Abstract
Human herpesvirus 8 (HHV-8) (or Kaposi's sarcoma [KS]-associated herpesvirus) is associated with all forms of KS. HHV-8 DNA load in peripheral blood mononuclear cells (PBMCs) of KS patients has been shown to correlate with the clinical stage of the disease. Studies have been done to assess the HHV-8 viral load in different sample types from KS patients and its clinical relevance. This paper describes the design and evaluation of a quantitative real-time (TaqMan) PCR assay for routine diagnosis of HHV-8 infection. The linear dynamic range was 5 to 5 × 106 copies of HHV-8 DNA (r2 > 0.99). The assay is very sensitive, specific, and easily reproducible (less than 2% variability) and can be used for different clinical samples, such as serum, plasma, and PBMCs. The question of which clinical sample, serum or plasma, is preferable for HHV8 DNA testing was addressed, using this newly developed real-time PCR assay. From 85 patients with diagnosed AIDS-KS, matched plasma and serum samples were collected. Of the 85 patients tested, 35 were positive for HHV-8 DNA in both plasma and serum (41%), 8 were positive in serum but not plasma, and 7 had detectable HHV-8 DNA only in plasma. The HHV-8 load was similar in both plasma and serum, and no significant difference was found. However, more inhibition was seen in the plasma samples with the use of a system quality control, seal herpesvirus type 1. Therefore, our results suggest that serum is the preferred material for HHV-8 load testing, since there is less possible hindrance in the amplification than with plasma.
Human herpesvirus 8 (HHV-8) (or Kaposi's sarcoma [KS]-associated herpesvirus) is associated with all forms of KS (24). Detection of herpesvirus DNA in leukocytes could possibly represent latent infection, while detectable DNA in serum or plasma is usually associated with disease. This is seen for the herpesviruses Epstein-Barr virus (EBV), cytomegalovirus (CMV), and HHV-8 (1, 7, 25). Accordingly, detection of HHV-8 in peripheral blood mononuclear cells (PBMCs) may reflect latent infection, while detection of HHV-8 in serum or plasma, non-cell-associated HHV-8, reflects active lytic replication. The finding that some PBMC samples of KS patients also contain HHV-8 RNAs that encode viral structural proteins provides evidence that viral gene expression and replication can occur in KS leukocytes (22, 23). The finding that plasma viremia (HHV-8) is an important event in KS pathogenesis (5) concurs with that. As a result, HHV-8 DNA load in plasma or serum may have predictive value for disease development and progression. However, the DNA load measured is not necessarily the same in the two types of sample. Boom et al. found that the CMV load in serum was often significantly lower (10-fold or more) than those observed in the corresponding plasma samples (3); however, others found similar loads in serum and plasma for CMV (21). Similar varicella-zoster virus DNA loads were found in serum and plasma samples of six patients by de Jong et al. (8). For EBV, a member of the gamma herpesviruses like HHV-8, similar loads were found in matched plasma and serum samples by Fan et al. (9).
The aim of this study was to design and establish a quantitative real-time (TaqMan) PCR assay for routine diagnosis of HHV-8 infection and to establish which clinical sample, serum or plasma, is preferable for HHV-8 DNA testing. From 85 patients with diagnosed AIDS-KS, plasma and serum samples were collected at the same visit and subsequently tested.
MATERIALS AND METHODS
Patients.
Specimens were collected from human immunodeficiency virus type 1-infected homosexual men with histologically confirmed KS when serum and plasma were available from the same date. In total, 170 matched samples were selected from 85 patients. All the samples were taken after diagnosis of KS. Aliquots (150 μl) of plasma or serum were isolated by guanidinium thiocyanate lysis, binding to silica particles, and washing and elution, with a final elution volume of 50 μl. Nucleic acids present in plasma and sera are equally isolated with this method (4).
TaqMan.
The amount of HHV-8 DNA present in the matched plasma and serum samples was determined by real-time quantitative PCR of open reading frame 65 (ORF 65). The primers were designed upstream from a known single-base-pair deletion or substitution site in ORF 65 that is found in multiple-myeloma patients but not in KS patients (17). Apart from this site, the region is well conserved in all sequenced HHV-8 subtypes circulating in different parts of the world. The principle of real-time PCR has been described elsewhere (12). In short, during each PCR cycle, a fluorogenic oligonucleotide probe is activated by 5′-to-3′ exonuclease activity of Taq polymerase after binding to a specific PCR product. The amount of fluorescence that occurs as a result is monitored (TaqMan PCR). The number of PCR cycles required to reach the threshold fluorescence (Ct) is determined for each sample, and the measured Ct value is compared to the values of standards with known DNA template concentrations to determine the starting template concentration in the sample. Because the Ct is determined during the exponential phase of PCR, the value of Ct has a linear relationship to the logarithm of the template DNA concentration.
The primers were designed with Primer Express software (PE Biosystems) and amplify a 68-bp region of the ORF 65 gene of HHV-8. The primer sequences were 5′-CCTCTGGTCCCCATTCATTG-3′ and 5′-CGTTTCCGTCGTGGATGAG-3′. The sequence of the fluorogenic probe was 5′-(6-carboxyfluorescein) CCGGCGTCAGACATTCTCACAACC (6-carboxytetramethylrhodamine)-3′. A BLAST search indicated that neither the primers nor the probe shares significant homology with other nucleotide sequences. PCR was done using Platinum Quantitative PCR Supermix UDG (Invitrogen/Life Technologies, Carlsbad, Calif.). Each PCR contained 10 μl of sample and 40 μl of PCR mixture consisting of Platinum Quantitative PCR Supermix UDG, 3.6 mM MgCl2, 0.9 μM forward and reverse primer, 0.2 μM TaqMan probe, and 1 μl of ROX reference dye (50× concentrated) (Invitrogen/Life Technologies). Following the activation of UDG (2 min, 50°C) and activation of Platinum Taq DNA polymerase (10 min, 95°C), 45 cycles (15 s, 95°C and 1 min, 60°C) were performed on an ABI 7700 sequence detection system (Perkin-Elmer Applied Biosystems). As a control for cross-contamination, samples consisting of distilled water were also subjected to the isolation method, and the extracts were tested for both ORF 65 and the quality control. The threshold cycle (Ct) for all these “no-template” samples was >45 cycles. The Ct for each sample of the standard curve was plotted against the input copy number. The value of Ct was determined by the first cycle number at which fluorescence was greater than the set threshold value. For accurate comparison of the samples, the threshold was the same for all the experiments. Linear regression was used to determine the copy number of the experimental samples. The HHV-8 copy number measured was converted to copies per milliliter of sample. The cutoff Ct value of <40 cycles was used to define a sample as positive.
Standardization.
An electron microscopy counted standard is not available for HHV-8, and therefore, a plasmid was used for standardization of the assay. A plasmid containing part of ORF 65 of HHV-8 (11) was used, and the concentration was determined by UV spectroscopy. Serial dilutions of this plasmid ranging from 5 to 5 × 106 copies were used to characterize the linearity, precision, specificity, and sensitivity of the TaqMan assay. Besides, serial dilution series with two primary effusion lymphoma cell lines, BCBL-1 and BCP-1, were included in order to study the linear relationship between the value of the threshold cycle (Ct) for the standards and the HHV-8 DNA copy number.
Quality control.
PhHV, a type 1 seal herpesvirus, was added to the clinical samples both as a control for the isolation method and as a control for the presence of PCR inhibitors (26). PhHV was chosen as a universal and nonhuman viral control because, like HHV-8, it is a member of the herpesvirus family, but it is highly unlikely that patients will be infected with PhHV, making it suitable as a quality control for the isolation and amplification. PhHV was added to the clinical sample before the isolation to the equivalent of a Ct value of approximately 25 in the real-time detection system used. The samples were subsequently tested for both HHV-8 and PhHV. All the samples were tested in duplicate.
RESULTS
Real-time assay.
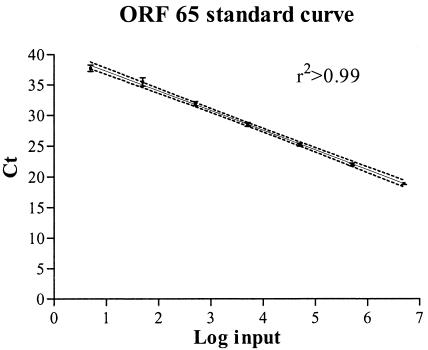
The Ct, the amplification cycle at which the fluorescence signal became detectable over the background, was linear over a range of at least 6 orders of magnitude of input DNA molecules, as shown by the dilution series of plasmid DNA in Fig. 1. Statistical analysis of 20 standard curves showed that the amplification is linear over a range of 5 to 5 × 106 molecules input with a detection limit of five molecules input (r2 > 0.99; P < 0.001). The detection rate was 100% when the copy number was ≥50 copies per well and 66% for 5 copies per well. The 95% confidence interval is very small, showing the robustness and precision of the assay. The ability of the assay to detect and quantify HHV-8 DNA in human cells, e.g., PBMCs, was first assessed with a dilution series of the cell lines BCBL-1 and BCP-1 (range, 1 to 1 × 104 cells). In the background of human DNA, a linear amplification of the integrated HHV-8 was seen (data not shown). The estimated HHV-8 copy number per cell was 36 for BCBL-1 and 39 for BCP-1, which is in agreement with the literature (20, 28). Dilution series of PBMCs infected with HHV-8 did not show any inhibition in the amplification when using up to 1.5 × 105 PBMCs (∼1 μg of DNA) as the maximum input (data not shown).
FIG. 1.
Standard curve for TaqMan PCR. Serial dilution of the HHV-8 plasmid ranging from 5 to 5 × 106 copies input. The error bars show the standard deviation of the Ct values. The solid line was obtained by linear regression analysis (r2 > 0.99; P < 0.001). The dotted line represents the 95% confidence interval for the regression.
A quantification method based on serial dilutions of a standard plasmid in water may not represent the DNA extracted from clinical samples and can lead to an overestimation of the sensitivity of the assay. To address this question, the plasmid was diluted in human DNA. The Ct values were not altered, and they fall within the standard deviation of the assay (data not shown).
Quality control.
A universal control was added to each clinical sample before the isolation step. This consisted of a seal herpesvirus (PhHV-1) that was added at a low concentration to each sample. This quality control was isolated and amplified in each sample, and this provided a measurement for the precision and reproducibility of the assay (mean Ct value, 25; coefficient of variation, 2.2% [data not shown]). If the Ct value for PhHV exceeded 26.5 (mean + 1.5 standard deviation), it was assumed that there was inhibition or that loss of sample had occurred. The results were considered invalid, and the sample was tested again. Of the 170 samples, 4 serum samples and 12 plasma samples had to be retested.
Patient samples.
Of the 85 patients tested, 35 (41%) were positive for HHV-8 DNA both in serum and in plasma. In 7 (8%) patients, HHV-8 DNA was found in plasma but not in serum, and 8 (9%) patients were positive for HHV-8 DNA in serum but not in plasma. Thirty-five patients had no detectable HHV-8 DNA in either plasma or serum. The results are shown in Table 1.
TABLE 1.
HHV-8 DNA load (log/ml) for matched plasma and serum samples of 85 AIDS-KS patients
| Sample type | No. (%) of patients positive for HHV-8 DNA | HHV-8 DNA load (log/ml)
|
|||
|---|---|---|---|---|---|
| Minimum | Median | Maximum | Mean | ||
| Either | |||||
| Serum | 43 (51) | 1.95 | 2.89 | 4.37 | 2.90 |
| Plasma | 42 (49) | 2.12 | 2.70 | 4.43 | 2.86 |
| Both | |||||
| Serum | 35 (41) | 1.95 | 2.98 | 4.37 | 3.00 |
| Plasma | 35 (41) | 2.18 | 2.75 | 4.43 | 2.95 |
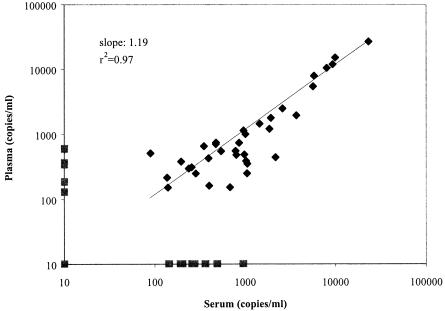
Quantitation of HHV-8 DNA in the clinical samples showed highly similar viral DNA loads. There was no significant difference in HHV-8 DNA load measured in the matched plasma and serum samples (Fig. 2). The difference in clinical sample was noticeable, however, when looking at the PhHV assay. There was more interference in the plasma samples than in the serum samples. Twelve plasma samples (14%) had to be repeated because of the high Ct value of the control, compared to four serum samples (5%). Inhibiting elements are more often coisolated with the plasma samples than with serum. The mean Ct values for the PhHV assay for the serum and plasma samples were 24.97 and 25.13, respectively. The difference was not significant (P = 0.618). The quality control also provided evidence that negative results were real negatives and not caused by inhibition.
FIG. 2.
HHV-8 DNA load in paired plasma and serum samples from 85 AIDS-KS patients. The slope and correlation coefficient were obtained using linear regression analysis.
DISCUSSION
The TaqMan assay developed for the quantification of HHV-8 DNA is sensitive, accurate, and robust, and it can be used with serum, plasma, and PBMCs. The prevalence of HHV-8 DNA in plasma or serum of persons with AIDS-KS in Europe or the United States ranges from 7 to 46% (2, 13, 18, 27). In our study, of the 85 patients tested, 35 were positive for HHV-8 DNA in both plasma and serum (41%). Looking at only plasma or serum, the number of positive patients rises to 43 (51%) for serum samples and 42 (49%) for the plasma samples. The HHV-8 load was similar in both plasma and serum, and no significant difference was found. However, there was more inhibition in the plasma samples. Plasma contains more PCR-inhibiting factors that are not always completely removed during isolation. Inhibition is detected by the addition of a quality control (PhHV). Although the isolation method used in this study is shown to remove inhibitory factors efficiently (19), our results show that there is still inhibition in some of the samples.
The fact that there is no difference found between the different clinical sample types is in agreement with the studies done on other herpesviruses (8, 9, 21). It is important to take into account that the amplicon measured with the assay is small; this is inherent in the design of the TaqMan system. As is stated by Boom et al. (3), there is a difference in the load in serum and in plasma if the amplicon used is large, probably due to fragmentation of the viral DNA. It can be hypothesized that the HHV-8 DNA load does not necessarily represent complete viral genomes but rather represents HHV-8 DNA fragments due to cell lysis or incomplete viral production.
In summary, this paper describes the development and validation of a real-time PCR assay for the quantification of cell-free and cell-associated HHV-8 DNA in laboratory and clinical samples. The sensitivity and linear range are similar to those of other real-time PCR assays described previously (6, 14, 28). With this assay, paired serum and plasma samples were tested to examine a possible variation in HHV-8 DNA load in the different clinical sample types, but none was found. However, as shown by the use of a quality control, there is more often inhibition in the plasma samples. It was speculated by Lo et al. (15, 16) that because clotting liberates nonspecific cellular DNA, plasma is the preferred sample compared to serum. However, Gallagher et al. (10) reported that serum was better as a source for EBV than plasma in a small Hodgkin's disease cohort. Our results also suggest that serum is the preferred clinical sample for HHV-8 DNA load measurements.
Acknowledgments
We thank Martin Schutten for the PhHV stock (Department of Virology, Erasmus MC, Rotterdam, The Netherlands) and Margreet Bakker for the sample selection.
REFERENCES
- 1.Barkholt, L. M., H. Dahl, M. Enbom, and A. Linde. 1996. Epstein-Barr virus DNA in serum after liver transplantation—surveillance of viral activity during treatment with different immunosuppressive agents. Transplant. Int. 9:439-445. [DOI] [PubMed] [Google Scholar]
- 2.Boivin, G., S. Cote, N. Cloutier, Y. Abed, M. Maguigad, and J. P. Routy. 2002. Quantification of human herpesvirus 8 by real-time PCR in blood fractions of AIDS patients with Kaposi's sarcoma and multicentric Castleman's disease. J. Med. Virol. 68:399-403. [DOI] [PubMed] [Google Scholar]
- 3.Boom, R., C. Sol, J. Weel, Y. Gerrits, M. de Boer, and P. Wertheim-van Dillen. 1999. A highly sensitive assay for detection and quantitation of human cytomegalovirus DNA in serum and plasma by PCR and electrochemiluminescence. J. Clin. Microbiol. 37:1489-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boom, R., C. J. Sol, M. M. Salimans, C. L. Jansen, P. M. Wertheim-van Dillen, and J. van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Broccolo, F., S. Bossolasco, A. M. Careddu, G. Tambussi, A. Lazzarin, and P. Cinque. 2002. Detection of DNA of lymphotropic herpesviruses in plasma of human immunodeficiency virus-infected patients: frequency and clinical significance. Clin. Diagn. Lab. Immunol. 9:1222-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broccolo, F., G. Locatelli, L. Sarmati, S. Piergiovanni, F. Veglia, M. Andreoni, S. Butto, B. Ensoli, P. Lusso, and M. S. Malnati. 2002. Calibrated real-time PCR assay for quantitation of human herpesvirus 8 DNA in biological fluids. J. Clin. Microbiol. 40:4652-4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brytting, M., W. Xu, B. Wahren, and V. A. Sundqvist. 1992. Cytomegalovirus DNA detection in sera from patients with active cytomegalovirus infections. J. Clin. Microbiol. 30:1937-1941.1323573 [Google Scholar]
- 8.de Jong, M. D., J. F. Weel, T. Schuurman, P. M. Wertheim-van Dillen, and R. Boom. 2000. Quantitation of varicella-zoster virus DNA in whole blood, plasma, and serum by PCR and electrochemiluminescence. J. Clin. Microbiol. 38:2568-2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan, H., S. A. Schichman, L. J. Swinnen, J. M. Nicholls, P. A. Eagan, M. Luther, and M. L. Gulley. 2001. Analytic validation of a competitive polymerase chain reaction assay for measuring Epstein-Barr viral load. Diagn. Mol. Pathol. 10:255-264. [DOI] [PubMed] [Google Scholar]
- 10.Gallagher, A., A. A. Armstrong, J. MacKenzie, L. Shield, G. Khan, A. Lake, S. Proctor, P. Taylor, G. B. Clements, and R. F. Jarrett. 1999. Detection of Epstein-Barr virus (EBV) genomes in the serum of patients with EBV-associated Hodgkin's disease. Int. J. Cancer. 84:442-448. [DOI] [PubMed] [Google Scholar]
- 11.Goudsmit, J., N. Renwick, N. H. T. M. Dukers, R. A. Coutinho, S. Heisterkamp, M. Bakker, T. F. Schulz, M. Cornelissen, and G. J. Weverling. 2000. Human herpesvirus 8 infections in the Amsterdam Cohort Studies (1984-1997): analysis of seroconversions to ORF65 and ORF73. Proc. Natl. Acad. Sci. USA 97:4838-4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 13.LaDuca, J. R., J. L. Love, L. Z. Abbott, S. Dube, A. E. Freidman-Kien, and B. J. Poiesz. 1998. Detection of human herpesvirus 8 DNA sequences in tissues and bodily fluids. J. Infect. Dis. 178:1610-1615. [DOI] [PubMed] [Google Scholar]
- 14.Lallemand, F., N. Desire, W. Rozenbaum, J. C. Nicolas, and V. Marechal. 2000. Quantitative analysis of human herpesvirus 8 viral load using a real-time PCR assay. J. Clin. Microbiol. 38:1404-1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lo, Y. M., L. Y. Chan, K. W. Lo, S. F. Leung, J. Zhang, A. T. Chan, J. C. Lee, N. M. Hjelm, P. J. Johnson, and D. P. Huang. 1999. Quantitative analysis of cell-free Epstein-Barr virus DNA in plasma of patients with nasopharyngeal carcinoma. Cancer Res. 59:1188-1191. [PubMed] [Google Scholar]
- 16.Lo, Y. M., M. S. Tein, T. K. Lau, C. J. Haines, T. N. Leung, P. M. Poon, J. S. Wainscoat, P. J. Johnson, A. M. Chang, and N. M. Hjelm. 1998. Quantitative analysis of fetal DNA in maternal plasma and serum: implications for noninvasive prenatal diagnosis. Am. J. Hum. Genet. 62:768-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma, H. J., N. N. Sjak-Shie, R. A. Vescio, M. Kaminsky, A. Mikail, M. Pold, K. Parker, M. Beksac, D. Belson, T. J. Moss, C. H. Wu, J. Zhou, L. Zhang, G. Chen, J. W. Said, and J. R. Berenson. 2000. Human herpesvirus 8 open reading frame 26 and open reading frame 65 sequences from multiple myeloma patients: a shared pattern not found in Kaposi's sarcoma or primary effusion lymphoma. Clin. Cancer Res. 6:4226-4233. [PubMed] [Google Scholar]
- 18.Marchioli, C. C., J. L. Love, L. Z. Abbott, Y. Q. Huang, S. C. Remick, N. Surtento-Reodica, R. E. Hutchison, D. Mildvan, A. E. Friedman-Kien, and B. J. Poiesz. 1996. Prevalence of human herpesvirus 8 DNA sequences in several patient populations. J. Clin. Microbiol. 34:2635-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niesters, H. G., J. van Esser, E. Fries, K. C. Wolthers, J. Cornelissen, and A. D. Osterhaus. 2000. Development of a real-time quantitative assay for detection of Epstein-Barr virus. J. Clin. Microbiol. 38:712-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Neill, E., J. L. Douglas, M. L. Chien, and J. V. Garcia. 1997. Open reading frame 26 of human herpesvirus 8 encodes a tetradecanoyl phorbol acetate- and butyrate-inducible 32-kilodalton protein expressed in a body cavity-based lymphoma cell line. J. Virol. 71:4791-4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patel, R., T. F. Smith, M. Espy, R. H. Wiesner, R. A. Krom, D. Portela, and C. V. Paya. 1994. Detection of cytomegalovirus DNA in sera of liver transplant recipients. J. Clin. Microbiol. 32:1431-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Polstra, A. M., J. Goudsmit, and M. Cornelissen. 2002. Development of real-time NASBA assays with molecular beacon detection to quantify mRNA coding for HHV-8 lytic and latent genes. BMC Infect. Dis. 2:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Polstra, A. M., J. Goudsmit, and M. Cornelissen. 2003. Latent and lytic HHV-8 mRNA expression in PBMCs and Kaposi's sarcoma skin biopsies of AIDS Kaposi's sarcoma patients. J. Med. Virol. 70:624-627. [DOI] [PubMed] [Google Scholar]
- 24.Schulz, T. F. 2001. KSHV/HHV8-associated lymphoproliferations in the AIDS setting. Eur. J. Cancer. 37:1217-1226. [DOI] [PubMed] [Google Scholar]
- 25.Secchiero, P., D. R. Carrigan, Y. Asano, L. Benedetti, R. W. Crowley, A. L. Komaroff, R. C. Gallo, and P. Lusso. 1995. Detection of human herpesvirus 6 in plasma of children with primary infection and immunosuppressed patients by polymerase chain reaction. J. Infect. Dis. 171:273-280. [DOI] [PubMed] [Google Scholar]
- 26.van Doornum, G. J., J. Guldemeester, A. D. Osterhaus, and H. G. Niesters. 2003. Diagnosing herpesvirus infections by real-time amplification and rapid culture. J. Clin. Microbiol. 41:576-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whitby, D., M. R. Howard, M. Tenant-Flowers, N. S. Brink, A. Copas, C. Boshoff, T. Hatzioannou, F. E. Suggett, D. M. Aldam, A. S. Denton, R. F. Miller, I. V. D. Weller, R. A. Weiss, R. S. Tedder, and T. F. Schulz. 1995. Detection of Kaposi sarcoma associated herpesvirus in peripheral blood of HIV-infected individuals and progression to Kaposi's sarcoma. Lancet 346:799-802. [DOI] [PubMed] [Google Scholar]
- 28.White, I. E., and T. B. Campbell. 2000. Quantitation of cell-free and cell-associated Kaposi's sarcoma-associated herpesvirus DNA by real-time PCR. J. Clin. Microbiol. 38:1992-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]