Abstract
Multidrug-resistant tuberculosis is an increasing health problem worldwide, especially in developing countries. The PCR-UHG-Rif assay, which detects mutations within the rpoB gene associated with rifampin resistance, was evaluated for its ability and reliability to detect and identify drug-resistant Mycobacterium tuberculosis in a developing country where tuberculosis is highly endemic.
Mycobacterium tuberculosis infection continues to be the major infectious cause of human morbidity and mortality in the world (28). The synergistic interaction between human immunodeficiency virus and tuberculosis (TB) infection has increased the prevalence of multidrug-resistant tuberculosis (MDR-TB) (1, 12, 19), which constitutes a significant obstacle in the control of TB (15).κ
Peru has the highest prevalence of drug-resistant TB in South America (4.5%) (17, 27). Active TB among young adults (ages 15 to 44) is as high as 800 per 100,000 in parts of the general population (8), with MDR-TB as high as 94% among patients with treatment failures (2).
Assays for rapid detection and drug susceptibility testing of M. tuberculosis are necessary for the effective control and prevention of TB and MDR-TB. Rapid detection of TB and MDR-TB could be accelerated by using DNA amplification techniques based on PCR and mutation detection assays (5, 9, 13, 22). Targeting rifampin resistance is a good strategy, since rifampin resistance is conferred by mutations within a short sequence in the rpoB gene of M. tuberculosis (20, 24). In addition, most of the rifampin-resistant strains are also resistant to isoniazid and, hence, are MDR-TB (6, 27).
The PCR-UHG-Rif assay, based on the amplification and detection of mutations in the rpoB gene, has been developed and evaluated in the United States for the rapid and sensitive detection of M. tuberculosis and its rifampin genotype directly from sputum specimens (23, 25). The sensitivity of this assay for the detection of M. tuberculosis and rifampin susceptibility in smear-negative sputum samples has not been established. The goals of the present study were (i) to confirm the ability of the PCR-UHG-Rif assay to determine rifampin resistance accurately by testing a sizable number of drug-resistant specimens, (ii) to determine its sensitivity for the detection of smear-negative TB, and (iii) to determine how well the assay performed in a laboratory in Peru, a developing country where TB and drug-resistant TB are highly endemic.
A total of 1,892 sputum samples were analyzed. Of these samples, 1,390 were obtained from 288 patients attending the Pulmonary Clinic at Maria Auxiliadora Hospital and 502 samples were from 106 patients attending the Infectious Diseases Clinic at Dos de Mayo Hospital in Lima, Peru. Two consecutive sputum samples were collected per patient before treatment was started (month 0) and at 1 month (month 1), 2 months (month 2), and 4 months (month 4) after starting treatment. Patients were treated for 2 months with isoniazid, rifampin, pyrazinamide, and ethambutol six times a week followed by 4 months of treatment with isoniazid and rifampin twice weekly, as recommended by the World Health Organization (26).
Sputum specimens were processed at the Infectious Diseases Laboratory, Universidad Peruana Cayetano Heredia, Lima, Perú. All specimens were coded and processed simultaneously by our routine technicians in a blinded fashion.
The specimens were homogenized, decontaminated, and concentrated by the N-acetyl-l-cysteine-NaOH method (14). From each decontaminated sputum, one smear was prepared, stained with Auramine O, and then graded (21); one slant of Lowenstein-Jensen (L/J) (Difco, Detroit, Mich.) was inoculated; the microscopic observation broth drug susceptibility assay (MODS) was carried out to detect the presence of M. tuberculosis (4, 18); a hemi-nested PCR assay, targeting insertion element IS6110, was performed to identify the presence of M. tuberculosis (4, 7, 10, 16); and the PCR-UHG-Rif assay was performed as previously described (23, 25).
M. tuberculosis isolates were tested for their susceptibility to rifampin and isoniazid by using a well-described colorimetric susceptibility test, the microplate Alamar Blue assay (MABA) (4, 11).
Data were analyzed using STATA package version 7.0 (Stata Corporation, College Station, Tex.).
A positive TB culture was considered the “gold standard” test. A total of 43.8% (828 of 1,892) of the samples were culture-positive by either broth (MODS) or L/J slope solid culture (Table 1). Of the culture-positive samples, 90.2% (747 of 828) were positive on L/J and 94.9% (786 of 828) were positive with MODS. The IS6110 PCR assay detected M. tuberculosis in 56.9% (1,076 of 1,892) of the samples, while the PCR-UHG-Rif assay detected the bacterium in 42.9% (812 of 1,892) of the samples. The sensitivity, specificity, and predictive values of the PCR-UHG-Rif assay compared to those of culture, auramine staining, and IS6110 PCR are shown in Tables 1, 2, and 3, respectively.
TABLE 1.
Comparison of the PCR-UHG-Rif assay to culture (gold standard) for detection of M. tuberculosis directly from sputum sediments
| PCR-UHG-Rif | Culturea
|
% Predictive value
|
||
|---|---|---|---|---|
| Positiveb | Negative | Positive | Negative | |
| Positivec | 695 | 117 | 85.6 | |
| Negative | 133 | 947 | 87.7 | |
| % Sensitivity-specificity | 83.9 | 89.0 | ||
NALC-OH-treated sputum sediments were cultured by using L/J and MODS.
Culture-positive samples were positive for growth in one or both culture systems.
Positive specimens were positive for the 193-bp M. tuberculosis-specific band on heteroduplex gel.
TABLE 2.
Comparison of the PCR-UHG-Rif assay to sputum smears (gold standard) for detection of M. tuberculosis directly from sputum sediments
| PCR-UHG-Rif | Smearsa
|
% Predictive value
|
||
|---|---|---|---|---|
| Positive | Negative | Positive | Negative | |
| Positiveb | 590 | 222 | 72.7 | |
| Negative | 1 | 1079 | 99.9 | |
| % Sensitivity-specificity | 99.8 | 82.9 | ||
Smears of sputum samples were prepared by using Auramine O staining (7).
The positive specimens were positive for the 193-bp M. tuberculosis-specific rpoB fragment band on heteroduplex gel.
TABLE 3.
Comparison of the PCR-UHG-Rif assay to the IS6110 PCR assay (gold standard) for the detection of M. tuberculosis directly from sputum sediments
| PCR-UHG-Rif |
IS6110 PCRa
|
% Predictive value
|
||
|---|---|---|---|---|
| Positive | Negative | Positive | Negative | |
| Positiveb | 754 | 58 | 92.9 | |
| Negative | 322 | 758 | 70.2 | |
| % Sensitivity-specificity | 70.1 | 92.9 | ||
The positive samples for the hemi-nested IS6110 PCR contained a 337-bp product on ethidium bromide-stained 2% agarose gel.
The positive specimens were positive for the 193-bp M. tuberculosis-specific rpoB fragment band on heteroduplex gel.
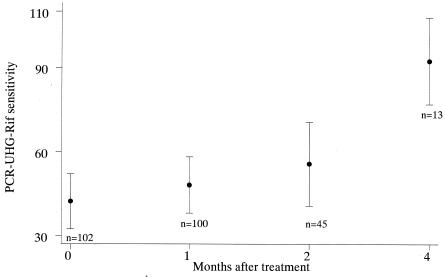
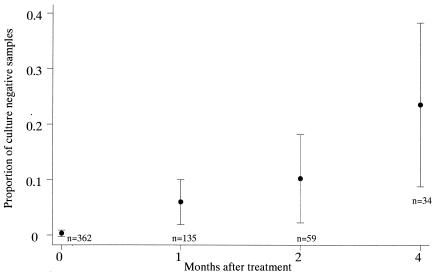
The PCR-UHG-Rif assay detected 99% of the smear-positive samples. However, it detected only 49% of the smear-negative, culture-positive samples. The sensitivity of the PCR-UHG-Rif (compared to that of culture as the gold standard) was significantly higher for smear-negative culture-positive samples collected 4 months after treatment than for samples collected before treatment (odds ratio [OR] = 16.47, P = 0.008) or 1 (OR = 13.0, P = 0.016) or 2 months after treatment (OR = 9.6, P = 0.037) (Fig. 1). In smear-negative cases, sensitivity of the PCR-UHG-Rif increased as the time after treatment increased, presumably because PCR-UHG-Rif detected nonviable M. tuberculosis killed by treatment (Fig. 2). Note that smear-negative cases are less likely to transmit TB (3).
FIG. 1.
Results sensitivity of the PCR-UHG-Rif assay compared to that for culture (by either L/J or MODS or both) among smear-negative sputum specimens. Each bar represents the 95% confidence interval for the sensitivity of the PCR-UHG-Rif test compared to that for culture.
FIG. 2.
Proportion of culture-negative samples among PCR-UHG-Rif-positive samples by time of collection of samples after treatment. Each bar represents the 95% confidence interval for the proportion of samples that were PCR-UHG-Rif-positive and culture-negative.
There was no significant difference in sensitivity or specificity of the PCR-UHG-Rif when the two hospital populations were examined separately (P = 0.79). No difference was observed when PCR-UHG-Rif sensitivity or specificity was analyzed according to patient HIV status (P = 0.16).
When the statistical analysis was performed in a cohort of 73 patients with all eight samples collected, no difference was observed in the performance of the PCR-UHG-Rif assay compared to that of the statistical analysis performed by using the complete set of data. In the cohort, in smear-negative samples the same trend of increased sensitivity of the PCR-UHG-Rif assay was maintained as the duration of treatment increased but was no longer statistically significant (data not shown).
When M. tuberculosis was detected, there was a high percentage of agreement in rifampin susceptibility between the MABA and the PCR-UHG-Rif assay (Table 4). Among samples positive by PCR-UHG-Rif, there was no difference in the proportion of sensitive and resistant results between culture-negative and culture-positive samples (data not shown).
TABLE 4.
Comparison of PCR-UHG-Rif and MABA for determination of rifampin susceptibility of M. tuberculosis in clinical specimensc
| PCR-UHG-Rif | MABAa
|
|
|---|---|---|
| Susceptible (%) | Resistant (%) | |
| Susceptibleb | 508 (97.1%) | 3 (1.8%) |
| Resistant | 15 (2.9%) | 162 (98.2%) |
Samples for which refampin MICs were >1.0 μg/ml as determined by MABA were considered resistant to rifampin (5, 19).
The samples showed a rifampin heteroduplex pattern with the PCR-UHG-Rif assay that was similar to that of M. tuberculosis H37Rv (ATTC 27294).
Agreement, 97.4%; κ, 0.9294; P, <0.0001.
The MICs were significantly higher (P < 0.001, Kruskal-Wallis test) for samples that were rifampin-susceptible by MABA and resistant by PCR-UHG-Rif (mean MIC = 0.317 μg, n = 15) than for samples that were susceptible to rifampin both by MABA and PCR-UHG-Rif (mean MIC = 0.080 μg, n = 508). Of the 165 samples that were rifampin-resistant with the MABA test, 162 samples were also resistant with PCR-UHG-Rif and, for 160 of these, rifampin MICs were >16 μg/ml. For two of the three samples resistant to rifampin by MABA but showing a sensitive genotype by PCR-UHG-Rif, rifampin MICs were >16 μg/ml; for the other sample, rifampin MICs were equal to 2 μg/ml.
To determine if the PCR-UHG-Rif assay was a good predictor of MDR-TB (resistance to at least isoniazid and rifampin) directly from sputum samples, the samples were also analyzed for isoniazid susceptibility using MABA and compared to rifampin susceptibility results obtained by MABA and PCR-UHG-Rif. There was a strong correlation between the results of these assays, as the agreement between susceptibility to rifampin and to isoniazid was 86% and 85%, respectively (Table 5).
TABLE 5.
Agreement between isoniazid susceptibility as determined by MABA and rifampin susceptibility as determined by MABA and PCR-UHG-Rif, respectively
| Isoniazid susceptibilityc | Rifampin susceptibility as determined by:
|
|||
|---|---|---|---|---|
| MABAa
|
PCR-UHG-Rifb
|
|||
| Susceptible | Resistant | Susceptible | Resistant | |
| Susceptible | 451 | 28 | 448 | 31 |
| Resistant | 72 | 137 | 63 | 146 |
Strains were considered resistant to rifampin when rifampin MICs were > 1.0 μg/ml as determined by MABA (5, 19). Agreement, 85.4%; κ, 0.6347; P, <0.0001.
Samples determined susceptible by PCR-UHG-Rif were those presenting the same heteroduplex pattern as M. tuberculosis H37Rv on acrylamide gels. Agreement, 86.3%; κ, 0.6624; P, <0.0003.
Detection of M. tuberculosis by culture and susceptibility testing in Peru is primarily based on L/J slants. More sophisticated techniques are not widely used in most developing countries because of the need for radioactive reagents, high costs, and the special equipment requirements.
This study demonstrated that the PCR-UHG-Rif assay was very effective for the simultaneous detection of smear-positive M. tuberculosis and susceptibility to rifampin directly from ethanol-fixed NALC-OH-treated sputum sediments, confirming previously published results (23). It also confirms that the assay is a good predictor of MDR-TB.
In addition, we demonstrated the utility of the PCR-UHG-Rif assay in a laboratory in a developing country where equipment, molecular expertise, and funding are limited. Excluding labor and equipment costs, the PCR-UHG-Rif assay can be performed for approximately $2.75 per specimen with standard PCR and electrophoresis equipment and polyacrylamide minigels. It should be emphasized that PCR equipment costs are similar to the cost of an enzyme-linked immunosorbent assay reader.
Because of its high sensitivity, specificity, and predictive values for smear-positive TB and MDR-TB, the incorporation of this assay as a routine diagnostic tool for testing M. tuberculosis susceptibility to rifampin should permit the detection of MDR-TB patients within 24 h of specimen acquisition. When used in a hospital setting, the detection of rifampin resistance by the PCR-UHG-Rif assay should facilitate early isolation and appropriate treatment of MDR-TB that may decrease nosocomial transmission of MDR-TB. Its use as a single test in smear-negative samples, however, is limited by its relatively low sensitivity for detecting tuberculosis in these cases.
Acknowledgments
We are grateful to Laynette Spring of the Laboratory Research Branch, Molecular Biology Research, National Hansen's Disease Program LSU-SVM, Baton Rouge, Louisiana; J. P. Castillo; J. B. Phu; and D. Sara for their technical assistance.
Other members of The Tuberculosis Working Group in Peru are Vivian Kawai, Giselle Soto, Patricia Sheen, Juliana Cordova, Patricia Fuentes, Pilar Navarro, Marco Ñavincopa, Richard Rodriguez, and Lilia Cabrera.
This study was funded in part by US-AID grant HRN-A-00-96-90006-00, Fogarty-NIH TB training grant D43TW 00010, ITREID grant 5 D43 TW00910, and Peruvian government grant CONCYTEC-TBC/Heteroduplex10. C.E. was funded by the Wellcome Trust.
REFERENCES
- 1.Barnes, P., A. B. Bloch, P. T. Davidson, and D. E. Snider, Jr. 1991. Tuberculosis in patients with immunodeficiency virus infection. N. Engl. J. Med. 324:1644-1650. [DOI] [PubMed] [Google Scholar]
- 2.Becerra, M. C., J. Freeman, J. Bayona, S. S. Shin, J. Y. Kim, J. J. Furin, B. Werner, A. Sloutsky, R. Timperi, M. E. Wilson, M. Pagano, and P. E. Farmer. 2000. Using treatment failure under effective directly observed short-course chemotherapy programs to identify patients with multidrug-resistant tuberculosis. Int. J. Tuber. Lung Dis. 4:108-114. [PubMed] [Google Scholar]
- 3.Behr, M. A., S. A. Warren, H. Salamon, P. C. Hopewell, A. Ponce de Leon, C. L. Daley, and P. M. Small. 1999. Transmission of Mycobacterium tuberculosis from patients smear-negative for acid-fast bacilli. Lancet 353:444-449. [DOI] [PubMed] [Google Scholar]
- 4.Caviedes, L., T. S. Lee, R. H. Gilman, P. Sheen, E. Spellman, E. H. Lee, D. E. Berg, and S. Montenegro-James. 2000. Rapid, efficient detection and drug susceptibility testing of Mycobacterium tuberculosis in sputum by microscopic observation of broth cultures. The Tuberculosis Working Group in Peru. J. Clin. Microbiol. 38:1203-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caws, M., and F. A. Drobniewski. 2001. Molecular techniques in the diagnosis of Mycobacterium tuberculosis and the detection of drug resistance. Ann. N. Y. Acad. Sci. 953:138-145. [DOI] [PubMed] [Google Scholar]
- 6.Cole, S. T., and A. Telenti. 1995. Drug resistance in Mycobacterium tuberculosis. Eur. Res. J. 8:701S-13S. [PubMed] [Google Scholar]
- 7.Dalovisio, J. R., S. Montenegro-James, S. A. Kemmerly, C. F. Genre, R. Chambers, D. Greer, G. A. Pankey, D. M. Failla, K. G. Haydel, L. Hutchinson, M. F. Lidney, B. M. Nunez, A. Praba, K. D. Eisenach, and E. S. Cooper. 1996. Comparison of the amplified Mycobacterium tuberculosis (MTB) direct test, Amplicor MTB PCR, and IS6110-PCR for detection of MTB in respiratory specimens. Clin. Infect. Dis. 23:1099-1106. [DOI] [PubMed] [Google Scholar]
- 8.Farmer, P. 2001. DOTS and DOTS-plus: not the only answer. Ann. N. Y. Acad. Sci. 953:165-184. [DOI] [PubMed] [Google Scholar]
- 9.Fluit, A. C., M. R. Visser, and F. J. Schmitz. 2001. Molecular detection of antimicrobial resistance. Clin. Microbiol. Rev. 14:836-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franchi, L. M., R. I. Cama, R. H. Gilman, S. Montenegro-James, and P. Sheen. 1998. Detection of Mycobacterium tuberculosis in nasopharyngeal aspirate samples in children. Lancet 352:1681-1682. [DOI] [PubMed] [Google Scholar]
- 11.Franzblau, S. G., R. S. Witzig, J. C. McLaughlin, P. Torres, G. Madico, A. Hernandez, M. T. Degnan, M. B. Cook, V. K. Quenzer, R. M. Ferguson, and R. H. Gilman. 1998. Rapid, low-technology MIC determination with clinical Mycobacterium tuberculosis isolates by using the microplate Alamar Blue assay. J. Clin. Microbiol. 36:362-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frieden, T. R., T. Sterling, A. Pablos-Mendez, J. O. Kilburn, G. M. Cauthen, and S. W. Dooley. 1993. The emergence of drug-resistant tuberculosis in New York City. N. Engl. J. Med. 328:521-526. [DOI] [PubMed] [Google Scholar]
- 13.Goyal, M., R. J. Shaw, D. K. Banerjee, R. J. Coker, B. D. Robertson, and D. B. Young. 1997. Rapid detection of multidrug-resistant tuberculosis. Eur. Respir. J. 10:1120-1124. [DOI] [PubMed] [Google Scholar]
- 14.Kent, P. T., and G. P. Kubica. 1985. Public health mycobacteriology: a guide for the level III laboratory. U.S. Department of Health and Human Services. Center for Disease Control, Atlanta, Ga.
- 15.Kochi, A., B. Vareldzis, and K. Styblo. 1993. Multidrug-resistant tuberculosis and its control. Res. Microbiol. 144:104-110. [DOI] [PubMed] [Google Scholar]
- 16.Montenegro, S. H., R. H. Gilman, P. Sheen, R. Cama, L. Caviedes, T. Hopper, R. Chambers, and R. A. Oberhelman. 2003. Improved detection of Mycobacterium tuberculosis in Peruvian children by use of a heminested IS6110 polymerase chain reaction assay. Clin. Infect. Dis. 36:16-23. [DOI] [PubMed] [Google Scholar]
- 17.Pablos-Mendez, A., M. C. Raviglione, A. Laszlo, N. Binkin, H. L. Rieder, F. Bustreo, D. L. Cohn, C. S. Lambregts-van Weezenbeek, S. J. Kim, P. Chaulet, and P. Nun. 1998. Global surveillance for antituberculosis-drug resistance, 1994-1997. World Health Organization-International Union against Tuberculosis and Lung Disease Working Group on Anti-Tuberculosis Drug Resistance Surveillance. N. Engl. J. Med. 338:1641-1649. [DOI] [PubMed] [Google Scholar]
- 18.Park, W. G., W. R. Bishai, R. E. Chaisson, and S. E. Dorman. 2002. Performance of the microscopic observation drug susceptibility assay in drug susceptibility testing for Mycobacterium tuberculosis. J. Clin. Microbiol. 40:4750-4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pozniak, A. 2001. Multidrug-resistant tuberculosis and HIV infection. Ann. N. Y. Acad. Sci. 953:192-198. [DOI] [PubMed] [Google Scholar]
- 20.Ramaswamy, S., and J. M. Musser. 1998. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber. Lung Dis. 79:3-29. [DOI] [PubMed] [Google Scholar]
- 21.Singh, N. P., and S. C. Parija. 1998. The value of fluorescence microscopy of auramine stained sputum smears for the diagnosis of pulmonary tuberculosis. Southeast Asian J. Trop. Med. Public Health 29:860-863. [PubMed] [Google Scholar]
- 22.Soini, H., and J. M. Musser. 2001. Molecular diagnosis of mycobacteria. Clin. Chem. 47:809-814. [PubMed] [Google Scholar]
- 23.Williams, D. L., L. Spring, T. P. Gillis, M. Salfinger, and D. H. Persing. 1998. Evaluation of a polymerase chain reaction-based universal heteroduplex generator assay for direct detection of rifampin susceptibility of Mycobacterium tuberculosis from sputum specimens. Clin. Infect. Dis. 26:446-450. [DOI] [PubMed] [Google Scholar]
- 24.Williams, D. L., C. Waguespack, K. Eisenach, J. T. Crawford, E. Portaels, M. Salfinger, C. M. Nolan, C. Abe, V. Sticht-Groh, and T. P. Gillis. 1994. Characterization of rifampin resistance in pathogenic mycobacteria. Antimicrob. Agents Chemother. 38:2380-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams, D. L., C. Limbers, L. Spring, S. Jayachandra, and T. Gillis. 1996. PCR-heteroduplex detection of rifampin-resistant Mycobacterium tuberculosis, p. 122-129. In D. Persing (ed.), PCR protocols for emerging infectious diseases. ASM Press, Washington, D.C.
- 26.World Health Organization. 1994. WHO Tuberculosis Program: framework for effective tuberculosis control. World Health Organization, Geneva, Switzerland.
- 27.World Health Organization. 1998. Anti-tuberculosis drug resistance in the world. WHO/IUATLD Global Project on Anti-Tuberculosis Drug Resistance Surveillance 1994-1997. World Health Organization, Geneva, Switzerland.
- 28.World Health Organization. 2001. Global tuberculosis control. World Health Organization, Geneva, Switzerland.