Abstract
The recent isolation of Wolbachia pipientis in the continuous cell line Aa23, established from eggs of a strain of the Asian tiger mosquito Aedes albopictus, allowed us to perform extensive characterization of the isolate. Bacterial growth could be obtained in C6/36, another A. albopictus cell line, at 28°C and in a human embryonic lung fibroblast monolayer at 28 and 37°C, confirming that its host cell range is broader than was initially thought. The bacteria were best visualized by Diff-Quik and May-Grünwald-Giemsa staining. Proteins from 213 to 18 kDa with two major protein bands of 65 and 25 kDa were observed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. By Western blotting with specific polyclonal mouse and rabbit antisera, dominant immunoreactive antigens were found at approximately 100, 80, and 30 kDa. The genome size was calculated to be 1,790 ± 17 kb by pulsed-field gel electrophoresis. The sequence of the citrate synthase gene (gltA) of W. pipientis was determined by gene walking. Its position in the phylogenetic tree constructed with gltA confirmed that found in a phylogenetic tree constructed with 16S rRNA genes and that it belongs in the α subgroup of the class Proteobacteria and that it is closely related to but independent from the genera Ehrlichia, Anaplasma, and Neorickettsia.
Bacteria of the genus Wolbachia are maternally inherited and are known to infect a wide range of arthropods and filarial nematodes (31, 37). The first description of Wolbachia was made in 1924, when it was detected in the ovaries of the mosquito Culex pipiens and classified as an unnamed Rickettsia (11). The bacterium was subsequently named Wolbachia pipientis (10), to honor Wolbach, who first made these observations. On the basis of its association with arthropods and its intracellular location, it was classified in the family Rickettsiaceae, tribe Wolbachieae (36). The advent of molecular tools for phylogenetic and taxonomic studies, especially 16S rRNA gene sequencing, has dramatically modified classification of the bacteria belonging to the family Rickettsiaceae (35, 36). W. pipientis was found to be in the α subgroup of the class Proteobacteria and closely related to the genus Rickettsia (7). On the basis of analysis of 16S rRNA genes, groESL, and surface protein genes, the members of the order Rickettsiales have recently been reorganized in the families Rickettsiaceae and Anaplasmataceae (7). W. pipientis was then included in the family Anaplasmataceae with the genera Ehrlichia, Neorickettsia, and Anaplasma. The increasing use of gene sequencing for the characterization of insect endosymbionts has demonstrated the extreme diversity of the Wolbachia genus (21). Phylogeny based on genes such as ftsZ and wsp has revealed the existence of four major clades within the genus: clades A and B in insects, mites, and crustaceans and clades C and D in filarial nematodes (1, 2, 38). However, no consensus opinion exists at present for description of members of these clades as species or subspecies (16). Since the original description of the genus Wolbachia, the inability to culture these endosymbionts outside of the invertebrate host has limited research on these bacteria. Until now, as no isolate of Wolbachia was maintained in vitro, extensive study of bacteria of the Wolbachia genus has not been performed. However, the recent establishment of a strain of W. pipientis in a cell line originating from Aedes albopictus (i.e., the Aa23 cell line) allows the production and study of significant numbers of bacteria (22). Here, we report on the culture, purification, and molecular characterization of this bacterium, which has also been named strain wAlbB on the basis of wsp sequencing (40).
MATERIALS AND METHODS
Strain cultivation.
Details of the isolation and routine culture of the W. pipientis strain in the Aa23 mosquito cell line are provided elsewhere (22). Briefly, W. pipientis was cultured in Aa23 cell monolayers grown in 150-cm2 cell culture flasks with 30 ml of a mixture (1:1; vol/vol) of Mitsuhashi-Maramorosh insect medium (Sigma, St. Louis, Mo.) and Schneider's insect medium (Sigma). The mixture was supplemented with 10% fetal bovine serum (Gibco, Cergy Pontoise, France). The culture flasks were incubated at 28°C. Medium was replaced every week. For the production of large amounts of bacteria, infected cells from one flask were harvested every 5 days and inoculated into three cell culture flasks with fresh medium (1:3 split). Aa23 cells were cured of infection with W. pipientis by adding 10 μg of doxycycline per ml in culture medium for three passages. For uninfected cells, all cell culture flasks were incubated at 28°C. Medium was replaced every week.
Subculture assays.
Propagation of W. pipientis was attempted on human embryonic lung (HEL) fibroblast monolayers (CCL-137; American Type Culture Collection, Manassas, Va.) at 28 and 37°C under previously described conditions (24), and in C6/36 cells (CRL-1660; American Type Culture Collection), another A. albopictus cell line grown in Leibowitz-15 medium with l-glutamine and l-amino acids (Gibco), 5% (vol/vol) fetal bovine serum, and 2% (vol/vol) tryptose phosphate (Gibco) at 28°C. For subculture in these cell lines, the bacteria were first grown in Aa23 cells for 10 days without medium replacement. The supernatant was then harvested, filtered through 0.80-μm-pore-size filters in order to remove intact cells, and then centrifuged at 9,000 × g for 30 min. The resulting pellet was resuspended in fresh culture medium (which varied according to the cell line used for subculture) and inoculated into a 25-cm2 cell culture flask. In order to objectively measure bacterial growth in HEL cells at 28°C, we also performed quantitative detection of the DNA of Wolbachia growing in cells. Inoculation of five 25-cm2 cell culture flasks containing HEL cells was performed as described above. From the day of inoculation and every 3 days, the flasks were tested for Wolbachia DNA quantification by a real-time PCR targeting the wsp gene under previously described conditions (8).
Purification of W. pipientis from Aa23 cells.
Aa23 cells infected with the bacterium were harvested from 20 150-cm2 cell culture flasks. The suspension was subjected to sonication, after which unlysed cells were removed by centrifugation at 100 × g for 15 min. The supernatant was layered onto a 25% (wt/vol) sucrose solution in phosphate-buffered saline (PBS). After centrifugation at 9,000 × g for 30 min at 4°C, the bacterium-containing pellet was resuspended in 2 ml of PBS and carefully layered onto a 25 to 45% (wt/vol in PBS) step density gradient (Gastrografine; Shering, Lys Lez Lannoy, France). This gradient was subjected to centrifugation at 130,000 × g for 1 h at 4°C; and the bacteria were harvested and washed twice in PBS, resuspended in sterile distilled water in the smallest possible volume, and then frozen at −80°C. To assess the purity of the bacteria, a PCR targeting the sequence of the 18S rRNA gene was performed before and after the purification of W. pipientis from Aa23 cells, as described previously (13).
Animal immunization.
Production of mouse and rabbit polyclonal antibodies was performed as follows. Six- to 8-week-old immunocompetent BALB/c mice were inoculated subcutaneously with 0.5 ml of a suspension of 1 mg of the bacterium obtained from purified bacteria and Freund's complete adjuvant. Inoculations were repeated on days 10, 20, and 30. On day 40, the mice were anesthetized and killed. Blood was sampled by intracardiac puncture, and serum was frozen at −80°C for further studies. Rabbits were immunized by intradermal inoculation of a total of 1 mg of purified bacteria and Freund's complete adjuvant. The rabbits were given a booster immunization on day 20, and serum sampled 10 days later was frozen at −80°C for further studies. Production of rabbit polyclonal antibodies was performed as described below.
Morphological studies. (i) Staining for light microscopy.
Infected cells and the supernatant were separately cytocentrifuged for staining with Gram, Gimenez, Diff-Quik (Dade Behring, Marburg, Germany), and May-Grünwald-Giemsa stains (4, 9, 24).
(ii) Electron microscopy.
Infected cells were fixed in 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.2) containing 0.1 M sucrose for 1 h at 4°C. Fixed cells were washed overnight with the same buffer and then postfixed for 1 h at room temperature with 1% osmium tetroxide in 0.1 M cacodylate buffer. Dehydration was performed through washes in increasing concentrations (25 to 100%) of acetone. The cells were then embedded in Araldite (Fluka, St. Quentin Fallavier, France). Thin sections were cut from blocks with embedded cells with an Ultracut microtome (Reichert-Leica, Marseille, France) and were poststained with a saturated solution of methanol-uranyl acetate and lead nitrate with sodium citrate in water before examination with a Jeol 1220 electron microscope (Jeol, Croissy sur Seine, France).
SDS-PAGE and Western blot immunoassay.
To characterize the total protein profile of W. pipientis, Renografin-purified preparations of bacterial suspensions of the Wolbachia strain cultured in Aa23 cells were spectrophotometrically adjusted to 1 mg · ml−1. Neorickettsia sennetsu (Miyayama strain), Ehrlichia chaffeensis (Arkansas strain), and Anaplasma phagocytophilum (Webster strain) were cultured under previously described conditions (26) and were resuspended in sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) sample buffer (0.625 M Tris [pH 8.0], 2% [wt/vol] SDS, 5% [vol/vol] 2-mercaptoethanol, 10% [vol/vol] glycerol, 0.002% [wt/vol] bromophenol blue) for protein immunoblotting. An aliquot of each was then heated for 10 min at 100°C, and then heated and unheated aliquots were loaded onto 10% polyacrylamide gels (18 cm by 20 cm by 1.5 mm). Proteins were electrophoretically resolved at a constant current of 60 mA/gel at 10°C until the dye front reached the bottom of the gel (15). Proteins were visualized with silver stain by a modified Blum procedure (18). For Western immunoblotting procedures, proteins were extracted and heated as described above for SDS-PAGE. A total of 10 μg of total protein was loaded per lane. Following electrophoresis, the proteins were electroblotted onto nitrocellulose membranes at 100 V for 1 h at 10°C. The membranes were incubated with 5% (wt/vol) nonfat dry milk in Tris buffer (10 mM Tris-HCl [pH 8.8], 150 mM NaCl, 0.1% Tween 20) overnight and were then incubated with either mouse or rabbit antiserum and mouse or rabbit preimmune serum diluted 1:100 in 0.5% (wt/vol) nonfat dry milk in PBS for 1 h at room temperature. After three 10-min washing steps in Tris buffer, the membranes were incubated with goat anti-mouse immunoglobulin G (IgG) plus IgM (Jackson ImmunoResearch Laboratories, Inc.) and with donkey anti-rabbit IgG (Jackson ImmunoResearch Laboratories, Inc.) diluted 1:200 in PBS with 0.5% (wt/vol) nonfat dry milk. After three washes in PBS, bound peroxidase was revealed by using 4-chloro-1-naphthol as the substrate.
Genomic studies. (i) PFGE.
Purified bacteria were suspended in PBS to an optical density of 1.1 to 1.2 at a λ value of 260 nm and were immediately used for preparation of pulsed-field gel electrophoresis (PFGE) plugs. Plug preparation, enzymatic digestion, and PFGE migration were performed as described previously (25). A 1% agarose gel was used for determination of the molecular weight of the restricted fragment. In order to resolve the low- and medium-molecular-weight DNA fragments, electrophoresis conditions of 6 V/cm with a ramped pulse time from 1 to 3 s for 8 h, 6 V/cm with a ramped pulse time from 1 to 5 s for 9 h, and then 6 V/cm with a ramped pulse time from 1 to 10 s for 10 h were used. Determination of medium-molecular-weight DNA fragments was performed at 5.7 V/cm with a ramped pulse time from 20 to 40 s for 10 h. A 0.6% agarose gel and a ramped pulse time of 5 to 120 s for 33 h at 4.5 V/cm were used for entire chromosomal DNA migration as well as for determination of the sizes of fragments with high molecular weights. Several restriction endonucleases (BssHII, SacII, SmaI, BstzI, NotI, XbaI, SfiI, SalI) were tested for their abilities to determine the genome sizes of Ehrlichia, Anaplasma, and Neorickettsia (26). Finally, it was found that BssHII, SacII, and SalI were suitable for the study.
(ii) Study of the citrate synthase gene (gltA).
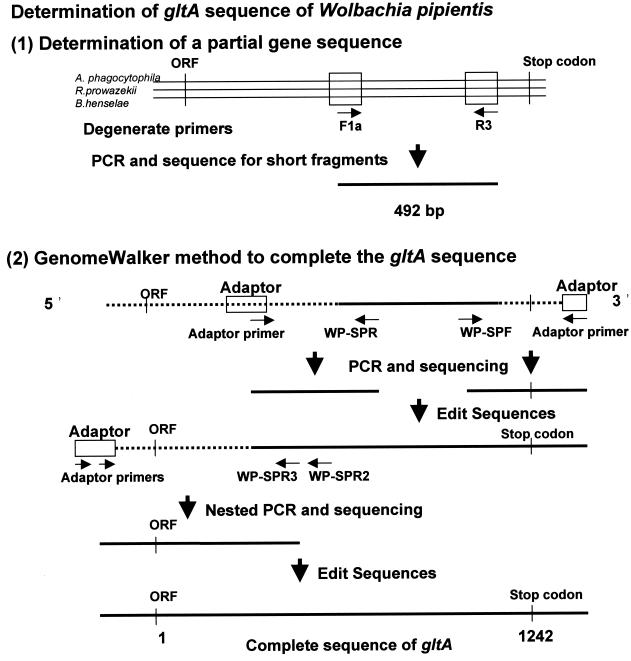
Genomic DNA was extracted from purified bacteria by using the QIAamp tissue kit (Qiagen GmbH, Hilden, Germany) according to the recommendations of the manufacturer. The strategy used to determine the gltA sequences of W. pipientis used in the present study is summarized in Fig. 1. First, a partial sequence of gltA was determined by PCR with degenerate primers F1 (5′-CAT-GAR-CAR-AAT-GCT-TC-3′) and R3 (5′-CNG-CCC-ANC-CAG-ACG-T-3′) and sequencing. Both primers were designed after alignment of the conserved region of gltA among A. phagocytophilum, Rickettsia prowazekii, and Bartonella henselae. For the amplification, the reaction mixture contained 50 pmol of each primer, 1.5 U of Taq DNA polymerase (Gibco BRL, Gaithersburg, Md.), 20 mM each deoxynucleoside triphosphate, 10 mM Tris-HCl, 50 mM KCl, 1.6 mM MgCl2, and 5 μl of template DNA with a final volume of 50 μl. The amplification was performed in a Peltier model PTC-200 thermal cycler (MJ Research, Inc., Watertown, Mass.) with the following program: an initial 5 min of denaturation at 95°C; 34 cycles of denaturation (95°C for 30 s), annealing (51°C for 30 s), and extension (72°C for 90 s); and 5 min of extension at 72°C. Distilled water and B. henselae DNA were included in the PCR as negative and positive controls, respectively. The amplification products were visualized on a 1% ethidium bromide-stained agarose gel after electrophoretic migration of the amplified material. A positive amplification product of the expected size (492 bp) was purified for sequence analysis. After determination of the partial sequence, the unknown sequences of both the 3′ and 5′ ends of the gene were amplified by PCR by using the Universal GenomeWalker kit (Clontech Laboratories, Palo Alto, Calif.). Briefly, genomic DNA was digested with EcoRV, DraI, PvuII, StuI, and ScaI. The DNA fragments were ligated with a GenomeWalker adaptor, which had one blunt end and one end with a 5′ overhang. The ligation mixture of the adaptor and the genomic DNA fragments was used as the template for PCR. This PCR was performed with an adaptor primer supplied by the manufacturer and W. pipientis gltA-specific primers to walk downstream on the DNA sequence (Table 1). For the amplification, 1.5 U of Elongase (Boehringer Manheim) was used with 10 pmol of each primer, 20 mM each deoxynucleoside triphosphate, 10 mM Tris-HCl, 50 mM KCl, 1.6 mM MgCl2, and 5 μl of template with a final volume of 50 μl. Distilled water was included in each PCR as a negative control. The following program was used for the amplification: an initial 2 min of denaturation at 94°C; 44 cycles of denaturation (94°C for 30 s), annealing (53°C for 60 s), and extension (68°C for 60 s); and 3 min of extension at 68°C. The PCR products for DNA sequencing were purified with QIAquick PCR purification kits (Qiagen) and were sequenced directly by using PCR primers when a single clear band was observed on the ethidium bromide-stained agarose gel. When multiple bands including bands of the expected sizes were obtained in a PCR, a Gel Extraction kit (Qiagen) was used to purify the expected bands from the gel. The fluorescent dideoxynucleotide technology (Perkin-Elmer, Applied Biosystems Division, Foster City, Calif.) was used for the DNA sequencing reactions. The sequenced fragments were separated, and data were collected on an ABI PRISM 310 Genetic Analyzer (Perkin-Elmer). The sequences collected were assembled and edited with the AutoAssembler program (version 1.4; Perkin-Elmer). To avoid editorial errors by use of the GenomeWalking method, the sequences of the citrate synthase-coding region, including open reading frames at the 5′ end and stop codons at the 3′ end, were confirmed by PCR with primers WP-M91F (5′-TGA-CCG-CTT-AAT-ACG-GTT-AAT-G-3′) and WP1274R (5′-TTG-AGG-ACG-AAT-TGT-GAA-TGA-3′) and sequenced. The gltA sequences of the following species were analyzed for levels of similarity and phylogenetic relationships: E. chaffeensis, Ehrlichia muris, an Ehrlichia sp. detected in Ixodes ovatus ticks, Ehrlichia canis, Ehrlichia ruminantium, A. phagocytophilum (composed of the former species Ehrlichia phagocytophila, Ehrlichia equi, and the human granulocytic ehrlichia agent), Anaplasma marginale, Anaplasma centrale, W. pipientis, N. sennetsu, Neorickettsia risticii, Neorickettsia helminthoeca, R. prowazekii, and B. henselae. Pairwise percent identities of the sequences with all gaps omitted were calculated by use of a program designed by H. Ogata (Ogata@igs.cnrs-mrs.fr), Institut de Génomique Structurale, Centre National de la Recherche Scientifique-Unité Mixte de Recherche, Marseille, France. Multiple-sequence alignment analysis, distance matrix calculation, and construction of phylogenetic trees were performed with the ClustalW program (version 1.8) (33) at the DNA Data Bank of Japan (Mishima, Japan [http://www.ddbj.nig.ac.jp/htmls/E-mail/clustalw-e.html]). The distance matrices for the aligned sequences with all gaps ignored were calculated by the Kimura two-parameter method (14), and the neighbor-joining method was used to construct phylogenetic trees (28). The stability of the tree obtained was estimated by bootstrap analysis with 1,000 replications by using the ClustalW program. Tree figures were generated with the Tree View program (version 1.61) (23). The same analyses of similarity and phylogenetic relationships were also performed for the deduced amino acid sequences of gltA and the 16S rRNA gene. The GenBank accession numbers of the gltA sequences used in this study are as follows: E. chaffeensis, AF304142; E. muris, AF304144; Ehrlichia sp. detected in I. ovatus ticks, AF304145; E. canis, AF304143; E. ruminantium, AF304146; A. phagocytophilum, AF304138; A. phagocytophilum (formerly E. equi), AF304137; A. phagocytophilum (formerly the human granulocytic ehrlichia agent), AF304136; A. marginale, AF304139; A. centrale, AF304141; N. sennetsu, AF304148; N. risticii, AF304147; N. helminthoeca, AF304149; R. prowazekii, M17149; B. henselae L38987; and Escherichia coli, J01619. The GenBank accession numbers of the 16S rRNA gene sequences used to calculate percent identities and construct phylogenetic trees are as follows: E. chaffeensis, M73222; Ehrlichia sp. detected in I. ovatus ticks, AF260591; E. muris, U15527; E. canis, M73221; E. ruminantium, AF069758; A. phagocytophilum, M73224; A. phagocytophilum (MRK strain), M73223; A. phagocytophilum (a Webster strain), U02521; A. marginale, M60313; A. centrale, AF283007; W. pipientis, AF179630; N. sennetsu, M73225; N. risticii, M21290; N. helminthoeca, U12457; R. prowazekii, M21789; and B. henselae, AJ223779.
FIG. 1.
Strategy for determination of citrate synthase gene (gltA) of the strain of W. pipientis. Primers F1a and R3 were created after alignment of gltA of A. phagocytophilum, R. prowazekii, and B. henselae. After determination of the partial sequence, the unknown sequences of both the 3′ and the 5′ ends of the gene were amplified by PCR with an adaptor primer provided in the Universal GenomeWalker kit and the strain-specific primers based on the partial sequence. Alignment and assembly of these sequences allowed the determination of the complete gltA sequence of W. pipientis. ORF, open reading frame.
TABLE 1.
Oligonucleotide primers and restriction genome libraries used for genome walking of the W. pipientis citrate synthase gene
| Primer | Sequence | Restriction genomic library |
|---|---|---|
| R1b | CGA-TGA-CCA-AAA-CCC-AT | EcoRV and DraI |
| WP-SPR1 | CTA-CCA-ATC-TGA-CAG-CTG-C | EcoRV and DraI |
| WP-SPR2 | GCT-GAC-AAA-TTT-GCA-AAA-CAT-G | EcoRV and DraI |
| WP-SPR3 | TGG-AAA-TGC-TTT-GAT-TAC-ATT-TG | EcoRV and DraI |
| F1b | GAT-CAT-GAR-CAR-AAT-GCT-TC | StuI |
| WP-SPF | GCA-AAA-AAA-CTT-GAA-GAA-ATA-GC | StuI |
Nucleotide sequence accession number.
The gltA sequence of W. pipientis has been deposited in the GenBank database under accession number AF332584.
RESULTS
Morphological studies.
Bacteria were best visualized by the Diff-Quik and May-Grünwald-Giemsa staining methods. On Gimenez staining, the bacteria stained poorly and appeared as pale pink structures. On Gram staining, the bacteria also stained poorly and always appeared to be gram negative. The bacteria appeared as small cocci. No obvious cytopathic effect was observed in Aa23 cells that remained persistently infected. The level of infection in chronically infected cells was variable within a single flask, with some cells containing high numbers of bacteria and others containing few detectable bacteria. Due to the presence of some heavily infected cells that coexisted with lightly infected or uninfected cells, it is likely that some cell lysis was compensated for by the concomitant multiplication of uninfected cells. The infection level within a single flask was also variable over time. When HEL and C6/36 cells were inoculated with W. pipientis, the cells became progressively filled with bacteria. After 2 weeks, the appearance on Giemsa staining was the same as that observed with Aa23 cells, and the cells remained constantly infected without an evident cytopathic effect. The same variations in levels of infection observed in Aa23 cells were observed in HEL and C6/36 cells. These infected HEL and C6/36 cells remained chronically infected for at least 3 months. Bacteria were seen in vacuoles within HEL cells by electron microscopy. For determination of the growth of W. pipientis in HEL cells at 28°C by PCR assay, a primary inoculum with approximately 103 DNA copies/ml was evaluated. The growth of W. pipientis was exponential, with a nearly 2-log increase in the bacterial load after a 9-day incubation of the cultures.
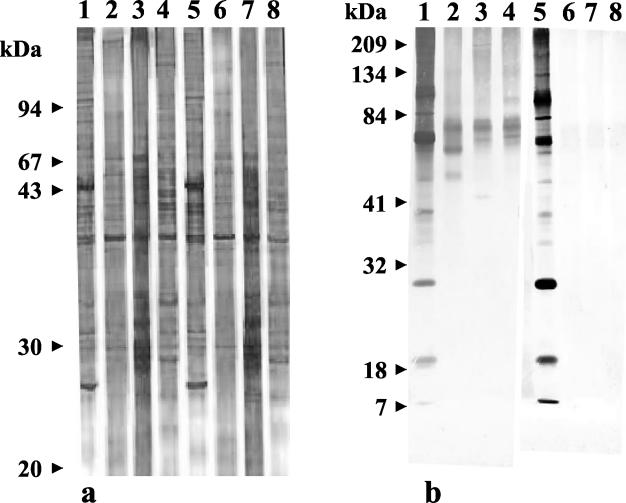
Purification of W. pipientis, SDS-PAGE analysis, and Western blot immunoassay (Fig. 2). The PCR targeting the 18S rRNA sequence was positive before purification and negative after purification. SDS-PAGE showed the presence of proteins from 18 to 213 kDa. The main difference between W. pipientis, A. phagocytophilum, N. sennetsu, and E. chaffeensis was the presence of two major bands of approximately 65 and 25 kDa for W. pipientis that were lacking for the other bacteria. By the immunofluorescence assay, the titers of antibodies to W. pipientis in rabbit and mouse sera were 3,200 and 1,600, respectively. Polyclonal rabbit serum recognized several W. pipientis antigens, with dominant bands of approximately 100, 80, and 30 kDa and several less intense bands of 96 and 18 kDa. There was no cross-reactivity with A. phagocytophilum, N. sennetsu, or E. chaffeensis. Polyclonal mouse serum recognized several W. pipientis antigens with dominant bands of approximately 100, 80, and 30 kDa and several less intense bands of 96 and 18 kDa. It also recognized a band of approximately 82 kDa, in common with A. phagocytophilum, N. sennetsu, and E. chaffeensis. No cross-reactivity was observed with mouse or rabbit preimmune serum (data not shown).
FIG. 2.
(a) Silver-stained SDS-polyacrylamide gel of whole-cell protein preparations of W. pipientis. Lanes 1 and 5, W. pipientis; lanes 2 and 6, A. phagocytophilum; lanes 3 and 7, E. chaffeensis; lanes 4 and 8, N. sennetsu; lanes 1 to 4, unheated samples; lanes 5 to 8, heated samples. (b) Western blot of the strain reacted with mouse and rabbit polyclonal antisera. Lanes 1 and 5, W. pipientis; lanes 2 and 6, A. phagocytophilum; lanes 3 and 7, E. chaffeensis; lanes 4 and 8, N. sennetsu; lanes 1 to 4, mouse polyclonal serum; lanes 5 to 8, rabbit polyclonal serum.
Genomic studies.
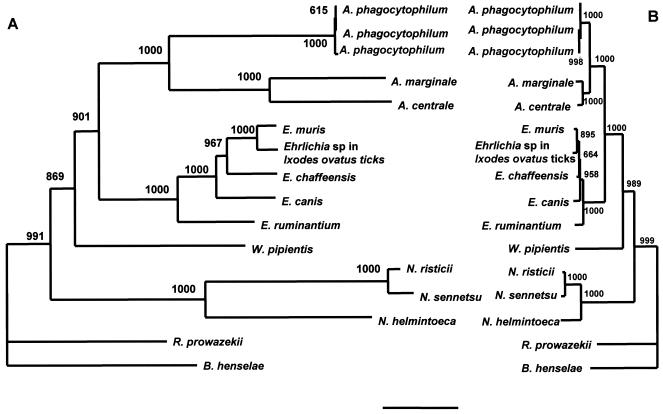
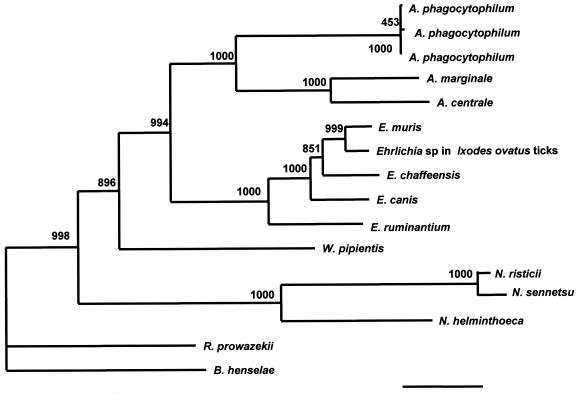
By PFGE, W. pipientis was digested only with the enzymes listed in Table 2, and the genome size was calculated to be 1,790 ± 17 kb. A 1,242-bp open reading frame extending from the ATG start codon down to the TAA stop codon of the gltA sequence was determined by the GenomeWalker PCR method. The G+C content of the gene was calculated to be 34 mol%. A phylogenetic tree constructed on the basis of the multiple-sequence alignments of the gltA nucleotide sequence was compared with the tree constructed on the basis of the multiple-sequence alignments of the 16S rRNA gene sequences (Fig. 3). A phylogenetic tree based on the deduced amino acid sequences was also constructed (Fig. 4). The positions of W. pipientis in the phylogenetic trees constructed by use of the gltA nucleotide sequences and amino acid sequences were almost the same as that in the tree derived from the 16S rRNA gene sequences.
TABLE 2.
Sizes of linearized DNA fragments after restriction endonuclease digestion for W. pipientis, as determined by PFGEa
| Size (kb) of linearized fragment(s)
| ||
|---|---|---|
| BssHII | SacII | SalI |
| 291b | 542 | 398 |
| 210b | 339 | 251b |
| 169 | 242 | 140 |
| 160 | 194 | 120b |
| 130 | 170 | 110b |
| 121 | 140 | 90 |
| 110 | 80 | 56 |
| 97 | 50 | 50 |
| 40 | 46 | |
| 10 | 31 | |
The total sizes after digestion with BssHI, SacII, and SalI were 1,789, 1,807, and 1,773 kb, respectively (mean ± standard deviation, 1,790 ± 17).
Two fragments.
FIG. 3.
Phylogenetic relationship of W. pipientis and closely related species based on the nucleotide sequences of the citrate synthase gene (A) and the 16S rRNA gene (B). The neighbor-joining method was used to construct the phylogenetic tree by using the ClustalW program. The scale bar represents 1% divergence. The numbers at the nodes are the proportions of 1,000 bootstrap resamplings that support the topology shown.
FIG. 4.
Phylogenetic relationship of W. pipientis and closely related species based on the deduced amino acid sequences of the citrate synthase gene. The neighbor-joining method was used to construct the phylogenetic tree by using the ClustalW program. The scale bar represents 1% divergence. The numbers at the nodes are the proportions of 1,000 bootstrap resamplings that support the topology shown.
DISCUSSION
Bacteria of the genus Wolbachia are associated with arthropods and nematode worms. These bacteria are responsible for sex manipulation in insects that causes a number of reproductive alterations in their arthropod hosts, including cytoplasmic incompatibility between strains and related species, male killing, parthenogenesis, and feminization (37). A variant strain detected in Drosophila melanogaster was demonstrated to proliferate massively in adults, leading to widespread degeneration of tissues followed by early death of the insect (17). The bacteria may be associated with nematodes and thus may be indirectly responsible for pathogenesis in human filariasis (27, 32).
Wolbachia is extremely widespread and has been found in up to 16% of insect species, including each of the major insect orders. A recent study that used a more sensitive “long” PCR detection method suggested that this value might be considerably higher (12). All nematode worms responsible for human diseases studied so far by PCR are infected with Wolbachia, with the possible exception of Loa loa worms (3, 31). Nematodes infected with Wolbachia bacteria are confined to the superfamily Filaroidea, raising the possibility that these parasites were infected via their arthropod hosts (31). No evidence for the recent transfer of bacteria between arthropods and nematodes has been found.
For a long time, Wolbachia research has been limited to laboratories with insect-rearing facilities due to the inability to cultivate these bacteria outside of invertebrate hosts. It was not until 1997 that a strain of W. pipientis was established in vitro. A new study has demonstrated that Wolbachia infections can be simply established and stably maintained in vitro by using standard tissue cell cultures in shell vials (6). Insect cell lines have been stably infected with Wolbachia isolates from Drosophila simulans, Cadra cautella, and Culex pipiens. These infections represent a phylogenetically diverse range of Wolbachia types from a broad range of invertebrate hosts. Wolbachia infections were realized in cell lines derived from A. albopictus (Aa23, C6/36), Spodoptera frugiperda (SF9), and D. melanogaster (S2). Another recent study has demonstrated that Wolbachia proliferates not only in a lepidopteran (Heliothis zea) cell line but also in a mammalian cell line obtained from a mouse (L929) (19). These successes in establishing infections have suggested that the Wolbachia host cell range is broader than was previously thought. These results were confirmed with the C6/36 and mammalian cell lines and were extended to the culture of Wolbachia in human cells.
The purification of bacteria was obtained by using a Renografin density gradient and allowed the characterization of whole-cell proteins by SDS-PAGE, the preparation of polyclonal antisera, the characterization of the proteins by Western blot immunoassay, and the determination of the W. pipientis genome size. By SDS-PAGE, the main differences between W. pipientis and the other genera of the family Anaplasmataceae was the presence of two major bands of approximately 65 and 25 kDa. By Western blotting, polyclonal mouse and rabbit antisera have low degrees of cross-reactivity with the other genera of the family Anaplasmataceae. By PFGE, the molecular size of W. pipientis chromosomal DNA is slightly larger than those of E. ruminantium (1,576 kb) (5), E. chaffeensis (1,226 kb), A. phagocytophilum (1,494 kb), and A. marginale and Rickettsia spp. (1,138 to 1,660 kb) (26) and differs considerably from those of N. sennetsu and N. risticii (about 880 kb) (26). This determination confirms the heterogeneity in the genome size for representative genomes of the family Anaplasmataceae. The molecular size of W. pipientis chromosomal DNA is nearly the same as those previously described for Wolbachia strains infecting Drosophila (1,400 to 1,600 kb), but is larger than those of Wolbachia strains infecting nematodes (950 and 1,100 kb) (30), confirming the large difference in genome sizes between these different groups.
With the availability of molecular biology-based techniques, it has become evident that bacteria of the Wolbachia group are more diverse than was previously believed. The use of gltA in this work helped to confirm the position of W. pipientis among the closely related α subgroup of the class Proteobacteria, but only determination of the sequence of this gene in other W. pipientis isolates will allow verification that it is a good tool for the distinction of several groups, as has previously been described with genes such as ftsZ and wsp (29). Nevertheless, if the data obtained with the ftsZ and wsp genes suggest the presence of different species in the genus Wolbachia, only the establishment of new strains followed by DNA-DNA hybridization and comparison with the W. pipientis wAlbB strain, the sole available strain of the species, will allow definition of novel species (34).
In the phylogenetic trees constructed by use of the 16S rRNA and gltA gene sequences, W. pipientis and the Wolbachia spp. belong to the α subgroup of Proteobacteria and are close to but independent from the genera Ehrlichia, Cowdria, and Anaplasma. W. pipientis and the Wolbachia spp. occupy a position intermediate between the two tick-transmitted genera (Ehrlichia and Anaplasma) and the helminth-borne genus (Neorickettsia). The deduced amino acid sequences of Wolbachia sp. outer membrane proteins exhibit similarity to those of the major outer membrane proteins of A. marginale, A. phagocytophilum, E. chaffeensis, E. canis, and E. ruminantium, corroborating the phylogenetic position of W. pipientis (20, 39). However, W. pipientis is not recognized as a vertebrate pathogen, since mammalian infections have never been documented.
In conclusion, our results show that the W. pipientis host range is broader than was initially thought. The ability to establish in vitro infections should now permit new approaches to the investigation of Wolbachia. Thus, an important step in the future will be the establishment of a nematode strain of W. pipientis in vitro, provided that the host range in nematodes is as large as that in arthropods.
Acknowledgments
We are indebted to S. O'Neill for providing infected Aa23 cells and H. Ogata for help with the phylogenetic study.
REFERENCES
- 1.Bandi, C., T. Anderson, C. Genchi, and M. Blaxter. 1998. Phylogeny of Wolbachia in filarial nematodes. Proc. R Soc. Lond. 265:2407-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bazzocchi, C., W. Jamnogluk, S. O'Neill, T. Anderson, C. Genchi, and C. Bandi. 2000. wsp gene sequences from the Wolbachia of filarial nematodes. Curr. Microbiol. 41:96-100. [DOI] [PubMed] [Google Scholar]
- 3.Brouqui, P., P. Fournier, and D. Raoult. 2001. Doxycycline and eradication of microfilaremia in patients with loiasis. Emerg. Infect. Dis. 7:604-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chapin, K. 1995. Clinical microscopy, p. 33-51. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 6th ed. American Society for Microbiology, Washington, D.C.
- 5.de Villiers, E., K. Brayton, E. Zweygarth, and B. Allsopp. 2000. Genome size and genetic map of Cowdria ruminantium. Microbiology 146:2627-2634. [DOI] [PubMed] [Google Scholar]
- 6.Dobson, S., E. Marsland, Z. Veneti, K. Bourtzis, and S. O'Neill. 2002. Characterization of Wolbachia host cell range via the in vitro establishment of infections. Appl. Environ. Microbiol. 68:656-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dumler, J., A. Barbet, C. Bekker, G. Dasch, G. Palmer, S. Ray, Y. Rikihisa, and F. Rurangirwa. 2001. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and “HGE agent” as subjective synonyms of Ehrlichia phagocytophila. Int. J. Syst. Evol. Microbiol. 51:2145-2165. [DOI] [PubMed] [Google Scholar]
- 8.Fenollar, F., M. Maurin, and D. Raoult. 2003. Wolbachia pipientis: growth kinetics and susceptibilities to 13 antibiotics determined by immunofluorescence and real-time PCR. Antimicrob. Agents Chemother. 47:1665-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gimenez, D. 1964. Staining rickettsiae in yolk-sac cultures. Stain Technol. 39:135-140. [DOI] [PubMed] [Google Scholar]
- 10.Hertig, M. 1936. The rickettsia, Wolbachia pipientis (gen. et sp. n.) and associated inclusions of the mosquito, Culex pipientis. Parasitology 28:453-486. [Google Scholar]
- 11.Hertig, M., and S. Wolbach. 1924. Studies on rickettsia-like microorganisms in insects. J. Med. Res. 44:329-374. [PMC free article] [PubMed] [Google Scholar]
- 12.Jeyaprakash, A., and M. Hoy. 2000. Long PCR improves Wolbachia DNA amplification: wsp sequences in 76% of sixty-three arthropod species. Insect Mol. Biol. 9:349-405. [DOI] [PubMed] [Google Scholar]
- 13.Johnson, K., and M. Whiting. 2002. Multiple genes and the monophyly of Ischnocera (Insecta: Phthiraptera). Mol. Phylogenet. Evol. 22:101-110. [DOI] [PubMed] [Google Scholar]
- 14.Kimura, M. 1980. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 15.Laemmli, U. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 16.La Scola, B., C. Bandi, and D. Raoult. 2003. Genus Wolbachia Hertig 1936. In R. Murray and D. Brenner (ed.), Bergey's manual of systematic bacteriology. The Williams & Wilkins Co., Baltimore, Md.
- 17.Min, K., and S. Benzer. 1997. Wolbachia normally a symbiont of Drosophila can be virulent causing degeneration and early death. Proc. Natl. Acad. Sci. USA 94:10792-10796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nesterenko, M., M. Tilley, and S. Upton. 1994. A simple modification of Blum's silver stained method allows for 30 minutes detection of proteins in polyacrylamide gel. J. Biochem. Biophys. Methods 28:239-242. [DOI] [PubMed] [Google Scholar]
- 19.Noda, H., T. Miyoshi, and Y. Koizumi. 2002. In vitro cultivation of Wolbachia in insect and mammalian cell lines. In Vitro Cell. Dev. Biol. 38:423-427. [DOI] [PubMed] [Google Scholar]
- 20.Ohashi, N., A. Unver, N. Zhi, and Y. Rikihisa. 1998. Cloning and characterization of multigenes encoding the immunodominant 30-kilodalton major outer membrane proteins of Ehrlichia canis and application of the recombinant protein for serodiagnosis. J. Clin. Microbiol. 36:2671-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Neill, S., R. Giordano, A. Colbert, T. Karr, and H. Robertson. 1992. 16S rRNA phylogenetic analysis of the bacterial endosymbionts associated with cytoplasmic incompatibility in insects. Proc. Natl. Acad. Sci. USA 89:2699-2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Neill, S., M. Pettigrew, S. Sinkins, H. Braig, T. Andreadis, and R. Tesh. 1997. In vitro cultivation of Wolbacha pipientis in an Aedes albopictus cell line. Insect Mol. Biol. 6:33-39. [DOI] [PubMed] [Google Scholar]
- 23.Page, R. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 24.Raoult, D., B. La Scola, M. Enea, P. Fournier, V. Roux, F. Fenollar, M. Galvao, and X. de Lamballerie. 2001. A flea-associated rickettsia pathogenic for humans. Emerg. Infect. Dis. 7:73-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roux, V., and D. Raoult. 1995. Inter- and intraspecies identification of Bartonella (Rochalimea) species. J. Clin. Microbiol. 33:1573-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rydkina, E., V. Roux, and D. Raoult. 1999. Determination of the genome size of Ehrlichia spp., using pulsed field gel electrophoresis. FEMS Microbiol. Lett. 176:73-78. [DOI] [PubMed] [Google Scholar]
- 27.Saint André, A., N. Blackwell, L. Hall, A. Hoerauf, N. Brattig, L. Volkmann, M. Taylor, L. Ford, A. Hise, J. Lass, E. Diaconu, and E. Pearlman. 2002. The role of endosymbiontic Wolbachia bacteria in the pathogenesis of river blindness. Science 295:1892-1895. [DOI] [PubMed] [Google Scholar]
- 28.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 29.Schulendburg, J., G. Hurst, T. Huigens, M. van Meer, F. Jiggins, and M. Majerus. 2000. Molecular evolution and phylogenetic utility of Wolbachia ftsZ and wsp gene sequences with special reference to the origin of male-killing. Mol. Biol. Evol. 17:584-600. [DOI] [PubMed] [Google Scholar]
- 30.Sun, L., J. Foster, G. Tzertzinis, M. Ono, C. Bandi, B. Slatko, and S. O'Neill. 2001. Determination of Wolbachia genome size by pulsed-field gel electrophoresis. J. Bacteriol. 183:2219-2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor, M., and A. Hoerauf. 1999. Wolbachia bacteria of filarial nematodes. Parasitol. Today 15:437-442. [DOI] [PubMed] [Google Scholar]
- 32.Taylor, M., and A. Hoerauf. 2001. A new approach to the treatment of filariasis. Curr. Opin. Infect. Dis. 14:727-731. [DOI] [PubMed] [Google Scholar]
- 33.Thompson, J., D. Higgins, and T. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wayne, L., D. Brenner, R. Colwell, P. Grimont, O. Kandler, M. Krichevsky, L. Moore, W. Moore, R. Murray, E. Stackebrandt, M. Starr, and H. Trüper. 1987. Report of the Ad Hoc Committee on Reconciliation of Approaches to Bacterial Systematics. Int. J. Syst. Bacteriol. 37:463-464. [Google Scholar]
- 35.Weisburg, W., M. Dobson, J. Samuel, G. Dasch, L. Mallavia, O. Baca, L. Mandelco, J. Sechrest, E. Weiss, and C. Woese. 1989. Phylogenetic diversity of the rickettsiae. J. Bacteriol. 171:4202-4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weiss, E., G. Dasch, and K. Chang. 1984. Wolbachia, p. 711-713. In N. R. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology. The Williams & Wilkins Co., Baltimore, Md.
- 37.Werren, J. 2001. Biology of Wolbachia. Annu. Rev. Entomol. 42:587-609. [DOI] [PubMed] [Google Scholar]
- 38.Werren, J., W. Zhang, and L. Guo. 1995. Evolution and phylogeny of Wolbachia: reproductive parasites of arthropods. Proc. R. Soc. London 261:55-71. [DOI] [PubMed] [Google Scholar]
- 39.Yu, X., J. W. McBride, and D. H. Walker. 1999. Characterization of the genus-common outer membrane proteins in Ehrlichia, p. 103-107. In D. Raoult and P. Brouqui (ed.), Rickettsiae and rickettsial diseases at the turn of the third millennium. Elsevier, Paris, France.
- 40.Zhou, W., F. Rousset, and S. O'Neill. 1998. Phylogeny and PCR-based classification of Wolbachia strains using wsp gene sequences. Proc. R. Soc. London 265:509-515. [DOI] [PMC free article] [PubMed] [Google Scholar]