Abstract
An essential step in the life cycle of human immunodeficiency virus type 1 (HIV-1) is integration of the double-stranded retroviral DNA into the genome of the host cell. HIV-1 integrase, the enzyme that inserts the vital DNA into the host chromosome, is an attractive and rational target for anti-AIDS drug design because it is essential for HIV replication and there are no known counterparts in the host cell. Inhibitors of this enzyme have a great potential to complement the therapeutic use of HIV protease and reverse transcriptase inhibitors. Natural products have provided a source of new drug candidates for anti-AIDS therapy. Dicaffeoylquinic acids, isolated from traditional medicinal plants, are a novel class of integrase inhibitors. These compounds are potent inhibitors of HIV-1 replication in cultured cell lines and catalytic activities of integrase in vitro. They are therefore promising compounds for developing new anti-AIDS drugs.
To understand how the inhibitors work and therefore design more potent and specific inhibitors, we have used molecular modeling techniques to investigate the binding modes of 3,4-dicaffeoylquinic acid. Our computational modeling study demonstrated that the inhibitor of this compound on HIV integrase is likely to proceed by two different but equivalent mechanisms with one bound to the active site region of the enzyme and another docked into the binding pocket located on the other side of the catalytic site. Our study will be of help to design new pharmaceuticals for the treatment of AIDS.
Keywords: HIV-1 Integrase, Anti-AIDS Drug, Molecular Modeling, Dicaffeoylquinic Acids
Introduction
Acquired immunodeficiency syndrome (AIDS) has been a serious, life-threatening health problem since it was first identified in 1981. Despite considerable investment and recent progress for treating this disease, anti-AIDS therapy still fails to be fully suppressive.1 It is the fourth greatest killer and the most quickly spreading disease in recent history.2,3 According to a report of the World Health Organization (WHO) and the Joint United Nations Programme on HIV and AIDS (UNAIDS), in 2007, an estimated 33.2 million people globally lived with the disease, and it killed an estimated 2.1 million people, including 330,000 children.4,5
Acquired immunodeficiency syndrome is a set of symptoms and infections resulting from the damage to the human immune system caused by the human immunodeficiency virus (HIV).6 Two major types of HIV have been identified, HIV-1 and HIV-2. HIV-1 is the cause of the worldwide epidemic. It encodes three enzymes: reverse transcriptase, protease, and integrase. Combination antiviral therapy with reverse transcriptase and protease inhibitors has shown the potential therapeutic efficacy of antiviral therapy for treatment of AIDS. However, the ability of HIV to rapidly evolve drug resistance, together with toxicity problems, requires the discovery and development of new classes of anti-AIDS drugs.7-9
The HIV-1 integrase, responsible for the integration of the newly synthesized double-stranded viral DNA into the host genomic DNA, is a new and important target of potential clinical relevance.10 For instance, two integrase inhibitors, raltegravir and elvitegravir, have been shown to be promising in clinical trials, and the first has been recently made available for clinical practice.11 HIV-1 integrase can be divided into three discrete domains, N-terminus, core, and C-terminus. The structure of a complex of its core domain with a novel inhibitor has been determined,12 which provide us a platform for the investigation and discovery of anti-AIDS agents.
Dicaffeoylquinic acids (DCQAs), isolated from traditional medicinal plants,13,14 are a novel class of integrase inhibitors. They are potent inhibitors of HIV-1 replication in cultured cell lines and catalytic activities of integrase in vitro. They represent promising lead compounds for developing new anti-AIDS drugs and offer a significant advance in the search for new HIV enzyme targets as they are both specific for HIV-1 integrase and relatively nontoxic.15
In our study, we used molecular modeling techniques to examine the binding mode of 3,4-DCQA in order to understand how and where the inhibitor binds to HIV-1 integrase.
Methodology
Protein and Ligand Structures
The coordinates of the core domain of HIV-1 integrase were taken from the Protein Data Bank (PDB, http://www.rcsb.org) with code 1QS4. The PDB file was modified to include only chain A of the core domain dimer, which was then converted from PDB format to the mol2 format using WinDock’s PMOL2Q module.16 Hydrogen atoms and chargers were added to the entire protein. The structure of 3,4-DCQA was first modeled using the ISIS Draw program (MDL ISIS Draw 2.5, MDL Information System Inc, 2002), then converted into a three-dimensional structure by ViewerLite software (ViewerLite 4.2, Accelrys Inc, 2001). Hydrogens were added and Gaisteiger charges were calculated. Energy minimization was performed with the molecular mechanics (MM) force field using the ArgusLab program (ArgusLab 4.0.1, www.arguslab.com). The integrase inhibitor included in the original PDB file, 5CITEP, was used as the comparison structure (Fig. 1).
Fig 1.
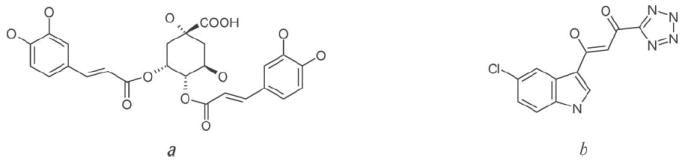
Chemical structures of 3,4-dicaffeoylquinic acid (a) and the inhibitor 5CITEP (b)
WinDock Docking Study
The docking study was performed with WinDock, a program developed in our laboratory,16 which uses the widely distributed DOCK searching engine17 to generate a set of spheres as the negative image of the protein binding site, and use an incremental construction and random conformation search method to dock flexible small molecules to macomolecular sites, and a Coulombic and Lennard-Jones grid-based scoring function to evaluate the binding affinity of ligands. WinDock’s SPHBOC module was used to determine the binding site and produce a set of spheres for binding site characterization. Contact scores and energy scores were calculated using an energy cutoff distance of 6.0 Å and a van der Waals repulsive exponent of 8.0 Å. Ligands were oriented to the spheres with a distance tolerance of 0.5 Å and minimum distance of 2.0 Å. A minimum anchor size of 50 was used with an internal energy repulsive exponent of 8.0 Å and clash overlap of 0.25 Å. All other parameters were left at their defaults.
Molegro Virtual Docker Docking Study
In order to investigate if there are other particular regions of the protein that are preferred by the ligand (blind docking), a docking calculation was performed using the Molegro Virtual Docker (MVD) program.18 The docking module of MVD is based on an evolution algorithm variant called differential evolution. Both structures of protein and ligand were uploaded into MVD. Bond orders, hybridizations, and hydrogen atoms were added, chargers were assigned, and flexible torsions of ligand were detected. A grid volume that was big enough to cover the entire surface of the protein was used for the docking calculation, while other parameters were at default.
Results
Prediction of 3,4-DCQA Binding to the Core Domain of HIV Integrase
The WinDock docking study revealed that the 3,4-DCQA was bound in the middle of the active site of subunit A of the enzyme. A schematic view of 3,4-DCQA bound at the active site of the enzyme is shown in Fig. 2a. This is consistent with the previous findings that the inhibition by DCQAs on integrase is directed toward conserved amino acids in the central core domain of integrase during catalysis.15
Fig 2.
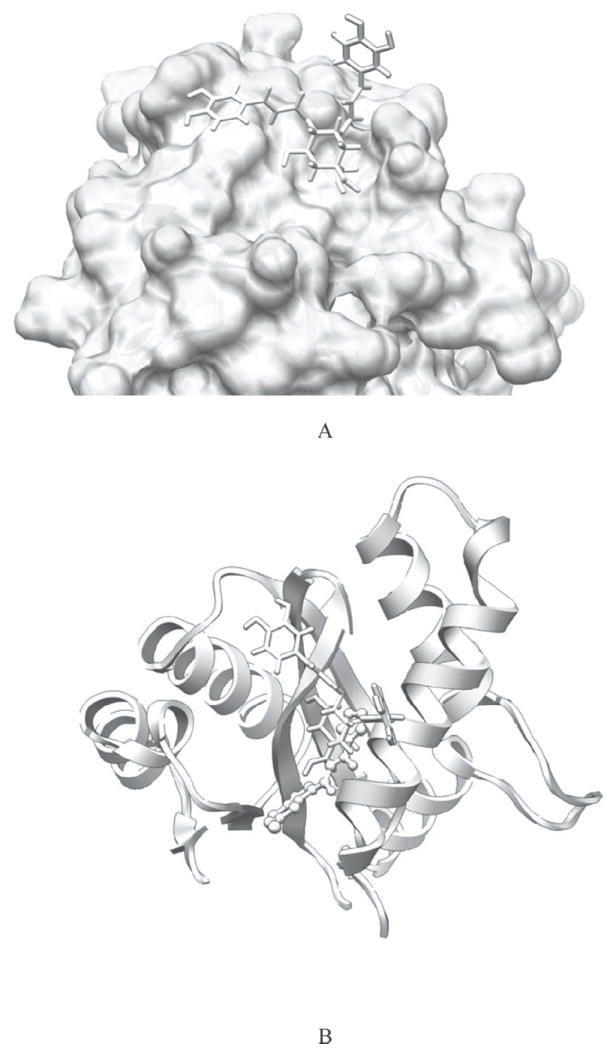
a) 3,4-DCQA is located in the active site of the protein b) 3,4-DCQA (stick representation) and the inhibitor 5CITEP (ball & stick representation) in the active site of the protein
Comparison of 3,4-DCQA Binding with the Inhibitor in the Complex
The comparison of 3,4-DCQA binding with the inhibitor 5CITEP is shown in Fig. 2b. They were all located in the active site between the three catalytic acidic residues, Asp64, Asp116, and Glu152. This suggests that 3,4-DCQA has the similar binding modes as the inhibitor 5CITEP. Both of these compounds interacted with the central core domain of integrase.
Comparison of Interface Residues between 3,4-DCQA and 5CITEP
The contacting residues (interface residues) for both 3,4-DCQA and the inhibitor 5CITEP are presented in Table 1. The 3,4-DCQA binding looks quite similar to the known inhibitor binding. There were 11 conserved amino acids in the binding site: Asp64, Cys65, Thr66, His67, Gly70, Lys71, Ile151, Glu152, Asn155, Lys156, and Lys159. Several residues that are known to be important for catalysis or DNA binding are involved in binding both ligands. They are all hydrogen-bonded to Asn155, Lys159, and Lys156. The difference is that 3,4-DCQA does not form a hydrogen bond to Gln148 as the inhibitor 5CITEP, but is close to the His67, Gly70, and Lys71. Since experiments have shown that several residues near the active site, including Thr143, Gln148, Lys156, and Lys159, are critical for binding viral DNA,19,20 and that Lys156 and Lys159 are also involved in 3,4-DCQA binding, it is tempting to speculate that the interactions between 3,4-DCQA and integrase at least partially mimic the DNA substrate/integrase interaction.
Table 1.
Comparison of interface residues between 3,4-DCQA and the inhibitor 5CITEP
| 3,4-DCQA
|
5CITEP
|
||||||
|---|---|---|---|---|---|---|---|
| Protein atom | Ligand atom | Protein atom | Ligand atom | ||||
| ASP64 | OD2 | C25 | 3.99 | ASP64 | OD2 | C12 | 3.35 |
| CYS65 | O | O10 | 3.36 | CYS65 | O | N4 | 4.73 |
| THR66 | CA | O10 | 3.03 | THR66 | OG1 | N2 | 2.75 |
| HIS67 | CD2 | C12 | 2.92 | ASP116 | CB | C6 | 4.23 |
| GLY70 | CA | O7 | 3.63 | GLN148 | NE2 | N9 | 3.63 |
| LYS71 | N | O7 | 4.55 | ||||
| ILE151 | O | O12 | 4.55 | ILE151 | CG2 | C12 | 3.61 |
| GLU152 | CA | O12 | 3.25 | GLU152 | OE2 | O2 | 2.58 |
| ASN155 | ND2 | O9 | 3.04 | ASN155 | ND2 | N4 | 3.49 |
| LYS156 | NZ | O1 | 3.03 | LYS156 | NZ | O1 | 3.27 |
| LYS159 | NZ | O6 | 2.88 | LYS159 | NZ | N1 | 2.82 |
Another Possible Binding Mode Predicted by Blind Docking
The blind docking study using MVD predicted another possible binding mode of 3,4-DCQA in which the compound docked into another binding pocket of HIV-1 integrase. The location of 3,4-DCQA was in close proximity to the active site of integrase and was located on the other side of the flexible loop from the catalytic residues, but was quite distant from the site where the inhibitor 5CITEP binds (Fig. 3). It seems that rather than preventing DNA binding to integrase directly, 3,4-DCQA might interact with the flexible loop, alters loop conformation, and affects the conformations of active site residues. Therefore, DCQAs inhibition of HIV-1 integrase is likely to proceed by an equivalent mechanism similar to another HIV integrase inhibitor, Y3.21
Fig 3.
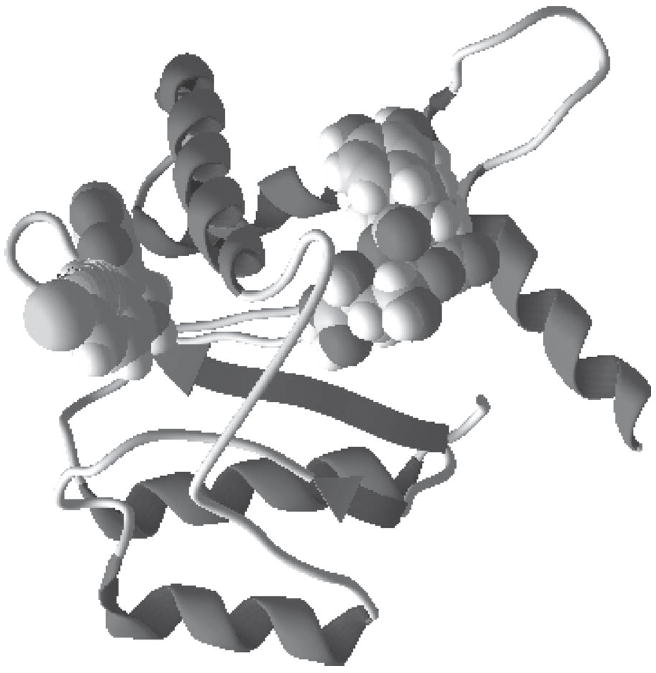
3,4-DCQA (space filling representation, right) docked into another binding pocket of HIV-1 integrase, compared with the inhibitor at the left (space filling representation, left)
Conclusions
Development of inhibitors that are specifically directed against additional targets, such as integrase, is a useful strategy for expanding the current combination therapy involving reverse transcriptase and protease inhibitors. Our goal in working with this class of inhibitors isolated from traditional medicinal plants was to provide leads to potential anti-AIDS drugs targeting HIV-1 integrase and develop a better understanding of the role of crucial target residues in the integrase binding site.
In the present study, molecular modeling techniques were applied in prediction of the binding mode of 3,4-DCQA with HIV-1 integrase.
Our WinDock docking study showed the 3,4-DCQA bound in the middle of the active site of the integrase and made similar close contacts with the protein, as the inhibitor, 5CITEP in the complex. There were 11 conserved amino acids in the binding site: Asp64, Cys65, Thr66, His67, Gly70, Lys71, Ile151, Glu152, Asn155, Lys156, and Lys159. This is consistent with the previous finding that the inhibition by DCQAs on integrase is directed toward conserved amino acids in the central core domain of integrase during catalysis. Analysis of the crystal structure of HIV-1 integrase revealed a cluster of lysine residues near the active site. Site-directed mutagenesis and photo-crosslinking studies have found that Lys156 and Lys159 are critical for the functional interaction of integrase with viral DNA. These two residues are also involved in 3,4-DCQA binding as the conserved amino acids. The interactions between 3,4-DCQA and integrase might at least partially mimic the DNA substrate/integrase interaction.
An alternative binding mode (from our blind docking study) indicated that the compound docked into another binding pocket of HIV-1 integrase, which is in close proximity to the active site of integrase, but is located on the other side of the flexible loop from the catalytic residues, quite distant from the site to which the inhibitor 5CITEP binds. It is possible for the compound to interact with the flexible loop, alter loop conformation, and therefore affect the conformations of active site residues.
Many compounds identified as integrase inhibitors, structurally belonging to different chemical classes, have been reported, such as aurintricarboxylic acid, cosalene analogues, DNA-binding agents, topoisomerase inhibitors, suramin, and bis-catechols.22 It has been shown that the action of DCQAs are different from these compounds. A majority of the compounds reported thus far are not selective for HIV-1 integrase while DCQAs are potent and selective inhibitors of the integrase.14,22 Therefore, DCQAs may prove important in understanding the function of HIV-1 integrase and thus leads to the discovery of more selective and potent inhibitors in the future.
AIDS is now a pandemic. In sub-Saharan Africa, it is now the leading cause of death.23-25 There is an urgent need to develop novel classes of anti-AIDS drugs. Molecular modeling of 3,4-DCQA with the core catalytic domain of HIV-1 integrase would provide useful information about how the compound works at the molecular level and may lead to the development of more potent and specific pharmaceuticals for the treatment of AIDS.
Acknowledgments
We wish to thank Guy M. Lingani for laboratory assistance. We are also thankful to Dr. Rene Thomsen and to Molegro ApS, Denmark, for giving us the opportunity to use the trial version of MVD. This work is supported by grant 2 G12 RR003048 from the RCMI Program, Division of Research Infrastructure, National Center for Research Resources, NIH.
References
- 1.Vitoria M, Granich R, Gilks CF, et al. The global fight against HIV/AIDS, tuberculosis, and malaria: current status and future perspectives. Am J Clin Pathol. 2009;131(6):844–848. doi: 10.1309/AJCP5XHDB1PNAEYT. [DOI] [PubMed] [Google Scholar]
- 2.Kallings LO. The first postmodern pandemic: 25 years of HIV/AIDS. J Intern Med. 2008;263(3):218–243. doi: 10.1111/j.1365-2796.2007.01910.x. [DOI] [PubMed] [Google Scholar]
- 3.Hazra R, Siberry GK, Mofenson LM. Growing up with HIV: children, adolescents, and young adults with perinatally acquired HIV infection. Annu Rev Med. 2009 Jul 21; doi: 10.1146/annurev.med.050108.151127. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 4.UNAIDS, WHO. AIDS epidemic update. 2007. [February 12, 2009];2007 December; http://data.unaids.org/pub/EPISlides/2007/2007_epiupdate_en.pdf.
- 5.Kilmarx PH. Global epidemiology of HIV. Curr Opin HIV AIDS. 2009;4(4):240–246. doi: 10.1097/COH.0b013e32832c06db. [DOI] [PubMed] [Google Scholar]
- 6.Weiss RA. How does HIV cause AIDS? Science. 1993;260(5112):1273–1279. doi: 10.1126/science.8493571. [DOI] [PubMed] [Google Scholar]
- 7.Matsushita S. Current status and future issues in the treatment of HIV-1 infection. Int J Hematol. 2000;72(1):20–27. [PubMed] [Google Scholar]
- 8.Prajapati DG, Ramajayam R, Yadav MR, Giridhar R. The search for potent, small molecule NNRTIs: A review. Bioorg Med Chem. 2009;17(16):5744–5762. doi: 10.1016/j.bmc.2009.06.060. [DOI] [PubMed] [Google Scholar]
- 9.Naider F, Anglister J. Peptides in the treatment of AIDS. Curr Opin Struct Biol. 2009;19(4):473–482. doi: 10.1016/j.sbi.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Semenova EA, Marchand C, Pommier Y. HIV-1 integrase inhibitors: update and perspectives. Adv Pharmacol. 2008;56:199–228. doi: 10.1016/S1054-3589(07)56007-2. [DOI] [PubMed] [Google Scholar]
- 11.Ceccherini-Silberstein F, Malet I, D’Arrigo R, Antinori A, Marcelin AG, Perno CF. Characterization and structural analysis of HIV-1 integrase conservation. AIDS Rev. 2009;11(1):17–29. [PubMed] [Google Scholar]
- 12.Goldgur Y, Craigie R, Cohen GH, et al. Structure of the HIV-1 integrase catalytic domain complexed with an inhibitor: a platform for antiviral drug design. Proc Natl Acad Sci USA. 1999;96(23):13040–13043. doi: 10.1073/pnas.96.23.13040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh IP, Bharate SB, Bhutani KK. Anti-HIV natural products. Current Science. 2005;89(2):269–290. [Google Scholar]
- 14.Robinson WE, Jr, Reinecke MG, Abdel-Malek S, Jia Q, Chow SA. Inhibitors of HIV-1 replication that inhibit HIV integrase. Proc Natl Acad Sci USA. 1996;93(13):6326–6331. doi: 10.1073/pnas.93.13.6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu K, Cordeiro ML, Atienza J, Robinson WE, Jr, Chow SA. Irreversible inhibition of human immunodeficiency virus type 1 integrase by dicaffeoylquinic acids. J Virol. 1999;73(4):3309–3316. doi: 10.1128/jvi.73.4.3309-3316.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu Z, Southerland W. WinDock: Structure-based drug discovery on Windows-based PCs. J Comp Chem. 2007;28(14):2347–2351. doi: 10.1002/jcc.20756. [DOI] [PubMed] [Google Scholar]
- 17.Ewing TJ, Makino S, Skillman AG, Kuntz ID. DOCK 4.0: search strategies for automated molecular docking of flexible molecule databases. J Comput Aided Mol Des. 2001;15(5):411–428. doi: 10.1023/a:1011115820450. [DOI] [PubMed] [Google Scholar]
- 18.Thomsen R, Christensen MH. MolDock: a new technique for high-accuracy molecular docking. J Med Chem. 2006;49(11):3315–3321. doi: 10.1021/jm051197e. [DOI] [PubMed] [Google Scholar]
- 19.Farnet CM, Wang B, Lipford JR, Bushman FD. Differential inhibition of HIV-1 preintegration complexes and purified integrase protein by small molecules. Proc Natl Acad Sci USA. 1996;93(18):9742–9747. doi: 10.1073/pnas.93.18.9742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jenkins TM, Esposito D, Engelman A, Craigie R. Critical contacts between HIV-1 integrase and viral DNA identified by structure-based analysis and photo-crosslinking. EMBO J. 1997;16(22):6849–6859. doi: 10.1093/emboj/16.22.6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lubkowski J, Yang F, Alexandratos J, Wlodawer A, Zhao H, Burke TR, Jr, Neamati N, Pommier Y, Merkel G, Skalka AM. Structure of the catalytic domain of avian sarcoma virus integrase with a bound HIV-1 integrase-targeted inhibitor. Proc Natl Acad Sci USA. 1998;95(9):4831–4836. doi: 10.1073/pnas.95.9.4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson WE, Jr, Cordeiro M, Abdel-Malek S, et al. Dicaffeoylquinic acid inhibitors of human immunodeficiency virus integrase: inhibition of the core catalytic domain of human immunodeficiency virus integrase. Mol Pharmacol. 1996;50(4):846–855. [PubMed] [Google Scholar]
- 23.Lawn SD. AIDS in Africa: the impact of coinfections on the pathogenesis of HIV-1 infection. J Infect. 2004;48(1):1–12. doi: 10.1016/j.jinf.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 24.Schneider MF, Gange SJ, Williams CM, Anastos K, Greenblatt RM, Kingsley L, Detels R, Muñoz A. Patterns of the hazard of death after AIDS through the evolution of antiretroviral therapy: 1984–2004. AIDS. 2005;19(17):2009–2018. doi: 10.1097/01.aids.0000189864.90053.22. [DOI] [PubMed] [Google Scholar]
- 25.Morgan D, Mahe C, Mayanja B, Okongo JM, Lubega R, Whitworth JA. HIV-1 infection in rural Africa: is there a difference in median time to AIDS and survival compared with that in industrialized countries? AIDS. 2002;16(4):597–603. doi: 10.1097/00002030-200203080-00011. [DOI] [PubMed] [Google Scholar]


