression was detected in the Hepa1c1c7 and H4IIE cells. Other examples of mouse and rat specific responses include forkhead box Q1 (FOXQ1, HomoloGene ID 7359) and ectonucleoside triphosphate diphosphohydrolase 2 (ENTPD2, HomoloGene ID 20333). FOXQ1 was up-regulated 5.9-fold in the Hepa1c1c7 cells while ENTPD2 was up-regulated 3.2-fold in the H4IIE cells. For both of these genes, the corresponding ortholog in the other two species did not exhibit significant differential expression. The species-specific responses of FMOD, FOXQ1 and ENTPD were verified using QRTPCR (Figure 6). These responses are consistent with previous reports of species-specific TCDD elicited hepatic gene expression characterized in mice and rats [10,11].
Figure 6.
QRTPCR verification of examples of species-specific orthologous gene expression responses identified from whole-genome microarray analysis at 24 hrs in the human HepG2, mouse Hepa1c1c7 and rat H4IIE cell lines. The same RNA used for microarray analysis was examined by QRTPCR. Representative orthologs that exhibited significant expression in only one species were verified; FMOD, ENTPD2 and FOXQ1 and were differentially expressed in only the HepG2, Hepa1c1c7 and H4IIE cells respectively. Fold changes were calculated relative to time-matched vehicle controls. Error bars represent the standard error of measurement for the average fold change and the asterisk (*) indicates p < 0.05.
Comparative analysis of the orthologous gene expression responses in HepG2, Hepa1c1c7 and H4IIE datasets identified 10 orthologs that were differentially expressed by TCDD across the three models. Further analysis indicated that not all of these responses were directionally conserved, i.e. pattern of expression was not the same in all species. For example, IER3 was induced 1.4-fold in Hepa1c1c7 cells at 4 hrs and 1.5-fold in H4IIE cells at 12 hrs, but repressed -1.9-fold in HepG2 cells at 24 hrs. Likewise, glutathione S-transferase alpha 5 (GSTA5, HomoloGene ID 74378) was induced 1.5-fold in Hepa1c1c7 cells and 4.9-fold in H4IIE cells, but down-regulated 2.5-fold in HepG2 cells. QRTPCR confirmed the divergent expression of GSTA5 in the rat and human cell lines, but found Hepa1c1c7 cells were relatively non-responsive (Figure 7A). Two other orthologs, cyclin D1 (CCND, HomoloGene ID 1334) and inhibitor of DNA binding 3 (ID3, HomoloGene ID 1633), also exhibited divergent expression between species, where the orthologs were down-regulated in the rodent cell lines but induced in HepG2 cells (Table 3). Species-specific and species-divergent responses have also been reported in vivo for TCDD elicited hepatic differential gene expression in mice and rats [10,11].
Figure 7.
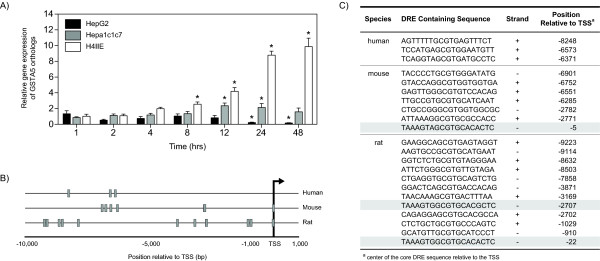
Comparative analysis of GSTA5 orthologs. (A) QRTPCR verification of the divergent expression of GSTA5 orthologs in human HepG2, mouse Hepa1c1c7 and rat H4IIE cell lines. Fold changes were calculated relative to time-matched vehicle controls. Error bars represent the standard error of measurement for the average fold change and the asterisk (*) indicates p < 0.05. (B) Gene regulatory regions (-10,000 to 1,000 bp relative to the transcription start site [TSS]) were computationally searched for the 5 bp DRE core (GCGTC) and indicated by the grey boxes. Each core was extended by the flanking 7 bp, and the resulting 19 bp sequence (C) was compared to a consensus DRE sequence using a position weight matrix developed from bona fide functional DREs. The shaded rows in (C) indicate the highly similar mouse and rat DRE sequences (Euclidean ≤ 3.0).
Functional Enrichment and Network Analysis
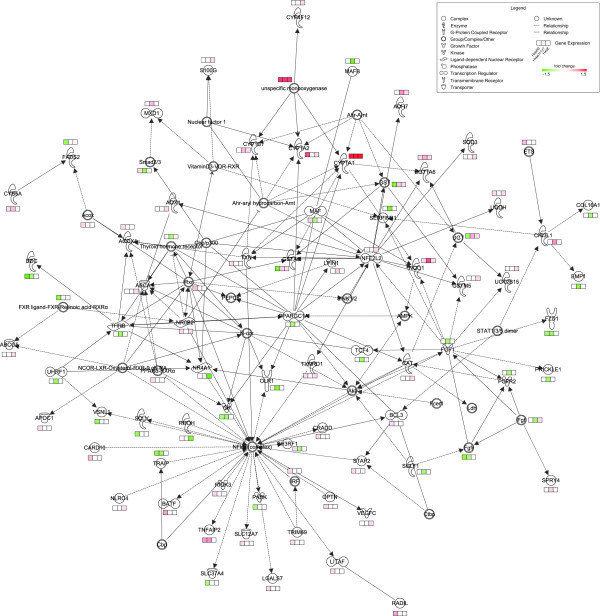
Microarray analysis identified a small subset of TCDD responsive genes in the entire genome of each species. Differentially expressed genes elicited by TCDD in each cell line were clustered for functional enrichment based on GO terms (enrichment score ≥ 1.5). Enriched HepG2, Hepa1c1c7, and H4IIE responses clusters included GO terms related to xenobiotic exposure (GO:0009410) and metabolism (GO:0006805) (Tables 4, 5, 6), indicating that these processes are conserved in all three cell lines, and consistent with reported in vivo studies using rats and mice[10-12]. However, other enriched GO terms, such as those related to lipid metabolism and transport, were only enriched in HepG2 and H4IIE cell lines. Genes functionally related to lipid metabolism were further analyzed using Ingenuity Pathway Analysis to identify a network of TCDD-responsive orthologs, and species-conserved and -specific responses (Figure 8). Although GO terms related to lipid metabolism were enriched HepG2 and H4IIE differential gene expression data sets, there were few orthologs that exhibited differential regulation in both cell lines based on the fold-change and statistical cutoffs used. In vivo rat studies also identified enrichment of genes related to lipid metabolism following TCDD treatment, suggesting that H4IIE cells may be predictive of TCDD-induced perturbations of this pathway rats.
Table 4.
Functional enrichment analysis of differentially regulateda genes elicited by TCDD in HepG2 cells using DAVID.
| Category | Term | Gene count | Fold enrichment | P-value |
|---|---|---|---|---|
| Enrichment Score: 2.99 | ||||
| GOTERM_BP_3 | GO:0009410 ~ response to xenobiotic stimulus | 10 | 8.03 | 1.54E-06 |
| GOTERM_BP_3 | GO:0006805 ~ xenobiotic metabolic process | 9 | 8.32 | 4.92E-06 |
| GOTERM_BP_3 | GO:0017144 ~ drug metabolic process | 3 | 3.46 | 2.13E-01 |
| GOTERM_BP_3 | GO:0006800 ~ oxygen and reactive oxygen species metabolic process | 4 | 1.12 | 7.00E-01 |
| Enrichment Score: 2.71 | ||||
| GOTERM_BP_3 | GO:0022403 ~ cell cycle phase | 40 | 1.80 | 4.36E-04 |
| GOTERM_BP_3 | GO:0048285 ~ organelle fission | 25 | 2.03 | 1.35E-03 |
| GOTERM_BP_3 | GO:0000278 ~ mitotic cell cycle | 34 | 1.70 | 3.07E-03 |
| GOTERM_BP_3 | GO:0022402 ~ cell cycle process | 45 | 1.48 | 8.10E-03 |
| Enrichment Score: 2.30 | ||||
| GOTERM_BP_3 | GO:0006629 ~ lipid metabolic process | 70 | 1.65 | 3.82E-05 |
| GOTERM_BP_3 | GO:0008610 ~ lipid biosynthetic process | 25 | 1.47 | 5.48E-02 |
| GOTERM_BP_3 | GO:0044255 ~ cellular lipid metabolic process | 37 | 1.34 | 6.10E-02 |
| Enrichment Score: 2.03 | ||||
| GOTERM_BP_3 | GO:0006790 ~ sulfur metabolic process | 15 | 2.41 | 3.58E-03 |
| GOTERM_BP_3 | GO:0006518 ~ peptide metabolic process | 9 | 3.20 | 6.37E-03 |
| GOTERM_BP_3 | GO:0051186 ~ cofactor metabolic process | 20 | 1.91 | 8.81E-03 |
| GOTERM_BP_3 | GO:0006519 ~ cellular amino acid and derivative metabolic process | 28 | 1.48 | 3.94E-02 |
| Enrichment Score: 1.63 | ||||
| GOTERM_BP_3 | GO:0005975 ~ carbohydrate metabolic process | 43 | 1.55 | 4.51E-03 |
| GOTERM_BP_3 | GO:0044262 ~ cellular carbohydrate metabolic process | 32 | 1.56 | 1.40E-02 |
| GOTERM_BP_3 | GO:0016051 ~ carbohydrate biosynthetic process | 12 | 2.09 | 2.79E-02 |
| GOTERM_BP_3 | GO:0005996 ~ monosaccharide metabolic process | 19 | 1.62 | 4.67E-02 |
| GOTERM_BP_3 | GO:0046165 ~ alcohol biosynthetic process | 6 | 2.52 | 8.71E-02 |
| Enrichment Score: 1.57 | ||||
| GOTERM_BP_3 | GO:0010817 ~ regulation of hormone levels | 17 | 2.11 | 6.67E-03 |
| GOTERM_BP_3 | GO:0034754 ~ cellular hormone metabolic process | 9 | 2.87 | 1.22E-02 |
| GOTERM_BP_3 | GO:0042446 ~ hormone biosynthetic process | 4 | 2.38 | 2.34E-01 |
aDifferentially regulated genes with P1(t) > 0.999 and |foldchange| > 1.4
Table 5.
Functional enrichment analysis of differentially regulateda genes elicited by TCDD in Hepa1c1c7 cells using DAVID.
| Category | Term | Gene count | Fold enrichment | P-value |
|---|---|---|---|---|
| Enrichment Score: 18.67 | ||||
| GOTERM_BP_3 | GO:0000278 ~ mitotic cell cycle | 43 | 5.95 | 7.67E-21 |
| GOTERM_BP_3 | GO:0022402 ~ cell cycle process | 52 | 4.50 | 1.15E-19 |
| GOTERM_BP_3 | GO:0022403 ~ cell cycle phase | 47 | 4.87 | 3.67E-19 |
| GOTERM_BP_3 | GO:0048285 ~ organelle fission | 36 | 6.14 | 6.31E-18 |
| Enrichment Score: 2.05 | ||||
| GOTERM_BP_3 | GO:0006805 ~ xenobiotic metabolic process | 4 | 11.20 | 4.67E-03 |
| GOTERM_BP_3 | GO:0009410 ~ response to xenobiotic stimulus | 4 | 8.96 | 9.05E-03 |
| GOTERM_BP_3 | GO:0006725 ~ cellular aromatic compound metabolic process | 9 | 2.75 | 1.66E-02 |
| Enrichment Score: 2.04 | ||||
| GOTERM_BP_3 | GO:0048522 ~ positive regulation of cellular process | 58 | 1.51 | 1.41E-03 |
| GOTERM_BP_3 | GO:0042127 ~ regulation of cell proliferation | 27 | 1.72 | 7.62E-03 |
| GOTERM_BP_3 | GO:0008284 ~ positive regulation of cell proliferation | 14 | 1.69 | 7.12E-02 |
| Enrichment Score: 1.62 | ||||
| GOTERM_BP_3 | GO:0051640 ~ organelle localization | 7 | 4.36 | 5.10E-03 |
| GOTERM_BP_3 | GO:0051651 ~ maintenance of location in cell | 4 | 5.38 | 3.68E-02 |
| GOTERM_BP_3 | GO:0051235 ~ maintenance of location | 4 | 4.07 | 7.35E-02 |
aDifferentially regulated genes with P1(t) > 0.999 and |foldchange| > 1.4
Table 6.
Functional enrichment analysis of differentially regulateda genes elicited by TCDD in H4IIE cells using DAVID.
| Category | Term | Gene count | Fold enrichment | P-value |
|---|---|---|---|---|
| Enrichment Score: 3.05 | ||||
| GOTERM_BP_3 | GO:0006805 ~ xenobiotic metabolic process | 7 | 34.21 | 3.52E-08 |
| GOTERM_BP_3 | GO:0009410 ~ response to xenobiotic stimulus | 7 | 31.47 | 6.09E-08 |
| GOTERM_BP_3 | GO:0006725 ~ cellular aromatic compound metabolic process | 6 | 5.07 | 6.28E-03 |
| GOTERM_BP_3 | GO:0017144 ~ drug metabolic process | 3 | 16.86 | 1.31E-02 |
| GOTERM_BP_3 | GO:0051186 ~ cofactor metabolic process | 6 | 3.61 | 2.44E-02 |
| GOTERM_BP_3 | GO:0046483 ~ heterocycle metabolic process | 5 | 1.78 | 3.03E-01 |
| GOTERM_BP_3 | GO:0044248 ~ cellular catabolic process | 8 | 1.39 | 3.41E-01 |
| Enrichment Score: 2.45 | ||||
| GOTERM_BP_3 | GO:0010035 ~ response to inorganic substance | 11 | 4.70 | 1.00E-04 |
| GOTERM_BP_3 | GO:0006800 ~ oxygen and reactive oxygen species metabolic process | 5 | 10.81 | 1.10E-03 |
| GOTERM_BP_3 | GO:0006979 ~ response to oxidative stress | 7 | 4.50 | 4.38E-03 |
| GOTERM_BP_3 | GO:0044248 ~ cellular catabolic process | 8 | 1.39 | 3.41E-01 |
| Enrichment Score: 1.59 | ||||
| GOTERM_BP_3 | GO:0006629 ~ lipid metabolic process | 14 | 2.49 | 3.37E-03 |
| GOTERM_BP_3 | GO:0006082 ~ organic acid metabolic process | 12 | 2.71 | 4.12E-03 |
| GOTERM_BP_3 | GO:0042180 ~ cellular ketone metabolic process | 12 | 2.68 | 4.57E-03 |
| GOTERM_BP_3 | GO:0044255 ~ cellular lipid metabolic process | 11 | 2.77 | 5.68E-03 |
| GOTERM_BP_3 | GO:0005996 ~ monosaccharide metabolic process | 7 | 4.23 | 5.88E-03 |
| GOTERM_BP_3 | GO:0016051 ~ carbohydrate biosynthetic process | 5 | 6.18 | 8.38E-03 |
| GOTERM_BP_3 | GO:0044262 ~ cellular carbohydrate metabolic process | 7 | 2.63 | 4.80E-02 |
| GOTERM_BP_3 | GO:0046165 ~ alcohol biosynthetic process | 3 | 8.03 | 5.25E-02 |
| GOTERM_BP_3 | GO:0005975 ~ carbohydrate metabolic process | 8 | 2.25 | 6.22E-02 |
| GOTERM_BP_3 | GO:0006091 ~ generation of precursor metabolites and energy | 5 | 2.70 | 1.11E-01 |
| GOTERM_BP_3 | GO:0008610 ~ lipid biosynthetic process | 4 | 1.71 | 4.09E-01 |
| GOTERM_BP_3 | GO:0044249 ~ cellular biosynthetic process | 16 | 1.04 | 5.74E-01 |
| Enrichment Score: 1.58 | ||||
| GOTERM_BP_3 | GO:0042493 ~ response to drug | 11 | 3.94 | 4.20E-04 |
| GOTERM_BP_3 | GO:0006869 ~ lipid transport | 4 | 3.91 | 8.07E-02 |
| GOTERM_BP_3 | GO:0010876 ~ lipid localization | 4 | 3.54 | 1.01E-01 |
| GOTERM_BP_3 | GO:0042592 ~ homeostatic process | 10 | 1.66 | 1.39E-01 |
| Enrichment Score: 1.54 | ||||
| GOTERM_BP_3 | GO:0044255 ~ cellular lipid metabolic process | 11 | 2.77 | 5.68E-03 |
| GOTERM_BP_3 | GO:0006721 ~ terpenoid metabolic process | 3 | 10.54 | 3.20E-02 |
| GOTERM_BP_3 | GO:0006766 ~ vitamin metabolic process | 3 | 4.75 | 1.29E-01 |
aDifferentially regulated genes with P1(t) > 0.999 and |foldchange| > 1.4
Figure 8.
Gene interaction network of TCDD-responsive orthologs functionally related to lipid metabolism. Molecules are represented by nodes connected by arrows to indicate either direct or indirect relationships (dotted lines). Colors in the gene expression boxes represent the direction and magnitude of the gene expression responses from the Agilent microarrays. The network visualizes species-conserved and -divergent gene expression responses elicited by TCDD in the human HepG2, mouse Hepa1c1c7 and rat H4IIE cell lines.
Despite the nature of these continuous cell lines and the dysregulation of genes related to cell cycle control and regulation, GO terms associated with these functions were not enriched across all three cell lines. TCDD-treated Human HepG2 and mouse Hepa1c1c7 cell lines identified Functional clusters associated with cell cycle control (GO:0022402) were in enriched TCDD-treated HepG2 and Hepa1c1c7 gene expression data sets, but not in the H4IIE data set. Species-differences in functional clustering of TCDD-elicited differentially expressed gene data sets further corroborate species-specific responses to TCDD.
Dioxin Response Element Analysis
Putative functional DREs are not equally distributed between species with more DREs associated with known human genes [17,18]. Within the human genome, there are 213,355 DRE cores in the proximal promoter regions of known genes (10kb upstream to 1kb downstream of a transcriptional start site [TSS]), with 139,289 and 100,614 DRE cores, in the mouse and rat genomes, respectively. To further investigate the divergent expression of CCND1, ID3 and GSTA5, their proximal promoter regions were searched for DRE cores. Human orthologs of CCND1 and ID3 contained 12 and 14 DRE cores, respectively, greater than the number found in either the mouse or rat genomes. In contrast, mouse and rat orthologs of GSTA5 each had 7 and 12 DRE cores, respectively, while the human only had 3 (Figures 7B and 7C). In each of these divergently regulated orthologs, the species with the greatest number of DRE cores within the regulatory region had the highest fold induction suggesting that species-specific regulons have important roles in regulating gene expression.
Each DRE core in the GSTA5 orthologs was extended by 7 bp on either end and assessed for sequence similarity by measuring the Euclidean distance between sequence pairs. Only the mouse and rat GSTA5 contained highly similar DRE sequences (Euclidian distance of ≤ 3.0; shaded rows in Figure 7C), and no human DRE sequences with high sequence similarity. Furthermore, the position of the conserved DRE sequence 5 bp upstream of the TSS in the mouse appears to be positionally conserved with the DRE sequence in rat the ortholog located 22 bp upstream of the TSS. Collectively, the disproportionate number of DREs between species and lack of sequence and spatially conserved DREs may account for the divergent regulation of human, mouse and rat GSTA5 orthologs.
Discussion
This study comprehensively and systematically compares the gene expression responses elicited by TCDD across human, mouse and rat cells. Incorporating both custom cDNA microarrays to profile the temporal responses and more comprehensive commercial oligonucleotide microarrays, a limited number of conserved responses between species were identified. In addition, divergent and a large number of species-specific responses were identified that may contribute to species-specific differences in sensitivity and toxicity. These results are consistent with the poor response correlations of orthologous genes between C57BL/6 mice, Sprague Dawley and Long-Evans rats [10,11]. Collectively, reported in vivo rodent comparisons, and the in vitro data presented in this study suggest there are significant differences in TCDD elicited gene expression between species, despite the conservation of the AhR [4] and its signaling pathway.
In vivo and in vitro studies examining TCDD-elicited global gene expression have demonstrated that AhR targets a limited portion of the genome [10-12,19-23]. In addition, PWM-based computational searches identified a low percentage of orthologs with conserved putative functional DREs within their regulatory regions (10kb upstream of the TSS and the 5'UTR) [17,18]. Our comparative in vitro microarray results corroborate these findings. Temporal analysis using custom cDNA microarrays found that TCDD elicited a response in 9.9% of the represented genes in the HepG2 cells, 5.2% in the Hepa1c1c7 cells and only 1.1% in the H4IIE cells. Similar results were obtained using whole-genome microarrays where 4.7%, 2.4% and 0.7% of the genes exhibiting differential expression in HepG2, Hepa1c1c7 and H4IIE cells, respectively, at 24 hrs.
All three cell lines differentially expressed a core set of conserved gene responses that included the induction of CYP1A1, NQO1 and UGT1A6, members of the AhR gene battery [24]. However, a significant number of responses were specific to a single species (Figures 3B and 3B), as reported in in vivo studies [10,11]. Moreover, these studies not only identified species-specific responses, but also orthologs with divergent responses (i.e. same gene up-regulated in one species and down-regulated in other). Comparisons of C57BL/6 mouse and Sprague Dawley rat responses found that 29% of the commonly regulated orthologs exhibited divergent regulation [11]. Similarly, GSTA5, CCND1 and ID3, exhibited divergent regulation across HepG2, Hepa1c1c7 and H4IIE cells (Table 3). Each of these genes exhibited the same pattern with Hepa1c1c7 and H4IIE cells having comparable profiles while HepG2 cells exhibited the divergent response. For example, GSTA5 was up-regulated in Hepa1c1c7 and H4IIE cells, but down-regulated in HepG2 cells (Table 3). In vivo gene expression responses of GSTA5 orthologs matched the in vitro responses in the Hepa1c1c7 and H4IIE cells; TCDD treated mice and rats exhibited 1.8- and 1.9-fold maximum induction, respectively [11,12]. QRTPCR verified the divergent response of GSTA5 in the H4IIE and HepG2 cells, but did not detect a significant induction in the Hepa1c1c7 cells (Figure 7A).
Computational analysis of GST5A found a disproportionate number of DRE cores within the regulatory region sequence of each species (Figure 7B and 7C). Sequence analysis also found DRE sequences with high similarity in the mouse and rat orthologs, but not in the human GSTA5. The identification of species-specific DREs is consistent with the divergent regulation of GSTA5 between these cell lines. Overall, differences in ortholog expression may contribute to differences in TCDD sensitivity and toxicity across species.
Although these continuous cells lines are similar in morphology and were derived from hepatomas, there are inherent differences that may bias the identification of species-conserved and -divergent responses. For example, HepG2 cells were originally derived from the liver biopsy of a 15-year old Caucasian male and therefore may not be representative of a mature adult liver [25]. Recent studies have compared basal gene expression of liver samples to primary human hepatocytes, HepG2 and HepaRG cells, another human hepatoma cell line. Although, HepaRG cells were most similar to primary hepatocytes and liver samples [26], toxicogenomic studies report that HepaRG and HepG2 gene expression responses retained common functional processes [27]. In addition, the HepaRG donor differed in age and sex compared to the HepG2 patient, and was also infected with hepatitis C, which may affect both basal and TCDD-elicited gene expression. HepG2 cells were more sensitive in terms of the magnitude of regulation, and also in the terms of the total number of differentially regulated genes, and may be a more sensitive model for assessing TCDD exposure [27].
Conclusion
Although a core set of conserved gene responses was identified, consistent with the role of AhR in mediating the adaptive metabolic responses, further evidence of differences in genome-wide gene expression profiles between species (i.e. species-specific regulons) is also presented. This is consistent with species-specific differences in TCDD sensitivity and toxicity [6,8,28,29], which are due to alterations in gene expression. Furthermore, there is a lack of conserved putative DREs within orthologous genes [18], and differences in genome-wide gene expression profiles has been reported between mice and rats in vivo [10,11]. Undoubtedly, the number of conserved responses and immaturity of the genome annotation, especially for the rat, limits the overall interspecies comparison [30]. Differences in AhR levels, co-activator availability, and protocols used in their isolation of these hepatoma cells may confound our comparisons. Nevertheless, the identification of numerous species-specific responses, evidence of divergent gene expression responses between species, and the discovery of distinct putative primary response sets in each cell line provides further compelling evidence that the effects of TCDD post-AhR binding are not conserved between species.
Methods
Cell Culture and Treatment
HepG2 (Dr. Trevor Archer, NIEHS, Research Triangle Park, NC), Hepa1c1c7 (Dr. Oliver Hankinson, University of California, Los Angeles, CA), and H4IIE (Dr. Niels Bols, University of Waterloo, Waterloo, Canada) cells were cultured in monolayers and treated with TCDD (S. Safe, Texas A&M University, College Station, TX) as previously described [16]. Briefly, cells were treated with either 10 nM TCDD or DMSO vehicle control for 1, 2, 4, 8, 12, 24 or 48 hrs for the time course studies. For co-treatment studies, cells were pretreated with 10 mg/ml cycloheximide (CHX; Sigma) for 1 hr and then treated for an additional 4 or 12 hrs with 10 nM TCDD or DMSO vehicle (Additional file 19). All treatment studies were performed in triplicate.
RNA Isolation
Cells were harvested with 2.0 mL of TRIzol® Reagent (Invitrogen) and total RNA isolated according to the manufacturer's protocol followed by an acid phenol:chloroform extraction. Isolated RNA was resuspended in The RNA Storage Solution (Ambion Inc., Austin, TX), quantified (A260), and assessed for purity by measuring the A260/A280 ratio and by visual inspection of 1.0 μg on a denaturing gel.
Microarray Experimental Design and Analysis
Gene expression changes were analyzed using custom human, mouse and rat cDNA microarrays as previously described [16,31,32]. Responses to CHX and TCDD co-treatment were also assayed with cDNA microarrays using a 2 × 2 factorial design (Additional file 20) [33]. Three replicates were performed with two independent labelings per sample (dye swap). In total, 42 cDNA microarrays were performed for each individual cell line. Additionally, 4 × 44 k Agilent Technologies whole-genome oligonucleotide microarrays (Santa Clara, CA), were used to profile the responses elicited by TCDD 24 hrs post-treatment according to the manufacturer's Two-Color Microarray-Based Gene Expression Analysis protocol Version 5.0.1, including dye swap labelings. Three replicates were performed for a total of 9 whole genome microarrays for each cell line. Microarray data quality was first assessed using a quality assurance protocol to ensure consistent high quality data throughout all studies prior to normalization and further analysis [34]. Microarray data were normalized using a semiparametric method [35], and statistically analyzed using an empirical Bayes methods [36]. The data were hierarchically clustered using the Euclidian distance in Cluster 3.0 [37] and visualized with Java Treeview [38]. Functional annotation clustering of Gene Ontology (GO) terms for differentially expressed genes was performed using DAVID (Database for Annotation, Visualization, and Integrated Discovery) [39]. Annotation clusters with an enrichment score ≥ 1.3 were considered significantly enriched. Networks of direct and indirect molecular interactions based on the whole-genome expression data were identified and visualized using the Ingenuity Pathway Analysis http://www.ingenuity.com.
Quantitative Real-Time PCR
The same total RNA samples isolated for microarray studies were used for QRTPCR as previously described [16]. The copy number of each unknown sample was standardized to the geometric mean of three house-keeping genes (β-actin, Gapd, Hprt or Rpl13). Official gene names and symbols, RefSeq and Entrez Gene IDs, forward and reverse primer sequences, and amplicon sizes are provided in Additional file 21. Data were analyzed by analysis of variance (ANOVA) followed by Tukey's post hoc test using SAS 9.1 (SAS Institute, Cary, NC). Differences between treatment groups were considered significant when p < 0.05.
Computational DREs Searches
The proximal promoter region (10 kb upstream and 1 kb downstream of a TSS) for a RefSeq corresponding to an individual gene were computationally searched for the substitute intolerant 5'-GCGTG-3' DRE core sequence and the position relative to the TSS were determined using the center of the 5 bp core (underlined). Each core was then extended by 7 bp upstream and downstream of the core to generate a 19 bp DRE core containing sequence, which were assigned a matrix similarity using a previously defined algorithm [18] and with an updated position weight matrix [17]. The 19 bp DRE core sequences from orthologous human, mouse and rat genes were hierarchically clustered in R http://www.R-project.org by measuring the Euclidean distance between pairs of sequences. DRE sequences that clustered together with a distance value ≤ 3.0 were characterized as orthologous DREs.
Authors' contributions
ED performed the cell culture experiments, microarrays, QRTPCR cross-species comparison of the microarray data, computational DRE promoter analysis and the initial preparation of the manuscript. Additional QRTPCR was performed AWL. LDB normalized the microarray data and provided input into the microarray study designs. TRZ oversaw the completion of the study. All the authors have given final approval of the version to be published.
Supplementary Material
HepG2 TCDD time course and cycloheximide cotreatment cDNA microarray data. A table containing the expression ratios relative to the time matched vehicle control for the time course study. Ratios for the cycloheximide studies are relative to the treatment condition. P1(t)-values represent posterior probabilities of activity on a per gene and time-point basis or treatment condition using the model-based t-value.
Hepa1c1c7 TCDD time course and cycloheximide cotreatment cDNA microarray data. A table containing the expression ratios relative to the time matched vehicle control for the time course study. Ratios for the cycloheximide studies are relative to the treatment condition. P1(t)-values represent posterior probabilities of activity on a per gene and time-point basis or treatment condition using the model-based t-value.
H4IIE TCDD time course and cycloheximide cotreatment cDNA microarray data. A table containing the expression ratios relative to the time matched vehicle control for the time course study. Ratios for the cycloheximide studies are relative to the treatment condition. P1(t)-values represent posterior probabilities of activity on a per gene and time-point basis or treatment condition using the model-based t-value.
Common differentially regulated orthologs elicited by TCDD in human HepG2 and mouse Hepa1c1c7 cells identified from the cDNA microarray time course studies. A table containing the expression ratios for significantly differentially regulated orthologs (|fold change| > 1.4 and P1(t) > 0.999).
Common differentially regulated orthologs elicited by TCDD in rat H4IIE and mouse Hepa1c1c7 cells identified from the cDNA microarray time course studies. A table containing the expression ratios for significantly differentially regulated orthologs (|fold change| > 1.4 and P1(t) > 0.999).
Common differentially regulated orthologs elicited by TCDD in human HepG2 and rat H4IIE cells identified from the cDNA microarray time course studies. A table containing the expression ratios for significantly differentially regulated orthologs (|fold change| > 1.4 and P1(t) > 0.999).
Putative human primary response genes and the number of DREs in their promoters. A table listing the putative primary response genes identified from the cycloheximide studies and the number of 5'-GCGTG-3' DRE cores in the region 10 kb upstream and 1 kb downstream of a TSS.
Putative mouse primary response genes and the number of DREs in their promoters. A table listing the putative primary response genes identified from the cycloheximide studies and the number of 5'-GCGTG-3' DRE cores in the region 10 kb upstream and 1 kb downstream of a TSS.
Putative rat primary response genes and the number of DREs in their promoters. A table listing the putative primary response genes identified from the cycloheximide studies and the number of 5'-GCGTG-3' DRE cores in the region 10 kb upstream and 1 kb downstream of a TSS.
Putative human secondary response genes and the number of DREs in their promoters. A table listing the putative secondary response genes identified from the cycloheximide studies and the number of 5'-GCGTG-3' DRE cores in the region 10 kb upstream and 1 kb downstream of a TSS.
Putative mouse secondary response genes and the number of DREs in their promoters. A table listing the putative secondary response genes identified from the cycloheximide studies and the number of 5'-GCGTG-3' DRE cores in the region 10 kb upstream and 1 kb downstream of a TSS.
Putative rat secondary response genes and the number of DREs in their promoters. A table listing the putative secondary response genes identified from the cycloheximide studies and the number of 5'-GCGTG-3' DRE cores in the region 10 kb upstream and 1 kb downstream of a TSS.
Whole-genome Agilent microarray data from HepG2 cells treated with TCDD for 24 hrs. A table containing the expression ratio relative to the time matched vehicle control. P1(t)-values represent posterior probabilities of activity on a per gene basis using the model-based t-value.
Whole-genome Agilent microarray data from Hepa1c1c7 cells treated with TCDD for 24 hrs. A table containing the expression ratio relative to the time matched vehicle control. P1(t)-values represent posterior probabilities of activity on a per gene basis using the model-based t-value.
Whole-genome Agilent microarray data from H4IIE cells treated with TCDD for 24 hrs. A table containing the expression ratio relative to the time matched vehicle control. P1(t)-values represent posterior probabilities of activity on a per gene basis using the model-based t-value.
Common differentially regulated orthologs elicited by TCDD in human HepG2 and mouse Hepa1c1c7 cells identified from the whole-genome Agilent microarrays at 24 hrs. A table containing the expression ratios for significantly differentially regulated orthologs (|fold change| > 1.4 and P1(t) > 0.999).
Common differentially regulated orthologs elicited by TCDD in rat H4IIE and mouse Hepa1c1c7 cells identified from the whole-genome Agilent microarrays at 24 hrs. A table containing the expression ratios for significantly differentially regulated orthologs (|fold change| > 1.4 and P1(t) > 0.999).
Common differentially regulated orthologs elicited by TCDD in human HepG2 and rat H4IIE cells identified from the whole-genome Agilent microarrays at 24 hrs. A table containing the expression ratios for significantly differentially regulated orthologs (|fold change| > 1.4 and P1(t) > 0.999).
Cell culture TCDD treatment and harvesting regimen. For the time course studies, cells were treated with 10 nM TCDD or 0.1% DMSO vehicle and harvested at 1, 2, 4, 8, 12, 24, or 48 hrs post-treatment. For the cycloheximide studies cells were treated with 10 mg/ml cycloheximide 1 hr and then treated with either 10 nM TCDD or 0.1% DMSO vehicle and for an additional 4 or 12 hrs (as indicated by *).
2 × 2 Factorial microarray experimental design used for the cycloheximide expression studies. The hybridization design used to identify putative primary and secondary gene expression responses elicited by TCDD. Each arrow represents one microarray where the arrow heads and tails refer to Cy5 and Cy3 dye labeling, respectively. Double-headed arrows indicate dye swaps (each sample labeled with Cy3 and Cy5 on different microarrays).
Gene symbols and primer sequences for QRTPCR. A list of genes and primer sequences used to verify cDNA and Agilent microarray responses.
Contributor Information
Edward Dere, Email: dereedwa@msu.edu.
Andrea W Lee, Email: andreale@msu.edu.
Lyle D Burgoon, Email: burgoonl@msu.edu.
Timothy R Zacharewski, Email: tzachare@msu.edu.
Acknowledgements
This work was supported by the National Institute of Environmental Health Sciences Superfund Basic Research Program [P42ES04911].
References
- Poland A, Knutson JC. 2,3,7,8-tetrachlorodibenzo-p-dioxin and related halogenated aromatic hydrocarbons: examination of the mechanism of toxicity. Annu Rev Pharmacol Toxicol. 1982;22:517–554. doi: 10.1146/annurev.pa.22.040182.002505. [DOI] [PubMed] [Google Scholar]
- Denison MS, Heath-Pagliuso S. The Ah receptor: a regulator of the biochemical and toxicological actions of structurally diverse chemicals. Bulletin of environmental contamination and toxicology. 1998;61:557–568. doi: 10.1007/PL00002973. [DOI] [PubMed] [Google Scholar]
- Hankinson O. The aryl hydrocarbon receptor complex. Annu Rev Pharmacol Toxicol. 1995;35:307–340. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- Schmidt JV, Bradfield CA. Ah receptor signaling pathways. Annu Rev Cell Dev Biol. 1996;12:55–89. doi: 10.1146/annurev.cellbio.12.1.55. [DOI] [PubMed] [Google Scholar]
- Olson H, Betton G, Robinson D, Thomas K, Monro A, Kolaja G, Lilly P, Sanders J, Sipes G, Bracken W. et al. Concordance of the toxicity of pharmaceuticals in humans and in animals. Regul Toxicol Pharmacol. 2000;32:56–67. doi: 10.1006/rtph.2000.1399. [DOI] [PubMed] [Google Scholar]
- Schwetz BA, Norris JM, Sparschu GL, Rowe UK, Gehring PJ, Emerson JL, Gerbig CG. Toxicology of chlorinated dibenzo-p-dioxins. Environ Health Perspect. 1973;5:87–99. doi: 10.2307/3428117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison MS, Vella LM, Okey AB. Structure and function of the Ah receptor for 2,3,7,8-tetrachlorodibenzo-p-dioxin. Species difference in molecular properties of the receptors from mouse and rat hepatic cytosols. J Biol Chem. 1986;261:3987–3995. [PubMed] [Google Scholar]
- Olson JR, Holscher MA, Neal RA. Toxicity of 2,3,7,8-tetrachlorodibenzo-p-dioxin in the golden Syrian hamster. Toxicol Appl Pharmacol. 1980;55:67–78. doi: 10.1016/0041-008X(80)90221-5. [DOI] [PubMed] [Google Scholar]
- Poland A, Glover E, Kende AS. Stereospecific, high affinity binding of 2,3,7,8-tetrachlorodibenzo-p-dioxin by hepatic cytosol. Evidence that the binding species is receptor for induction of aryl hydrocarbon hydroxylase. J Biol Chem. 1976;251:4936–4946. [PubMed] [Google Scholar]
- Boutros PC, Yan R, Moffat ID, Pohjanvirta R, Okey AB. Transcriptomic responses to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in liver: comparison of rat and mouse. BMC Genomics. 2008;9:419. doi: 10.1186/1471-2164-9-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boverhof DR, Burgoon LD, Tashiro C, Sharratt B, Chittim B, Harkema JR, Mendrick DL, Zacharewski TR. Comparative toxicogenomic analysis of the hepatotoxic effects of TCDD in Sprague Dawley rats and C57BL/6 mice. Toxicol Sci. 2006;94:398–416. doi: 10.1093/toxsci/kfl100. [DOI] [PubMed] [Google Scholar]
- Fletcher N, Wahlström D, Lundberg R, Nilsson CB, Nilsson KC, Stockling K, Hellmold H, Håkansson H. 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) alters the mRNA expression of critical genes associated with cholesterol metabolism, bile acid biosynthesis, and bile transport in rat liver: a microarray study. Toxicol Appl Pharmacol. 2005;207:1–24. doi: 10.1016/j.taap.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Lusska A, Wu L, Whitlock JP. Superinduction of CYP1A1 transcription by cycloheximide. Role of the DNA binding site for the liganded Ah receptor. J Biol Chem. 1992;267:15146–15151. [PubMed] [Google Scholar]
- Ma Q. Induction and superinduction of 2,3,7,8-tetrachlorodibenzo-rho-dioxin-inducible poly(ADP-ribose) polymerase: role of the aryl hydrocarbon receptor/aryl hydrocarbon receptor nuclear translocator transcription activation domains and a labile transcription repressor. Arch Biochem Biophys. 2002;404:309–316. doi: 10.1016/S0003-9861(02)00339-9. [DOI] [PubMed] [Google Scholar]
- Joiakim A, Mathieu PA, Elliott AA, Reiners JJ. Superinduction of CYP1A1 in MCF10A cultures by cycloheximide, anisomycin, and puromycin: a process independent of effects on protein translation and unrelated to suppression of aryl hydrocarbon receptor proteolysis by the proteasome. Molecular Pharmacology. 2004;66:936–947. doi: 10.1124/mol.66.4.. [DOI] [PubMed] [Google Scholar]
- Dere E, Boverhof DR, Burgoon LD, Zacharewski TR. In vivo-in vitro toxicogenomic comparison of TCDD-elicited gene expression in Hepa1c1c7 mouse hepatoma cells and C57BL/6 hepatic tissue. BMC Genomics. 2006;7:80. doi: 10.1186/1471-2164-7-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dere E, Forgacs AL, Zacharewski TR, Burgoon LD. Genome-Wide Computational Analysis of Dioxin Response Element Location and Distribution in the Human, Mouse, and Rat Genomes. Chem Res Toxicol. 2011. [DOI] [PMC free article] [PubMed]
- Sun YV, Boverhof DR, Burgoon LD, Fielden MR, Zacharewski TR. Comparative analysis of dioxin response elements in human, mouse and rat genomic sequences. Nucleic Acids Res. 2004;32:4512–4523. doi: 10.1093/nar/gkh782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin B, Kim G, Park DW, Ryu D-Y. Microarray analysis of gene regulation in the Hepa1c1c7 cell line following exposure to the DNA methylation inhibitor 5-aza-2'-deoxycytidine and 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol In Vitro. 2004;18:659–664. doi: 10.1016/j.tiv.2004.02.006. [DOI] [PubMed] [Google Scholar]
- McHale CM, Zhang L, Hubbard AE, Zhao X, Baccarelli A, Pesatori AC, Smith MT, Landi MT. Microarray analysis of gene expression in peripheral blood mononuclear cells from dioxin-exposed human subjects. Toxicology. 2007;229:101–113. doi: 10.1016/j.tox.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Puga A, Maier A, Medvedovic M. The transcriptional signature of dioxin in human hepatoma HepG2 cells. Biochem Pharmacol. 2000;60:1129–1142. doi: 10.1016/S0006-2952(00)00403-2. [DOI] [PubMed] [Google Scholar]
- Silkworth JB, Carlson EA, McCulloch C, Illouz K, Goodwin S, Sutter TR. Toxicogenomic analysis of gender, chemical, and dose effects in livers of TCDD- or aroclor 1254-exposed rats using a multifactor linear model. Toxicol Sci. 2008;102:291–309. doi: 10.1093/toxsci/kfm313. [DOI] [PubMed] [Google Scholar]
- Xiong KM, Peterson RE, Heideman W. Aryl hydrocarbon receptor-mediated down-regulation of sox9b causes jaw malformation in zebrafish embryos. Molecular Pharmacology. 2008;74:1544–1553. doi: 10.1124/mol.108.050435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebert DW, Roe AL, Dieter MZ, Solis WA, Yang Y, Dalton TP. Role of the aromatic hydrocarbon receptor and [Ah] gene battery in the oxidative stress response, cell cycle control, and apoptosis. Biochem Pharmacol. 2000;59:65–85. doi: 10.1016/S0006-2952(99)00310-X. [DOI] [PubMed] [Google Scholar]
- Ogino M, Nagata K, Yamazoe Y. Selective suppressions of human CYP3A forms, CYP3A5 and CYP3A7, by troglitazone in HepG2 cells. Drug Metab Pharmacokinet. 2002;17:42–46. doi: 10.2133/dmpk.17.42. [DOI] [PubMed] [Google Scholar]
- Hart SN, Li Y, Nakamoto K, Subileau E-a, Steen D, Zhong X-b. A comparison of whole genome gene expression profiles of HepaRG cells and HepG2 cells to primary human hepatocytes and human liver tissues. Drug Metab Dispos. 2010;38:988–994. doi: 10.1124/dmd.109.031831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennen DGJ, Magkoufopoulou C, Ketelslegers HB, van Herwijnen MHM, Kleinjans JCS, van Delft JHM. Comparison of HepG2 and HepaRG by whole-genome gene expression analysis for the purpose of chemical hazard identification. Toxicol Sci. 2010;115:66–79. doi: 10.1093/toxsci/kfq026. [DOI] [PubMed] [Google Scholar]
- Bickel M. Polychlorinated persistent compounds. Experientia. 1982;38:879–882. doi: 10.1007/BF01953634. [DOI] [PubMed] [Google Scholar]
- Vos JG, Moore JA, Zinkl JG. Toxicity of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in C57B1/6 mice. Toxicol Appl Pharmacol. 1974;29:229–241. doi: 10.1016/0041-008X(74)90060-X. [DOI] [PubMed] [Google Scholar]
- Boverhof DR, Zacharewski TR. Toxicogenomics in risk assessment: applications and needs. Toxicol Sci. 2006;89:352–360. doi: 10.1093/toxsci/kfj018. [DOI] [PubMed] [Google Scholar]
- Kim S, Dere E, Burgoon LD, Chang C-C, Zacharewski TR. Comparative analysis of AhR-mediated TCDD-elicited gene expression in human liver adult stem cells. Toxicol Sci. 2009;112:229–244. doi: 10.1093/toxsci/kfp189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwekel JC, Forgacs AL, Burgoon LD, Williams KJ, Zacharewski TR. Tamoxifen-elicited uterotrophy: cross-species and cross-ligand analysis of the gene expression program. BMC medical genomics. 2009;2:19. doi: 10.1186/1755-8794-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YH, Speed T. Design issues for cDNA microarray experiments. Nat Rev Genet. 2002;3:579–588. doi: 10.1038/nrg863. [DOI] [PubMed] [Google Scholar]
- Burgoon L, Eckel-Passow J, Gennings C, Boverhof D, Burt J, Fong C, Zacharewski T. Protocols for the assurance of microarray data quality and process control. Nucleic Acids Res. 2005;33:e172. doi: 10.1093/nar/gni167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel J, Gennings C, Therneau T, Burgoon L, Boverhof D, Zacharewski T. Normalization of two-channel microarray experiments: a semiparametric approach. Bioinformatics. 2005;21:1078–1083. doi: 10.1093/bioinformatics/bti105. [DOI] [PubMed] [Google Scholar]
- Eckel J, Gennings C, Chinchilli V, Burgoon L, Zacharewski T. Empirical bayes gene screening tool for time-course or dose-response microarray data. J Biopharm Stat. 2004;14:647–670. doi: 10.1081/BIP-200025656. [DOI] [PubMed] [Google Scholar]
- de Hoon MJL, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HepG2 TCDD time course and cycloheximide cotreatment cDNA microarray data. A table containing the expression ratios relative to the time matched vehicle control for the time course study. Ratios for the cycloheximide studies are relative to the treatment condition. P1(t)-values represent posterior probabilities of activity on a per gene and time-point basis or treatment condition using the model-based t-value.
Hepa1c1c7 TCDD time course and cycloheximide cotreatment cDNA microarray data. A table containing the expression ratios relative to the time matched vehicle control for the time course study. Ratios for the cycloheximide studies are relative to the treatment condition. P1(t)-values represent posterior probabilities of activity on a per gene and time-point basis or treatment condition using the model-based t-value.
H4IIE TCDD time course and cycloheximide cotreatment cDNA microarray data. A table containing the expression ratios relative to the time matched vehicle control for the time course study. Ratios for the cycloheximide studies are relative to the treatment condition. P1(t)-values represent posterior probabilities of activity on a per gene and time-point basis or treatment condition using the model-based t-value.
Common differentially regulated orthologs elicited by TCDD in human HepG2 and mouse Hepa1c1c7 cells identified from the cDNA microarray time course studies. A table containing the expression ratios for significantly differentially regulated orthologs (|fold change| > 1.4 and P1(t) > 0.999).
Common differentially regulated orthologs elicited by TCDD in rat H4IIE and mouse Hepa1c1c7 cells identified from the cDNA microarray time course studies. A table containing the expression ratios for significantly differentially regulated orthologs (|fold change| > 1.4 and P1(t) > 0.999).
Common differentially regulated orthologs elicited by TCDD in human HepG2 and rat H4IIE cells identified from the cDNA microarray time course studies. A table containing the expression ratios for significantly differentially regulated orthologs (|fold change| > 1.4 and P1(t) > 0.999).
Putative human primary response genes and the number of DREs in their promoters. A table listing the putative primary response genes identified from the cycloheximide studies and the number of 5'-GCGTG-3' DRE cores in the region 10 kb upstream and 1 kb downstream of a TSS.
Putative mouse primary response genes and the number of DREs in their promoters. A table listing the putative primary response genes identified from the cycloheximide studies and the number of 5'-GCGTG-3' DRE cores in the region 10 kb upstream and 1 kb downstream of a TSS.
Putative rat primary response genes and the number of DREs in their promoters. A table listing the putative primary response genes identified from the cycloheximide studies and the number of 5'-GCGTG-3' DRE cores in the region 10 kb upstream and 1 kb downstream of a TSS.
Putative human secondary response genes and the number of DREs in their promoters. A table listing the putative secondary response genes identified from the cycloheximide studies and the number of 5'-GCGTG-3' DRE cores in the region 10 kb upstream and 1 kb downstream of a TSS.
Putative mouse secondary response genes and the number of DREs in their promoters. A table listing the putative secondary response genes identified from the cycloheximide studies and the number of 5'-GCGTG-3' DRE cores in the region 10 kb upstream and 1 kb downstream of a TSS.
Putative rat secondary response genes and the number of DREs in their promoters. A table listing the putative secondary response genes identified from the cycloheximide studies and the number of 5'-GCGTG-3' DRE cores in the region 10 kb upstream and 1 kb downstream of a TSS.
Whole-genome Agilent microarray data from HepG2 cells treated with TCDD for 24 hrs. A table containing the expression ratio relative to the time matched vehicle control. P1(t)-values represent posterior probabilities of activity on a per gene basis using the model-based t-value.
Whole-genome Agilent microarray data from Hepa1c1c7 cells treated with TCDD for 24 hrs. A table containing the expression ratio relative to the time matched vehicle control. P1(t)-values represent posterior probabilities of activity on a per gene basis using the model-based t-value.
Whole-genome Agilent microarray data from H4IIE cells treated with TCDD for 24 hrs. A table containing the expression ratio relative to the time matched vehicle control. P1(t)-values represent posterior probabilities of activity on a per gene basis using the model-based t-value.
Common differentially regulated orthologs elicited by TCDD in human HepG2 and mouse Hepa1c1c7 cells identified from the whole-genome Agilent microarrays at 24 hrs. A table containing the expression ratios for significantly differentially regulated orthologs (|fold change| > 1.4 and P1(t) > 0.999).
Common differentially regulated orthologs elicited by TCDD in rat H4IIE and mouse Hepa1c1c7 cells identified from the whole-genome Agilent microarrays at 24 hrs. A table containing the expression ratios for significantly differentially regulated orthologs (|fold change| > 1.4 and P1(t) > 0.999).
Common differentially regulated orthologs elicited by TCDD in human HepG2 and rat H4IIE cells identified from the whole-genome Agilent microarrays at 24 hrs. A table containing the expression ratios for significantly differentially regulated orthologs (|fold change| > 1.4 and P1(t) > 0.999).
Cell culture TCDD treatment and harvesting regimen. For the time course studies, cells were treated with 10 nM TCDD or 0.1% DMSO vehicle and harvested at 1, 2, 4, 8, 12, 24, or 48 hrs post-treatment. For the cycloheximide studies cells were treated with 10 mg/ml cycloheximide 1 hr and then treated with either 10 nM TCDD or 0.1% DMSO vehicle and for an additional 4 or 12 hrs (as indicated by *).
2 × 2 Factorial microarray experimental design used for the cycloheximide expression studies. The hybridization design used to identify putative primary and secondary gene expression responses elicited by TCDD. Each arrow represents one microarray where the arrow heads and tails refer to Cy5 and Cy3 dye labeling, respectively. Double-headed arrows indicate dye swaps (each sample labeled with Cy3 and Cy5 on different microarrays).
Gene symbols and primer sequences for QRTPCR. A list of genes and primer sequences used to verify cDNA and Agilent microarray responses.