Abstract
The stability of the genotypic marker IS6110, used to define the epidemiology of Mycobacterium tuberculosis, is one of the most important factors influencing the interpretation of DNA fingerprint data. We propose that evolved strains should be considered together with clustered strains to represent chains of ongoing transmission. For the present study we used a large set of fingerprint data for strains collected between 1992 and 1998 from residents of a community with a high incidence of tuberculosis in Cape Town, South Africa. Interstrain genetic distances were calculated by counting the banding pattern mismatches in the IS6110 DNA fingerprints of different isolates. These data demonstrate that the propensity to change by one or two bands is independent of the IS6110 copy number. Hence, the genetic distance between pairs of isolates can be simply expressed as the number of differences in the banding patterns. From this foundation, a data set which identifies newly evolved strains has been generated. Inclusion of these evolved strains into various molecular epidemiological calculations significantly increased the estimate of ongoing transmission in this study setting. The indication is that nearly all cases of tuberculosis in this community are due to ongoing transmission. This has important implications for tuberculosis control, as it indicates that the control measures used at present are unable to reduce the level of transmission. This technique may also be applicable to the study of low-incidence tuberculosis outbreaks as well as the analysis of epidemiological data from other disease epidemics.
The discovery of repeated sequences, such as the transposable element IS6110, in the genome of Mycobacterium tuberculosis has facilitated the accurate genotypic classification of the disease-causing organism infecting an individual within the context of a community (16). Restriction fragment length polymorphism (RFLP) data have become the primary, standardized tool for study of the molecular epidemiology of M. tuberculosis, greatly enhancing the understanding of the tuberculosis epidemic in different settings (2, 7, 9, 14, 19). The interpretation of these data remains complex, however, and is dependent on a thorough understanding of the stability of IS6110 as a marker of relatedness (5, 6, 12, 21, 23).
It is generally accepted that in order for a marker to be useful for epidemiological tracking, its rate of evolution must be low enough that strains from epidemiologically linked cases will have identical RFLP patterns (recent transmission) (2, 7, 14), while the rate of evolution must be sufficiently rapid to enable the discrimination of strains from closely related cases from those from more distantly related cases (i.e., transmission versus reactivation). Strains with unique RFLP patterns therefore reflect reactivations of dormant infections, strains from an outside community, or strains transmitted from unidentified sources (2, 7, 14). This scenario, while convenient, is an oversimplification, as it ignores the possibility that as an organism multiplies within its host it may give rise to clonal variants of itself (6, 11, 22, 23), characterized by minor changes in the IS6110-based RFLP banding pattern. These evolved strains may, in turn, be transmitted to new hosts (12, 21), leading over time to increasing genetic diversity in the bacterial population.
By assuming a constant rate of mutation, genetic distance (GD), defined as the number of mutational events separating two strains, may be regarded as an indicator of the evolutionary time since their divergence from a common ancestor. Thus, a high degree of similarity between two strains implies close temporal coupling. Conversely, with a longer evolutionary time since divergence, the probability of accumulation of mutations will be higher and therefore the GD will be greater. Studies of the stability of IS6110 fingerprints have demonstrated a half-life on the order of 2 to 3.2 years (6, 11, 23). Those investigators concluded that this rate of change is sufficiently low to facilitate epidemiological tracking. However, the calculated rate is also high enough to significantly influence the interpretation of relatedness in molecular epidemiological studies, especially those in which the study duration is similar to or longer than the half-life of the marker system. Given this nonzero rate of IS6110 fingerprint variation, clonal variants that appear within a relatively short time frame and whose patterns differ by a few bands may represent recently evolved strains and therefore may be regarded as constituting an ongoing transmission chain (12, 21, 23). In a recent study (21) we found evidence for this and reported that the manifestation of evolution was associated with transmission. We suggested that these events probably reflect the overall evolutionary dynamics of the bacterial population in the study setting. Failure to account for recent evolution in assessing epidemiological transmission may therefore hinder our understanding of the factors driving the epidemic. This is the case for most algorithms used at present, which regard clonal variants to be part of independent transmission chains or reactivation events (2, 7, 14, 19).
In the study described here we have examined the effect of evolution within transmission chains on the interpretation of M. tuberculosis molecular epidemiological data. We used a systematic approach based on interstrain GDs to group strains into molecular superclusters representing chains of ongoing transmission. Analysis of these data indicates that the rate of evolution of M. tuberculosis strains remains constant and is largely independent of the IS6110 copy number. We estimated the amount of ongoing transmission to be at least 20% higher than that predicted by more established methods (14). Our results show that the incorporation of evolution in the algorithms quantifying the extent of ongoing transmission may have a profound influence on our understanding of the dynamics of the disease, with consequent implications for epidemiological control strategies.
MATERIALS AND METHODS
This study forms part of a larger, long-term molecular epidemiological project which was approved by the ethics committee of Stellenbosch University.
Study population.
From mid-1992 to December 1998, M. tuberculosis isolates were collected from patients residing in and attending health care clinics in two adjacent suburbs in Cape Town, South Africa (3). Approximately 350 new bacteriologically confirmed adult cases of tuberculosis per 100,000 population are reported in this community each year. Prior to 1996, all patients were treated at one of the primary care clinics by directly observed therapy, although there was no systematic surveillance for cure rates. In 1996 the World Health Organization directly observed therapy strategy was implemented with all its attendant requirements, resulting in the availability of cure rates.
DNA fingerprinting.
Each isolate was classified by DNA fingerprinting by the internationally standardized protocol (16, 20). The resulting autoradiographs were scanned and analyzed with the GelCompar II program (Applied Maths, Sint-Martens-Latem, Belgium). M. tuberculosis isolates with less than six IS6110 bands were excluded from the study, as it has previously been shown that the IS6110 banding patterns in these strains show very little diversity (21), precluding their use in epidemiological tracking (14). A total of 168 isolates suspected of being cross-contaminants (17) or identified as nontuberculosis mycobacteria were excluded from the study.
GD.
The RFLP fingerprints were aligned by use of the GelCompar program to maximize the number of matching bands between each fingerprint pair, with tolerance parameters allowing a 6% shift in each pattern as a whole and a 0.4% variance in individual band positions. This yielded 332 strains (as defined by distinct IS6110 patterns) with more than five IS6110 elements from 708 disease cases. Cases from the same patient were considered distinct if the IS6110 patterns of the strains differed by more than four bands. An exhaustive pairwise comparison of each IS6110 banding pattern was performed by using a band-matching algorithm (GelCompar II) to generate a GD matrix. This consisted of an N-by-N table of the number of mismatched IS6110 bands between each pair of strains. The results were imported into a Microsoft Access database as a table of strain pairs with their corresponding GDs. On the basis of the assumption that recent evolutionary events are represented by a maximum of four banding pattern differences (6, 21), strain pairs with GDs of less than or equal to four were assigned a putative transmission status of source or secondary on the basis of the order of appearance in the community. These assignments were made subsequent to the application of the following filters. (i) Strains occurring only in patients who were <12 years of age or who did not present with pulmonary tuberculosis were excluded as possible sources, as they were considered unlikely to transmit the bacillus (1). (ii) Strain pairs for which the time interval between the last case caused by the source strain and the first case caused by the secondary strain was greater than a defined interval (2 or 5 years) were excluded. These intervals were arbitrarily chosen to represent the minimum and maximum periods within which progression to active disease would be considered recent infection. For each remaining secondary strain, the source with the nearest GD (NGD; i.e., the one that was the most similar) was selected as the most probable candidate. When two or more possible source strains had the same NGD, candidacy was equally apportioned between them.
To assess the propensity of the result of RFLP analysis with the IS6110 probe to change as a function of the number of insertion elements present in the genome, we determined the number of variant strains produced by source strains, which were categorized by the number of IS6110 bands, as a proportion of all new cases attributable to those source strains.
Estimation of recent transmission.
To calculate the extent of ongoing transmission, strain pairs were linked together into transmission chains on the basis of common source or secondary strain type by using a custom-written Perl script (the source code is available at http://www.sun.ac.za/med_biochem/). This process was performed for each maximum NGD in the range of 0 to 4. Isolates belonging to strain pairs with NGDs less than or equal to the chosen limit were grouped into superclusters. The ongoing transmission rate was determined by the formula (N − S)/T, where N is the number of cases grouped in superclusters, S is the number of superclusters, and T is the total number of cases in the study. Because the probability of detecting a primary index case diminishes with increasing temporal proximity to the study commencement date, we formulated an alternative estimate of ongoing transmission in which the calculation of S was limited to those superclusters initiated after the first 18 months of the study. Thus, ongoing transmission is defined as (NE + NL − SL)/T, where NE is the number of cases in superclusters initiated within the first 18 months of the study, and NL and SL are the number of cases in superclusters initiated after the first 18 months of the study and the number of superclusters into which they are grouped, respectively.
RESULTS
During the period from mid-1992 to the end of 1998, 1,630 M. tuberculosis isolates were cultured from 866 patients residing in and attending the health care clinics in two adjacent suburbs of Cape Town, South Africa. Of these, 164 isolates were excluded, as the cultures were contaminated, lost viability, or were subsequently found to contain nontuberculosis mycobacteria. The remaining 807 patients corresponded to 849 disease cases, as some patients reported with multiple, consecutive infections. This represented approximately 70% of all bacteriologically confirmed cases among residents in the community. The isolates representing these cases were subjected to RFLP analysis by the internationally standardized method in combination with the IS6110 probe. A total of 342 different IS6110 banding patterns were identified. Ten of these banding patterns, representing 140 cases, possessed less than six IS6110 insertions and were excluded from further analysis, as it has previously been shown that the genotypes of these strains are extremely stable and are therefore unsuitable for epidemiological tracking (14).
Pairwise analysis of the 332 high-copy-number IS6110 banding patterns was used to quantify the number of differences between all possible strain pairs. From this data set a total of 3,019 strain pairs with less than four band differences were identified, of which 1,168 fulfilled the case inclusion criteria and reflected NGD pairs. In this study, we have assumed that NGDs of 1 to 4 reflect recent evolutionary events, as previous studies have shown that up to four IS6110 banding pattern changes may occur during recent transmission (12, 21) or persistent disease (11, 22, 23).
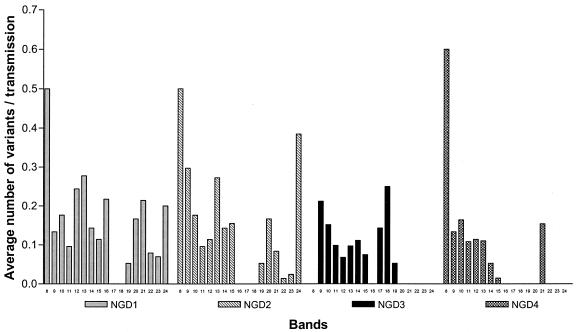
Analysis of these strain pairs showed that the propensity to evolve by either one or two mutational events (NGD = 1 or 2) appears to be independent of the number of IS6110-hybridizing bands present in the source strain (Fig. 1). This result differs from previous assumptions, which have suggested that the rate of change was proportional to the number of IS6110 elements in the source strain (13, 15). IS6110-mediated mutational events generating three or four banding pattern changes (NGD = 3 or 4) were also found to occur at a constant frequency in strains with 8 to 16 IS6110 insertions (Fig. 1). However, such events were largely absent from strains with 17 to 25 IS6110 insertions (Fig. 1), suggesting that multiple transposition events do not occur or are selected against in strains very high IS6110 copy numbers.
FIG. 1.
Numbers of variant strains produced as a proportion of observed transmission events from source cases with a defined number of IS6110 bands. Variant strains were produced by the loss or the gain of IS6110-hybridizing bands. Values are for strain pairs with NGDs equal to 1, 2, 3, and 4. The data shown are for a maximum interstrain interval of 2 years.
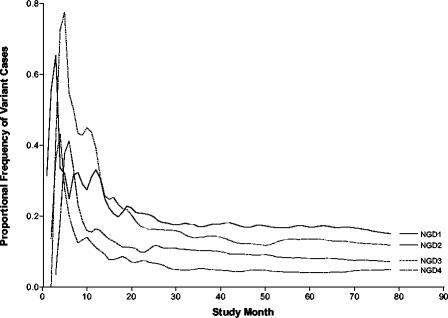
The absence of a clear correlation between propensity to change and the IS6110 copy number implies a simple relationship between mutational events (NGD = up to 4) and time, which is independent of the IS6110 copy number. This is demonstrated in Fig. 2, which shows that the production of variant strains as a proportion of all new cases is constant for the different NGD values. However, it is also clear that there was a considerable amount of instability in the first 18 months, suggesting that this early phase reflects a lead-in period in which the data are incomplete. For this reason, data from this period were excluded from rate calculations.
FIG. 2.
Frequency of appearance of new variant strains in the community as a proportion of observed transmissions over time elapsed since the study epoch. The data shown are for a maximum interstrain interval of 2 years and are plotted as a 5-month moving average.
The number of variant strains that appeared in the community and that could be linked by NGD to an identifiable source strain divided by the total number of observed transmissions from month 19 to the end of the study period is a measure of the rate of variant strain production. The estimates of these rates are given in Table 1. The rates at which variant strains were produced were 13.8 and 10.0% of transmissions for NGDs of 1 and 2, respectively, by use of a 2-year interstrain interval.
TABLE 1.
Estimates of the rate of variant strain production as a proportion of new cases due to transmission appearing per month
| NGD | Ratea | R2 |
|---|---|---|
| 1 | 0.1384 | 0.9997 |
| 2 | 0.0998 | 0.9983 |
| 3 | 0.0644 | 0.9930 |
| 4 | 0.0445 | 0.9988 |
The rate was calculated as the number of variant strains that could be linked by NGD to an identifiable source strain divided by the total number of observed transmissions from month 19 to the end of the study.
By using NGD as a measure of recent evolutionary events that were occurring during the process of transmission, it was possible to define genotypically related groups of strains (superclusters) representing ongoing transmission chains. From these calculations, if a 2-year maximum interstrain interval is assumed, 54 and 140 isolates previously classified as unique were included in superclusters for NGDs of 1 and 4, respectively. By using the formula (N − S)/T (14), the extent of recent transmission ranged from 66 to 89% (Table 2). However, these values may be underestimates due to the possible erroneous assignment of primary index status to cases in the early phase of the study. To circumvent this problem we propose the formula (NE + NL − SL)/T, in which it is assumed that the source cases can be defined only in superclusters initiated after the first 18 months of the study. Using this formula, we estimate the extent of recent transmission to be between 73 and 94%.
TABLE 2.
Degree of superclustering and standard and alternative calculations of recent transmission for various allowable ranges of NGD for 2- and 5-year maximum interstrain intervals
| NGD | Interstrain interval (yr) | No. of superclustered cases | No. of superclusters | Percent
|
||
|---|---|---|---|---|---|---|
| Superclustering (N/T) | Recent transmission
|
|||||
| (N − S)/T | (NE + NL − SL)/T | |||||
| 0 | 497 | 109 | 70.2 | 54.8 | 63.0 | |
| 0→1 | 2 | 552 | 85 | 78.0 | 66.0 | 72.9 |
| 5 | 836 | 92 | 84.9 | 75.5 | 81.4 | |
| 0→2 | 2 | 589 | 71 | 83.1 | 73.2 | 79.4 |
| 5 | 957 | 81 | 90.1 | 82.5 | 87.5 | |
| 0→3 | 2 | 616 | 59 | 87.0 | 78.7 | 84.5 |
| 5 | 1,096 | 77 | 93.9 | 87.3 | 92.4 | |
| 0→4 | 2 | 638 | 55 | 90.1 | 82.3 | 87.9 |
| 5 | 1,087 | 64 | 94.9 | 89.3 | 93.8 | |
DISCUSSION
The simplistic interpretation of DNA fingerprinting data disregards the fact that the genome is in a state of flux (23) and is evolving at a rate that will influence the interpretation of molecular epidemiological data (6, 11, 12, 21, 23).
In this study we have used an algorithmic method to explore the implications of IS6110-based RFLP pattern evolution on the understanding of an epidemic in a community with a high incidence of tuberculosis. Closely related M. tuberculosis strains were linked by NGD. In contrast to previous studies (13), this method of analysis shows that IS6110 evolution is independent of the number of IS6110-hybridizing bands present in the source strain. Consequently, GD is purely a measure of the number of band mismatches. However, a number of assumptions have been made in the interest of simplification in the calculation of GD. First, the loss and the gain of bands have been assumed to occur at the same rates, and therefore, a loss or a gain was assigned an equal GD. Since >60% of IS6110 fingerprint changes occur by replicative transposition, a more refined method might assign a higher GD to band loss. Second, a band shift has been counted as two events, i.e., a combination of a loss and a gain. The true frequency of this type of event is obscured by multiple events but is probably sufficiently rare to validate this assumption.
Analysis of the NGD data shows that closely related variant strains are appearing in the community at a constant annual rate. Thus, for a maximum NGD of 1 or 2 (by use of a 2-year interstrain interval), we found that 14 to 24% of transmission events produced variant strains. This value is similar to an estimate obtained in a previous study (21), in which we found that approximately 18.6% of transmission events within households generated a variant strain. The high proportion of newly evolved variant strains confirms that the M. tuberculosis strain population is diversifying at a constant rate in the study setting and that the process of its evolution is linked to transmission, significantly influencing molecular epidemiological calculations.
Consequently, studies depending on the stratification of cases according to genetic identity will underestimate the extent of transmission or incorrectly group cases for risk factor analysis. The factoring of NGD into clustering calculations suggests that transmission estimates may be 20% higher than those predicted by previously accepted formulae (14). However, the accuracy of this calculation is influenced by a number of factors. (i) The number of source cases initiating transmission chains affects the calculation. To minimize the overestimation of the number of primary index cases, we proposed an alternative formula in which it is assumed that clusters identified in the first 18 months of the study were initiated by source cases occurring prior to the onset of the study. (ii) The estimate assumes a 100% sample recovery. In this study only 70% of cases were included, and therefore, possible source or secondary cases may have been missed, leading to an underestimate of transmission (10). (iii) At present, there are few data on the extent of M. tuberculosis transmission in areas surrounding the study community. As this region experiences an extremely high incidence of disease, it is probable that patients may have been infected by sources outside of the community. (iv) While we have demonstrated the propensity of strains to change, this study also shows that most of the transmitted strains remain identical to their source strains and persist in the community for extended periods. By use of the present methodology, it is not possible to differentiate transmission from reactivation of such strains, possibly leading to an overestimate of transmission. Considering these limitations, we propose that the calculations presented here probably represent a conservative estimate of the true extent of disease due to transmission. Mathematical modeling predicts that ±95% of cases should correspond to transmission, given the high infection pressure in this community (18; P. B. Fourie, J. Lancaster, K. Weyer, and N. Beyers [Medical Research Council of South Africa] unpublished data; E. Vynnycky [London School of Hygiene & Tropical Medicine], personal communication, 2002).
Depending on the parameters chosen, this study estimates that the proportion of the local epidemic due to ongoing transmission is between 66 and 94%. This is considerably higher than the 55% estimated by using genetic identity as a measure of the rate of transmission. From these results, we conclude that the epidemic is predominantly driven by transmission and not, as indicated by conventional calculations, by reactivation of dormant infections. A positive implication of this conclusion is that interventions which target transmission have the potential to dramatically influence the epidemic. In a setting of passive case finding, largely based on positive smear microscopy results, it is hypothesized that the majority of transmission events occur prior to diagnosis and treatment. This is a component of the epidemic which is not targeted by the present World Health Organization directly observed therapy strategy. On the basis of these results, it is envisaged that interventions which interrupt transmission should coincide with a decrease in GD-based superclustering (8). We suggest that GD should prove to be a useful tool in the analysis of longitudinal molecular epidemiological data which will aid in determining the efficacies of M. tuberculosis control strategies with a focus on reducing transmission.
While the present study focused on a community with a high prevalence of tuberculosis, we believe that the approach presented here may also be relevant to communities in which the incidence of tuberculosis is lower. Given the sizeable evolutionary rates reported in studies conducted in such areas (6, 12, 23), a GD-based analysis of the data may well be expected to produce an estimate of recent transmission significantly different from that obtained by conventional calculations. The rapid diversification of M. tuberculosis isolates in the New York City outbreak of strain W provides further weight to this argument (4). We believe that the evidence presented above indicates the need for a similar study to be done in a community with a low incidence of tuberculosis. In addition, we suggest that this technique is not limited to the study of M. tuberculosis but may also prove to be useful in the analysis of epidemiological data from other disease epidemics.
Acknowledgments
This work was made possible by grants from the GlaxoSmithKline Action TB Initiative, the Sequella Global Tuberculosis Foundation, and the National Research Foundation (THRP).
E. Engelke, S. Carlini, and M. De Kock are thanked for technical assistance.
REFERENCES
- 1.Agrons, G. A., R. I. Markowitz, and S. S. Kramer. 1993. Pulmonary tuberculosis in children. Semin. Roentgenol. 28:158-172. [DOI] [PubMed] [Google Scholar]
- 2.Alland, D., G. E. Kalkut, A. R. Moss, R. A. McAdam, J. A. Hahn, W. Bosworth, E. Drucker, and B. R. Bloom. 1994. Transmission of tuberculosis in New York City. An analysis by DNA fingerprinting and conventional epidemiologic methods. N. Engl. J. Med. 330:1710-1716. [DOI] [PubMed] [Google Scholar]
- 3.Beyers, N., R. P. Gie, H. L. Zietsman, M. Kunneke, J. Hauman, M. Tatley, and P. R. Donald. 1996. The use of a geographical information system (GIS) to evaluate the distribution of tuberculosis in a high-incidence community. S. Afr. Med. J. 86:40-44. [PubMed] [Google Scholar]
- 4.Bifani, P. J., B. B. Plikaytis, V. Kapur, K. Stockbauer, X. Pan, M. L. Lutfey, S. L. Moghazeh, W. Eisner, T. M. Daniel, M. H. Kaplan, J. T. Crawford, J. M. Musser, and B. N. Kreisworth. 1996. Origin and interstate spread of a New York City multidrug-resistant Mycobacterium tuberculosis clone family. JAMA 275:452-457. [PubMed] [Google Scholar]
- 5.Cave, M. D., K. D. Eisenach, G. Templeton, M. Salfinger, G. Mazurek, J. H. Bates, and J. T. Crawford. 1994. Stability of DNA fingerprint pattern produced with IS6110 in strains of Mycobacterium tuberculosis. J. Clin. Microbiol. 32:262-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Boer, A. S., M. W. Borgdorff, P. E. de Haas, N. J. Nagelkerke, J. D. van Embden, and D. van Soolingen. 1999. Analysis of rate of change of IS6110 RFLP patterns of Mycobacterium tuberculosis based on serial patient isolates. J. Infect. Dis. 180:1238-1244. [DOI] [PubMed] [Google Scholar]
- 7.Glynn, J. R., J. Bauer, A. S. de Boer, M. W. Borgdorff, P. E. Fine, P. Godfrey-Faussett, E. Vynnycky, et al. 1999. Interpreting DNA fingerprint clusters of Mycobacterium tuberculosis. Int. J. Tuberc. Lung Dis. 3:1055-1060. [PubMed] [Google Scholar]
- 8.Jasmer, R. M., J. A. Hahn, P. M. Small, C. L. Daley, M. A. Behr, A. R. Moss, J. M. Creasman, G. F. Schecter, E. A. Paz, and P. C. Hopewell. 1999. A molecular epidemiologic analysis of tuberculosis trends in San Francisco, 1991-1997. Ann. Intern. Med. 130:971-978. [DOI] [PubMed] [Google Scholar]
- 9.Maguire, H., J. W. Dale, T. D. McHugh, P. D. Butcher, S. H. Gillespie, A. Costetsos, H. Al Ghusein, R. Holland, A. Dickens, L. Marston, P. Wilson, R. Pitman, D. Strachan, F. A. Drobniewski, and D. K. Banerjee. 2002. Molecular epidemiology of tuberculosis in London 1995-7 showing low rate of active transmission. Thorax 57:617-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murray, M. 2002. Sampling bias in the molecular epidemiology of tuberculosis. Emerg. Infect. Dis. 8:363-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niemann, S., E. Richter, and S. Rusch-Gerdes. 1999. Stability of Mycobacterium tuberculosis IS6110 restriction fragment length polymorphism patterns and spoligotypes determined by analyzing serial isolates from patients with drug-resistant tuberculosis. J. Clin. Microbiol. 37:409-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niemann, S., S. Rusch-Gerdes, E. Richter, H. Thielen, H. Heykes-Uden, and R. Diel. 2000. Stability of IS6110 restriction fragment length polymorphism patterns of Mycobacterium tuberculosis strains in actual chains of transmission. J. Clin. Microbiol. 38:2563-2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salamon, H., M. A. Behr, J. T. Rhee, and P. M. Small. 2000. Genetic distances for the study of infectious disease epidemiology. Am. J. Epidemiol. 151:324-334. [DOI] [PubMed] [Google Scholar]
- 14.Small, P. M., P. C. Hopewell, S. P. Singh, A. Paz, J. Parsonnet, D. C. Ruston, G. F. Schecter, C. L. Daley, and G. K. Schoolnik. 1994. The epidemiology of tuberculosis in San Francisco. A population-based study using conventional and molecular methods. N. Engl. J. Med. 330:1703-1709. [DOI] [PubMed] [Google Scholar]
- 15.Tanaka, M. M., and N. A. Rosenberg. 2001. Optimal estimation of transposition rates of insertion sequences for molecular epidemiology. Stat. Med. 20:2409-2420. [DOI] [PubMed] [Google Scholar]
- 16.van Embden, J. D., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, and T. M. Shinnick. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Rie, A., R. Warren, M. Richardson, T. C. Victor, R. P. Gie, D. A. Enarson, N. Beyers, and P. D. van Helden. 1999. Exogenous reinfection as a cause of recurrent tuberculosis after curative treatment. N. Engl. J. Med. 341:1174-1179. [DOI] [PubMed] [Google Scholar]
- 18.Vynnycky, E., M. W. Borgdorff, D. van Sooligen, and P. E. Fine. 2003. Annual Mycobacterium tuberculosis infection risk and interpretation of clustering statistics. Emerg. Infect. Dis. 9:176-183. [DOI] [PubMed] [Google Scholar]
- 19.Warren, R., J. Hauman, N. Beyers, M. Richardson, H. S. Schaaf, P. Donald, and P. van Helden. 1996. Unexpectedly high strain diversity of Mycobacterium tuberculosis in a high-incidence community. S. Afr. Med. J. 86:45-49. [PubMed] [Google Scholar]
- 20.Warren, R. M., M. Richardson, S. L. Sampson, G. D. van der Spuy, W. Bourn, J. H. Hauman, H. Heersma, W. Hide, N. Beyers, and P. D. van Helden. 2001. Molecular evolution of Mycobacterium tuberculosis: phylogenetic reconstruction of clonal expansion. Tuberculosis (Edinburgh) 81:291-302. [DOI] [PubMed] [Google Scholar]
- 21.Warren, R. M., G. D. van der Spuy, M. Richardson, N. Beyers, C. Booysen, M. A. Behr, and P. D. van Helden. 2002. Evolution of the IS6110-based restriction fragment length polymorphism pattern during the transmission of Mycobacterium tuberculosis. J. Clin. Microbiol. 40:1277-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warren, R. M., G. D. van der Spuy, M. Richardson, N. Beyers, M. W. Borgdorff, M. A. Behr, and P. D. van Helden. 2002. Calculation of the stability of the IS6110 banding pattern in patients with persistent Mycobacterium tuberculosis disease. J. Clin. Microbiol. 40:1705-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yeh, R. W., D. L. Ponce, C. B. Agasino, J. A. Hahn, C. L. Daley, P. C. Hopewell, and P. M. Small. 1998. Stability of Mycobacterium tuberculosis DNA genotypes. J. Infect. Dis. 177:1107-1111. [DOI] [PubMed] [Google Scholar]