Abstract
Adult T-cell leukemia/lymphoma (ATLL), an aggressive neoplasm etiologically associated with human T-lymphotropic virus type-1 (HTLV-1), is resistant to treatment. In this study, we examined the effects of a new inhibitor of deacetylase enzymes, LBH589, on ATLL cells. LBH589 effectively induced apoptosis in ATLL-related cell lines and primary ATLL cells and reduced the size of tumors inoculated in SCID mice. Analyses, including with a DNA microarray, revealed that neither death receptors nor p53 pathways contributed to the apoptosis. Instead, LBH589 activated an intrinsic pathway through the activation of caspase-2. Furthermore, small interfering RNA experiments targeting caspase-2, caspase-9, RAIDD, p53-induced protein with a death domain (PIDD) and RIPK1 (RIP) indicated that activation of RAIDD is crucial and an event initiating this pathway. In addition, LBH589 caused a marked decrease in levels of factors involved in ATLL cell proliferation and invasion such as CCR4, IL-2R and HTLV-1 HBZ-SI, a spliced form of the HTLV-1 basic zipper factor HBZ. In conclusion, we showed that LBH589 is a strong inducer of apoptosis in ATLL cells and uncovered a novel apoptotic pathway initiated by activation of RAIDD.
Keywords: LBH589, apoptosis, adult T-cell leukemia, caspase-2, RAIDD
Introduction
There is evidence that gene expression governed by epigenetic changes is crucial to the pathogenesis of cancer.1 Histone deacetylases are enzymes involved in the remodeling of chromatin, and have a key role in the epigenetic regulation of gene expression. In addition, the activity of non-histone proteins can be regulated through histone deacetylase-mediated hypoacetylation.2 Deacetylase inhibitors (DACi) induce the hyperacetylation of non-histone proteins, as well as nucleosomal histones resulting in the expression of repressed genes involved in growth arrest, terminal differentiation and/or apoptosis among cancer cells.3, 4 Several classes of DACi have been found to have potent anticancer effects in preclinical studies.2 Despite a consensus on the importance of apoptosis to the effect of these drugs, different apoptotic mechanisms have been reported.2 For instance, even the contribution of caspases to DACi-induced apoptosis is quite controversial.5, 6, 7, 8
There are two general apoptotic pathways, the receptor (extrinsic) pathway and the mitochondrial (intrinsic) pathway.9 The extrinsic pathway requires activation of caspase-8, which results in activation of the zymogens of executioner caspases such as caspase-3, eventually leading to cleavage of poly ADP-ribose polymerase (PARP). Proteolytic caspase-8 can further activate BH3-interacting domain death agonist (BID), and cleaved BID moves to the mitochondria where it triggers the release of cytochrome-. In the intrinsic pathway, death signals lead to changes in the mitochondrial outer membrane's permeability, subsequently releasing cytochrome-, which forms an apoptosome with caspase-9, eventually activating executioner caspases.10
LAQ824 and the more potent and newly developed DACi analog LBH589 are now under clinical investigation as a therapeutic agent for several kinds of cancer.2, 4 One unique feature of these drugs is their role in the regulation of heat shock protein 90kDa-α (HSP90) and degradation of HSP90 client proteins such as bcr–abl in chronic myelogenous leukemia and fms-related tyrosine kinase 3 in acute myeloid leukemia.11, 12, 13, 14, 15
Adult T-cell leukemia/lymphoma (ATLL) is a neoplasm of mature T-lymphocyte origin etiologically associated with human T-lymphotropic virus type-1 (HTLV-1) and is known to be resistant to standard anticancer therapies.16 Previous findings suggest that the viral protein HTLV-1 Tax interferes with most DNA repair mechanisms, preventing cell cycle arrest and apoptosis, and contributing to the early stages of ATLL.17, 18 In addition, the novel viral protein HTLV-1 basic zipper factor (HBZ) and its spliced form (HBZ SP1RNA or HBZ-SI), which are encoded by the minus-strand RNA of the HTLV-1 genome, have been identified recently.19, 20, 21, 22, 23 These proteins are thought to be functional and expected to be closely involved in the late stages of ATLL.24 ATLL is generally classified into four clinical subtypes: acute, chronic, smoldering and lymphoma. Although several approaches have been reported, combination chemotherapy is still the treatment of choice for newly diagnosed acute and lymphoma-type ATLL. Patients with aggressive ATLL have a median survival period of 13 months, indicating limitations in the treatment of ATLL.25, 26 In this study, we examined the effects of LBH589 on ATLL cells in vitro and in vivo, and investigated the pathways or factors contributing to the LBH589-induced ATLL cell death.
Materials and methods
Cell preparation
The ATLL-derived cell lines ST1, KOB, LM-Y1, LM-Y2, KK1 and SO4 were established in our laboratory from ATLL patients27, 28 and maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum and 0.5 U/ml of interleukin-2 (kindly provided by Takeda Pharmaceutical Company, Ltd., Osaka, Japan). The HTLV-1-infected T-cell lines MT2 and HuT102,29, 30 human T-cell leukemia cell line Jurkat and erythromyeloblastoid cell line K562 were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum. The ST1, KOB, LM-Y1, MT2 and HuT102 cells have the wild-type p53.31 Primary leukemia cells from eight patients with acute-type ATLL were also analyzed. The diagnosis of ATLL and preparation of peripheral blood mononuclear cells from patients with ATLL and normal healthy donors were described previously.28 Each patient's sample contained more than 80% leukemia cells at the time of analysis. After approval by the Ethics Committee at Nagasaki University Hospital, all materials from patients were obtained with informed consent.
Chemicals and cell proliferation assay
Chemicals used in this study were LBH589 (kindly provided by Novartis Pharma AG., Basel, Switzerland), SAHA (Cayman Chemical, Ann Arbor, MI, USA), MG132 (Biomol Research Laboratories, Plymouth Meeting, PA, USA), LY294002 (Biomol), soluble tumor necrosis factor related apoptosis-inducing ligand (TRAIL) (Biomol), staurosporine (Merck, Darmstadt, Germany), the pan-caspase inhibitor z-VAD-fmk, the caspase-2 inhibitor Z-VDVAD-fmk and the caspase-9 inhibitor Z-LEHD (MBL, Nagoya, Japan). The cell proliferation assay (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt assay) was performed with a Cell Titer 96 AQueos Cell Proliferation Assay kit (Promega, Madison, WI, USA) according to the manufacturer's directions.
In vivo experiments using SCID mice
Five-week-old female C.B-17/Icr-SCID mice obtained from Ryukyu Biotec (Urasoe, Japan) were maintained in containment level 2 cabinets and provided with autoclaved food and water ad libitum. The mice were engrafted with 107 HuT102 cells by subcutaneous injection in the post-auricular region and randomly placed into two cohorts of five animals each that received vehicle or LBH589. Treatment was initiated on the day after cell injection. LBH589 was dissolved in distilled water at a concentration of 2 mg/ml, and 28 mg/kg body weight of LBH589 was administered by oral gavage three times a week. Control mice received the same volume of the vehicle only. Body weight and tumor numbers and size were monitored once a week. All mice were sacrificed on day 28, then the tumors were dissected out, and their weight was measured. After tumors were fixed for paraffin embedding and tissue sectioning, DNA fragmentation was evaluated by fluorescent TUNEL (TAKARA BIO INC., Shiga, Japan) as recommended by the manufacturer. All these experiments were performed according to the Guidelines for Animal Experimentation of the University of the Ryukyus and were approved by the Animal Care and Use Committee of the same university.
Flow cytometric analysis
To evaluate apoptotic changes and the permeability of the mitochondrial outer membrane, we used an Annexin-V and PI Kit (Bender Medsystems, Vienna, Austria) and a Mitochondrial Membrane Potential Assay Kit (5, 5′, 6, 6′-tetrachloro-1, 1′, 3, 3′-tetraethylbenzimidazol-carbocyanine iodide detection) (Cayman Chemical), respectively. Activities of caspase-8 and 9 were determined by fluorometric assay (MBL) according to the manufacturer's instructions. The cell-surface expression of death receptors (DRs), CCR4 and IL-2R was examined by flow cytometric analysis using anti-tumor necrosis factor-R1 (MBL), anti-CD95 (BD Biosciences, San Diego, CA, USA), anti-DR5 (Alexis Biochemicals, San Diego, CA, USA), anti-CCR4 (BD) and anti-CD25 (BD) monoclonal antibodies. Mouse IgG1 (DAKO, Kyoto, Japan) was used as a negative control. All experiments were performed using a FACSCalibur flowcytometer and Cellquest software (BD).
Preparation of whole-cell lysate, nuclear, mitochondrial and cytosolic fractions
Cells were harvested after treatment and whole-cell lysate was prepared as described previously.32 Nuclear extracts from cells were prepared for NF-κB transcription assays using a nuclear/cytosol fractionation kit (BioVision, San Diego, CA, USA), according to the instructions. Similarly, cytosolic fractions were prepared for western blotting by using a mitochondria/cytosol fractionation kit (BioVision).
Western blotting, immunoprecipitation and antibodies
Western blotting was performed as described previously.31 The analysis was performed using antibodies to p53 (DO-1), MDM2 (Ab-1), FADD, PUMA and NOXA (Merck), phospho-AKT, AKT, caspase-8, 9 and 3, cleaved caspase-9, cleaved PARP, BID, BAX, Bcl-xL, cytochrome-c, p21 and acetylated-Lysin (Cell Signaling Technology, Beverly, MA, USA), p53-induced protein with a death domain (PIDD) (LifeSpan Biosciences, Seattle, WA, USA), caspase-2 (11B4) and c-FLIP (Dave-2) (Alexis), RAIDD and TRADD (MBL), survivin (R&D systems Inc., Minneapolis, MN, USA), acetylated-histone-H3 and -H4, and Bcl-2 (Upstate Biotechnology, Waltham, MA, USA), RIP and XIAP (BD), HBZ (kindly provided by Dr JM Mesnard) and HBZ-SI,22 Tax,31 acetylated-tubulin, α-tubulin and β-actin (Sigma Chemicals, St Louis, MO, USA). In the immunoprecipitation assay, protein A-Sepharose beads (Sigma), HSP90 antibody (StressGen Biotechnologies Corporation, Victoria, BC, Canada) and RAIDD antibody (LifeSpan Biosciences) were used.
Immunohistochemistry
After tumors were fixed for paraffin embedding and tissue sectioning, immunohistochemical staining for acetylated-histone-H3 and -H4 was performed. The deparaffinized slides were pretreated with DAKO Target Retrival Solution (pH 9) (DAKO), and heated in a water bath at 95 °C for 40 min. For all stains, the endogenous peroxidase was quenched by 3% H2O2 for 15 min. Sections were then placed in 0.5% nonfat dry milk for 30 min at room temperature. The primary antibodies used were anti-acetylated-histone-H3 and -H4 (Cell Signaling). They were allowed to react for 1 h at room temperature, and then the DAKO EnVision+Dual Link System-HRP (DAKO) was applied using diaminobenzidine as the chromogen, following the manufacturer's directions.
Proteasome activity assay
The proteolytic activity of the 20S proteasome was evaluated with the Biomol AK-740 QuantiZyme Assay System (Biomol) following the manufacturers' instructions, which detects the release of the free 7-amino-4-methylcoumarin fluorophore, upon cleavage of the fluorogenic peptide Suc-LLVYAMC.
DNA microarray analysis
Total RNA from cells was extracted using ISOGEN (Nippon Gene, Toyama, Japan) and purified with a Message Clean kit (GenHunter Corp., Brookline, MA, USA). The RNA's integrity was assessed using an Agilent 2100 Bioanalyzer (Agilent, Palo Alto, CA, USA). Double-stranded cDNA and biotinylated cRNA were synthesized using a T7-poly-T primer and the BioArray RNA labeling kit (Enzo, Farmingdale, NY, USA), respectively. The labeled RNA was then fragmented and hybridized to HU-133A oligonucleotide arrays (Affymetrix, Santa Clara, CA, USA). The arrays were scanned using the Gene Array Scanner and analyzed using the DNA-Chip Analyzer.
Real-time quantitative RT-PCR
After total RNA was prepared as described above, Real-time RT-PCR for HBZ, HBZ-SI and Tax were performed using a LightCycler Technology System (Roche Diagnostics, Basel, Switzerland) as described previously.22, 28, 33 Similarly, PCRs for caspase-2, caspase-9, RAIDD, PIDD and PBGD were performed on a Roche LC480 (Roche) with LightCycler 480 Probes Master mix (Roche) according to the directions. We designed specific sets of primers and/or purchased probes as summarized in Supplementary Table 1.
NF-κB transcription factor assay
Nuclear extracts from cells were prepared as described above. Activities of NF-κB p50 and p65 were investigated using an NF-κB transcription factor assay kit (Chemicon, Temecula, CA, USA) as recommended by the manufacturer.
Transfection, luciferase assay and small interfering RNA experiments
Transfection was performed with a Cell line Nucleofector kit V and the Nucleofector system (Lonza, Cologne, Germany). The transfection programs for ST1 (O-17) and HuT102 (O-16) were determined so that the viability of cells and the transfection efficiency would be compatible (data not shown). Luciferase assays using reporter plasmid pG13-Luc were described previously.31 The plasmid Myr-AKT (pcDNA3 myr-HA-Akt1) was obtained from Addgene Inc. (Cambridge, MA, USA).34 Empty pcDNA3 or Myr-AKT vectors were used for the AKT-transfection experiment. Cells (5 × 106) were resuspended in 100 μl of Cell line Nucleofector solution V and mixed with 2 μg of plasmid DNA. Twenty-four hours after transfection, the cells were cultured with or without LBH589 and processed for flow cytometric analysis or western blotting. We prepared three different small interfering RNA (siRNA) against each target. Caspase-2: Silencer Select Validated siRNA s2412 (#1), s2410 (#2) and s2411 (#3), caspase-9: Silencer Select siRNA s2430 (#1), s2428 (#2) and s2429 (#3), RAIDD: Silencer Select siRNA s16656 (#1), s16654 (#2) and s16655 (#3), PIDD: Silencer Select siRNA s30843 (#1), s30844 (#2) and s226845 (#3), RIP: Silencer Select Validated siRNA s16651 (#1), s11652 (#2) and s16653 (#3), and control siRNA (Silencer negative control #1) (Applied Biosystems, Foster City, CA, USA). After evaluating the effect of each siRNA by monitoring the target's mRNA and protein (Supplementary Figure 2), we eventually selected a set of #1 siRNA against each target. Cells were used 24 h after transfection and each siRNA experiment was performed in triplicate.
Results
LBH589 causes acetylation of histones and non-histone proteins in ATLL cells
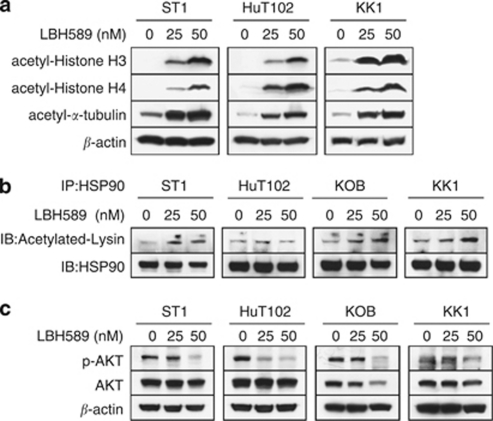
Hydroxamate-based DACi including LBH589 induce hyperacetylation of histones H3/H4 and α-tubulin.2, 4 Recent reports also indicated that LAQ824 and LBH589 can cause acetylation of HSP90 and the disruption of its chaperone function, resulting in repression of HSP90 client proteins including AKT.12, 35 We first confirmed that LBH589 was very effective in increasing the acetylation of histones H3/H4 and α-tubulin in ATLL-related cell lines (Figure 1a). As expected, LBH589 also caused acetylation of HSP90 in ST1, KOB and KK1 cells and significantly reduced the expression of phospho-AKT (Figures 1b and c).
Figure 1.
LBH589 causes acetylation of histones and non-histone proteins in adult T-cell leukemia/lymphoma (ATLL) cells. Cells were treated with either vehicle or the indicated concentrations of LBH589 for 24 h. Whole-cell lysate was prepared, and (a and c) western blotting was performed. (b) For each sample, 100 μg of cell lysate was used for immunoprecipitation with a monoclonal antibody to heat shock protein 90kDa α (HSP-90), and western blotting was performed.
LBH589 induces apoptosis in ATLL cells
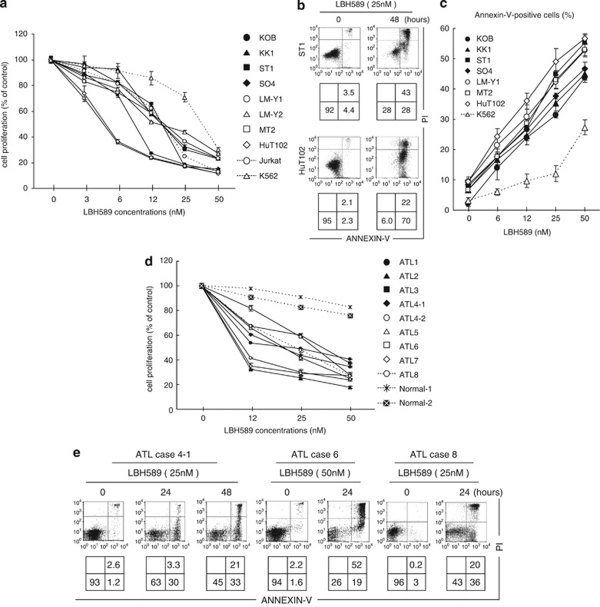
Next, we examined the effect of LBH589 on the growth of 8 ATLL-related cell lines, Jurkat cells and K562 cells. LBH589 caused ∼60% inhibition of cell growth at a concentration of 25 n in the ATLL-related cell lines and Jurkat cells, and about 30% inhibition in the K562 cells (Figure 2a). ST1, HuT102 and MT2 cells were especially sensitive with more than 60% of their growth inhibited at 12 n. To determine the details of LBH589-mediated cell death, we performed Annexin-V/PI staining. After 48-h treatment with 25 n of LBH589, Annexin-V-positive cells in the ST1 and HuT102 cell lines increased from 8 and 4% to 71 and 92%, respectively (Figure 2b). Likewise, more than 30% of cells were positive for Annexin-V after 24-h treatment with 25 n of LBH589 in the other cell lines except K562, in which the proportion was 9.5% (Figure 2c). These results indicate that ATLL-related cell lines are highly sensitive to LBH589-induced apoptosis. Similar to the results for the cell lines, LBH589 significantly inhibited cell proliferation in all primary ATLL cell samples examined. In contrast, normal peripheral blood mononuclear cells were less harmed than ATLL cells (Figure 2d). Annexin-V/PI staining confirmed that LBH589 induces apoptotic cell death in primary ATLL cell samples (Figure 2e).
Figure 2.
LBH589 induces apoptosis in adult T-cell leukemia/lymphoma (ATLL)-related cell lines and primary ATLL cells. (a and d) Cell lines (3–5 × 105/ml), primary ATLL cells or normal peripheral blood mononuclear cell (PBMCs) (7–10 × 105/ml) were treated with either vehicle or the indicated concentrations of LBH589 for 48 h and cell proliferation (% against cells cultured without LBH589) was evaluated by MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt) assay. (b, c and e) After cells were treated with the indicated concentrations and time course, Annexin-V/PI staining was performed. (b and e) Percentages of intact cells (Annexin-V− PI−), early apoptotic cells (Annexin-V+ PI−) and late apoptotic or necrotic cells (Annexin-V+ PI+) are indicated in the lower panels. (c) Percentages of Annexin-V-positive cells were evaluated. (a, c and d) Experiments were performed in triplicate and results were expressed as the mean±s.d. In ATLL case 4, samples from peripheral blood mononuclear cells (4–1) and pleural effusion (4–2) were investigated.
Treatment of subcutaneous tumors with LBH589
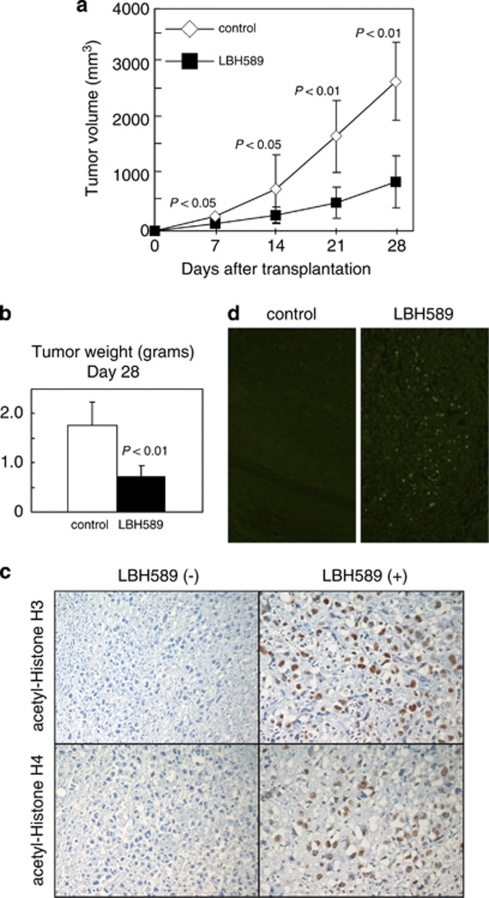
We examined whether LBH589 is also effective against ATLL cells in the in vivo setting, by using SCID mice transplanted with HuT102 cells. Ten mice were inoculated, five of which were treated with LBH589 and five of which were left untreated. LBH589 reduced the volume of tumors more than 70% (Figure 3a). The mean tumor weight of LBH589-treated mice was significantly lower than that of control mice (Figure 3b). Immunohistochemical staining confirmed that LBH589 was very effective in increasing the acetylation of histones H3/H4 in tumors of treated mice (Figure 3c). We further confirmed by a TUNEL assay that LBH589 caused evident apoptosis in transplanted tumors (Figure 3d).
Figure 3.
LBH589 reduces tumors inoculated in SCID mice. HuT102 cells (107 per mouse) were injected subcutaneously into SCID mice. The mice (five per group) were treated with either vehicle or LBH589. Treatment was initiated on the day after inoculation. Tumor volume and weight were monitored on the indicated days after the injection of cells. (a) Serial changes in tumor volume in treated and untreated mice. Data are the mean±s.d. for five mice each. Mann–Whitney's U-test was used to compare results with control values. (b) Tumors removed from untreated mice and LBH589-treated mice on day 28 after cell inoculation were weighed. (c) Immunohistochemical staining shows the acetylation of histones H3/H4 in tumors of treated mice. (d) TUNEL assays show apoptotic cells in tumors from mice treated with LBH589 compared with the control mice. Magnification, × 40.
Analysis of the extrinsic pathway in LBH589-induced apoptosis
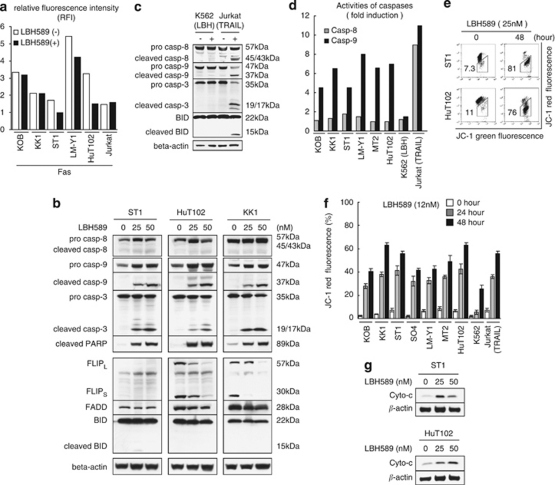
DACi are reported to activate the extrinsic pathway, in many cases, in cooperation with (DRs).2 Among typical DRs, DR5 was expressed in ATLL-related cell lines28 but tumor necrosis factor-R1 mostly was not (not shown) and there was no change after LBH589 treatment (not shown). LBH589 rather reduced Fas expression in ST1, LMY1 and HuT102 cells (Figure 4a). In typical DR-mediated apoptosis (Jurkat+TRAIL), both extrinsic and intrinsic pathways are activated including cleavage of BID, which was not observed in K562 cells treated with 12 n of LBH589 (Figure 4c). In ATLL cell lines, western blotting revealed no bands of cleaved caspase-8 on treatment with LBH589 (Figure 4b). This was accompanied by slight changes in FADD and BID expression (Figure 4b). In contrast, LBH589 reduced the expression of FLIP proteins in HuT102 and KK1 cells (Figure 4b). Fluorometric analysis eventually showed that LBH589 little activated caspase-8 in ATLL-related cell lines in contrast to caspase-9 (Figure 4d). These results suggest that LBH589 does not activate the extrinsic pathway in ATLL-related cell lines.
Figure 4.
Analysis of the apoptotic pathway in LBH589-induced cell death. Cells were treated with either vehicle or the indicated concentrations of LBH589 for 24–48 h. After cells were harvested, flow cytometric analysis (FCM) (a, d, e and f) or western blotting (b and g) was performed. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) (100 ng/ml)-treated Jurkat cells were used as a positive control of typical apoptosis and LBH589 (12 n)-treated K562 cells were used as a negative control (c, d and f). (a) Changes in expression of Fas were indicated using RFI (the ratio of mean fluorescence intensity for specific staining to that for control staining). (d) Activities of caspases. Cells were incubated with the IETD-FMK for caspase-8, conjugated to FITC (FITC-IETD-FMK) or the LEHD-FMK for caspase-9, conjugated to FITC (FITC-LEHD-FMK) and analyzed using FCM. Comparison of the fluorescence intensity in the treated sample with that of the untreated control allows determination of the fold increase in activity of each caspase. (e and f) Mitochondrial membrane permeability. Cells were incubated with the JC-1 dye and analyzed using FCM. The percentage of cells with low JC-1 red fluorescence was evaluated. (b, c and g) Western blotting was performed using whole-cell lysate (b and c) or a cytosolic fraction (g) prepared as described in Materials and methods.
LBH589 induces apoptosis in ATLL cells by activating the intrinsic pathway
Next, we investigated the changes in permeability of the mitochondrial membrane in cells treated with LBH589 by using the 5, 5′, 6, 6′-tetrachloro-1, 1′, 3, 3′-tetraethylbenzimidazol-carbocyanine iodide dye. The percentage of cells with decreased red fluorescence was increased from 7.3 to 81% and from 11 to 76% in ST1 and HuT102 cells, respectively (Figure 4e). Time course analyses of these changes are shown in Figure 4f. Furthermore, the release of cytochrome- from mitochondria to the cytosol was detected by western blotting (Figure 4g). The activation of caspase-9 was confirmed by the appearance of cleaved caspase-9 and by a fluorometric analysis, which showed a three–eightfold increase after treatment with LBH589 (Figures 4b and d). The bands of cleaved caspase-3 and cleaved PARP, as a result of apoptosis, were clearly observed (Figure 4b). Collectively, these results suggest that the major mechanism of LBH589-induced apoptosis is activation of the intrinsic pathway.
Contribution of caspase-9 and/or AKT in LBH589-induced apoptosis
Interestingly, the band for pro-caspase-9 did not decrease in intensity in spite of its consumption but rather increased on treatment with LBH589 (Figure 4b). AKT has been shown to downregulate the expression of caspase-9 via direct phosphorylation36 and LBH589 significantly reduced the expression of phospho-AKT (Figure 1c). Then, we used a typical PI3K/AKT inhibitor, LY294002, to investigate the impact of AKT's inactivation in LBH-induced apoptosis. ATLL cell lines with phospho-AKT actually underwent apoptosis and showed a decrease in phospho-AKT on treatment with LY294002. In this setting, however, the expression of caspase-9 was not upregulated (not shown). Furthermore, we performed transfection experiments with myr-AKT. The expression levels of phospho-AKT were not decreased by LBH589 in myr-AKT-transfected HuT102 cells (Supplementary Figure 1B). In this setting, we found that the percentage of Annexin-V-positive cells was decreased in myr-AKT cells (Supplementary Figure 1A). Expression levels of cleaved caspase-3 in LBH589-treated myr-AKT cells were moderately lower than those in mock-transfected cells (Supplementary Figure 1B). However, expression levels of other key molecules including caspase-9 were not altered (Supplementary Figure 1B). These results suggest that the LBH589-induced decrease in phospho-Akt levels is not an event upstream of the LBH589-induced accumulation of caspase-9 but partly contributes to the pro-apoptotic effects of LBH589.
Microarray analysis
We performed a DNA microarray analysis using HuT102 and LM-Y1 cells and compared gene expression profiles between cells treated with LBH589 and cells left untreated focusing on how the intrinsic pathway is activated during LBH589-induced apoptosis (Table 1). Among commonly observed changes, an upregulation of cytochrome- expression and a downregulation of p53 expression were remarkable. An upregulation of caspase-9 expression and a downregulation of FAS expression, were in accord with results of western blotting and flow cytometric analysis, respectively (Figures 4a and b). Taking into consideration the results of the microarray analysis, we performed further studies focusing on three different groups; ATLL-characteristic, p53-related and caspase-2-related proteins.
Table 1. Microarray analysis of HuT102 cells.
| Symbol | Gene |
Fold change |
|
|---|---|---|---|
| HuT102 | LM-Y1 | ||
| SPP1 | Secreted phosphoprotein 1 (osteopontin, bone sialoprotein I, early T-lymphocyte activation-1) | 6.4 | 9.4 |
| DHRS2 | Dehydrogenase/reductase (SDR family) member-2 | 6.2 | 2.5 |
| SERPINB2 | Serpin peptidase inhibitor, clade B (ovalbumin), member-2 | 5.6 | 3.8 |
| CASP9 | Caspase 9, apoptosis-related cysteine peptidase | 3.9 | 1.7 |
| BTG2 | BTG family, member-2 | 3.4 | 2.2 |
| CYCS | Cytochrome-, somatic | 3.4 | 2.1 |
| CLU | Clusterin | 3.1 | 2.1 |
| CTSB | Cathepsin B | 3.1 | 3.5 |
| GADD45B | Growth arrest and DNA damage inducible, beta | 2.9 | 3.6 |
| PHLDA1 | Pleckstrin homology-like domain, family A, member-1 | 2.9 | 2.0 |
| EFHC1 | EF-hand domain (C-terminal) containing 1 | 2.8 | 2.6 |
| IFI6 | Interferon, alpha-inducible protein 6 | 2.7 | 2.2 |
| NLRP1 | NLR family, pyrin domain containing 1 | 2.7 | 1.1 |
| LOH11CR2A | Loss of heterozygosity, 11, chromosomal region 2, gene A | 2.6 | 2.3 |
| RARRES3 | Retinoic acid receptor responder (tazarotene induced) 3 | 2.5 | 2.5 |
| ARHGAP20 | Rho GTPase activating protein 20 | 2.3 | 1.7 |
| BCL10 | B-cell CLL/lymphoma 10 | 2.3 | 1.5 |
| BTG1 | B-cell translocation gene 1, anti-proliferative | 2.3 | 1.4 |
| ACVR1C | Activin A receptor, type IC | 2.2 | 1.0 |
| FASLG | Fas ligand (TNF superfamily, member-6) | 2.2 | — |
| RECK | Reversion-inducing-cysteine-rich protein with kazal motifs | 2.2 | — |
| SIAH2 | Seven in absentia homolog 2 (Drosophia) | 2.2 | — |
| APAF1 | Apoptotic peptidase-activating factor-1 | 2.1 | 1.5 |
| ATM | Ataxia telangiectasia mutated (includes complementation groups A, C and D) | 2.1 | — |
| CRADD | CASP2 and RIPK1 domain containing adaptor with death domain | 2.1 | 1.6 |
| RIPK1 | Receptor (TNFRSF)-interacting serine-threonine kinase-1 | 2.1 | 1.3 |
| LTA | Lymphotoxin alpha (TNF superfamily, member-1) | −6.1 | — |
| IL10 | Interleukin-10 | −4.4 | — |
| TNFRSF8 | Tumor necrosis factor receptor superfamily, member-8 | −4.2 | — |
| CCR4 | Chemokine (C-C motif) receptor 4 | −3.8 | −2.6 |
| IL2RA | Interleukin-2 receptor, alpha | −3.8 | −2.6 |
| TP53 | Tumor protein p53 (Li-Fraumeni syndrome) | −3.5 | −1.5 |
| IKZF1 | IKAROS family zinc finger-1 (Ikaros) | −3.4 | −2.1 |
| CCNB1 | Cyclin B1 | −3.3 | −3.8 |
| FAS | Fas (TNF receptor superfamily, member-6) | −3.3 | −1.6 |
| MKI67 | Antigen identified by monoclonal antibody Ki-67 | −3.2 | −4.4 |
| CDC2 | Cell division cycle 2, G1 to S and G2 to M | −3.2 | — |
| PLK1 | Polo-like kinase 1 (Drosophila) | −3.0 | −1.4 |
| SULF1 | Sulfatase 1 | −3.0 | — |
| BIRC5 | Baculoviral IAP repeat-containing 5 (survivin) | −2.9 | −3.8 |
| CDK2AP1 | CDK2-associated protein-1 | −2.9 | −1.1 |
| AVEN | Apoptosis, caspase activation inhibitor | −2.8 | — |
| MCM5 | Minichromosome maintenance complex component-5 | −2.8 | −3.5 |
| CDC25A | Cell division cycle 25 homolog A (Schizosaccharomyces pombe) | −2.7 | −4.0 |
| MYBL2 | v-myb Myeloblastosis viral oncogene homolog (avian)-like-2 | −2.7 | −4.0 |
| MYC | v-myc Myelocytomatosis viral oncogene homolog (avian) | −2.7 | −1.9 |
| BUB1B | BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) | −2.6 | −4.1 |
| CDCA3 | Cell division cycle associated-3 | −2.6 | −3.5 |
| FAIM | Fas apoptotic inhibitory molecule | −2.6 | −2.1 |
| ATF5 | Activating transcription factor-5 | −2.5 | −3.3 |
| WEE1 | WEE1 homolog (Schizosaccharomyces pombe) | −2.5 | −2.8 |
| BCAT1 | Branched chain aminotransferase 1, cytosolic | −2.4 | −2.1 |
| CCNA2 | Cyclin A2 | −2.4 | −3.8 |
| STEAP3 | STEAP family member 3 | −2.4 | −1.3 |
| ZAK | Sterile alpha motif and leucine zipper containing kinase AZK | −2.4 | −3.2 |
| CCNB2 | Cyclin B2 | −2.3 | −3.8 |
| FBXO5 | F-box protein-5 | −2.3 | −2.6 |
| HMGB1 | High-mobility group box 1 | −2.3 | −2.7 |
| NEK6 | NIMA (never in mitosis gene a)-related kinase-6 | −2.3 | −3.2 |
| RYR1 | Ryanodine receptor 1 (skeletal) | −2.3 | — |
| SFRS2 | Splicing factor, arginine/serine-rich-2 | −2.3 | −1.7 |
| AURKA | Aurora kinase A | −2.2 | −3.0 |
| BCLAF1 | BCL2-associated transcription factor 1 | −2.2 | −1.9 |
| CXCR4 | Chemokine (C-X-C motif) receptor-4 | −2.2 | −1.7 |
| E2F8 | E2F transcription factor-8 | −2.2 | −3.4 |
| CDC45L | CDC45 cell division cycle 45-like (S. cerevisiae) | −2.1 | −4.3 |
| CSE1L | CSE1 chromosome segregation 1-like (yeast) | −2.1 | −1.8 |
| HK2 | Hexokinase-2 | −2.1 | — |
| NME1 | Non-metastatic cells 1, protein (NM23A) expressed in | −2.1 | −1.3 |
Abbreviation: TNF, tumor necrosis factor. HuT102 and LM-Y1 cells were treated with either vehicle or 50 n of LBH589 for 24 h and DNA microarray analyses were performed. Among genes with changes in expression of at least 2.1-fold (log 2 ratio) in either direction in HuT102 cells, we picked those with known functions related to apoptosis, the cell cycle and cell proliferation.
Analysis of proteins characteristic of ATLL cells
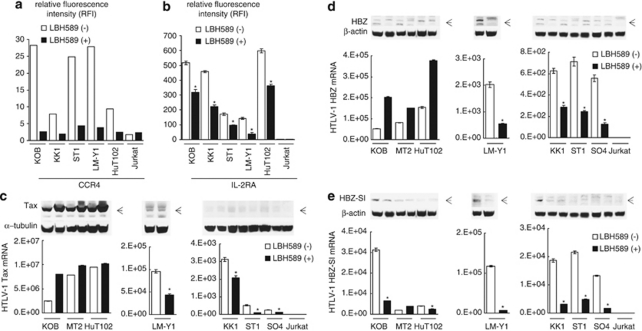
We first focused on CCR4 and IL-2R, which were remarkably affected by LBH589 (Table 1). A majority of ATLL cells consistently express CCR4, which contributes to tumor cell progression and/or invasion.37 ATLL-related cell lines had high levels of CCR4, which were dramatically decreased by LBH589 treatment (Figure 5a). IL-2R is also known to be overexpressed in ATLL cells and is often used as a diagnostic marker for ATLL.17 LBH589 repressed the expression of IL-2R ∼50% in the ATLL-related cell lines though levels were still extremely high (Figure 5b). Importantly, IL-2R is a typical gene whose promoter is activated by HTLV-1 Tax. Then, we investigated whether the expression of Tax mRNA is also affected by LBH589. The effect depended on the cells examined. In cells with high Tax mRNA levels (KOB, MT2 and HuT102), the expression tended to increase with LBH589 treatment, while in cells with relatively low Tax mRNA levels (LMY1, KK1 ST1 and SO4), it tended to decrease (Figure 5c). These results were consistent with those of western blotting (Figure 5c upper panel). The behavior of the HBZ mRNA was similar to that of the Tax mRNA; cells with high Tax levels tended to show an increase in HBZ mRNA on LBH589 treatment, while cells with lower Tax levels tended to exhibit a decrease (Figure 5d). In contrast, LBH589 significantly repressed HBZ-SI mRNA expression in ATLL-related cell lines except MT2 cells (Figure 5e). Protein levels of HBZ and HBZ-SI were not high in any cells examined and the expression profiles were approximately the same as those for the mRNA (Figures 5d and e). As NF-κB is constitutively activated in ATLL cells dependent on and/or independent of HTLV-1 Tax38, 39 and the repression of NF-κB is an important mechanism in DACi-induced ATLL cell death,40, 41 we examined the activities of NF-κB. However, repression of NF-κB by LBH589 was not commonly observed in ATLL-related cell lines (not shown).
Figure 5.
Effect of LBH589 on proteins characteristic of adult T-cell leukemia/lymphoma (ATLL) cells. Cells were treated with either vehicle or 50 n of LBH589 for 24 h. After cells were harvested, flow cytometric analysis (FCM), real time quantitative RT-PCR and western blotting were performed. (a and b) Changes in expression of CCR4 and IL-2R were evaluated by FCM. The results were indicated using RFI. (c–e) Real-time quantitative RT-PCR and western blotting against HTLV-1 Tax, HBZ and HBZ-SI were performed as described in materials and methods. RT-PCRs were carried out in duplicate and the average value was used as the absolute amount of each mRNA. The cells were divided into three groups according to the amount of Tax mRNA; cell lines with high Tax mRNA levels (KOB, MT2 and HuT102), moderate levels (LM-Y1) and low levels (KK1 ST1 and SO4). The results of western blotting are also shown in each panel. (b–e) Results were expressed as the mean±s.d. for three independent experiments and were also analyzed using Student's t-test. *P<0.01.
LBH589-induced apoptosis in ATLL cells does not depend on the p53 pathway
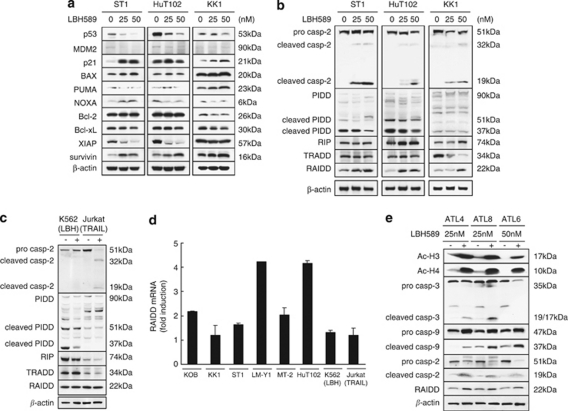
Although p53 is an important inducer of the intrinsic apoptotic pathway, its gene expression was significantly decreased by LBH589 treatment (Table 1). To verify this phenomenon, we performed a luciferase assay with pG13-Luc using ST1, HuT102, KOB and KK1 cells and found that activation of p53 had not occurred (not shown). Unexpectedly, LBH589 reduced the expression of p53 in ST1 and KK1 as well as HuT102 cells without any accumulation of MDM2, which degrades p53 (Figure 6a). In contrast, the band of p21 was increased in intensity in these cell lines irrespective of their p53 status. Among apoptosis-related proteins downstream of p53, we found an increase in survivin, a decrease in XIAP, and no change in BAX, PUMA and Bcl-2 on treatment with LBH589 in ST1 or HuT102 cells. These results suggest that typical Bcl-2 family members are not involved in the permeabilization of the mitochondrial membrane in LBH589-induced apoptosis. Interestingly, LBH589 increased the expression of Bax and PUMA and decreased that of Bcl-2 and Bcl-xL in KK1 cells carrying a non-functional p53 mutation. These results indicate that LBH589 induced apoptosis in ATLL-related cell lines in a p53-independent manner.
Figure 6.
LBH589 activates RAIDD and caspase-2, but not p53. Whether the p53 pathway or caspase-2-related factors contribute to the LBH589-induced cell death was investigated by using ST1 and HuT102 (p53 wild type) cells, and KK1 (p53 mutated) cells. (a, b and e) Cells were treated with either vehicle or the indicated concentrations of LBH589 for 24 h. After cells were harvested, western blotting was performed. (c) Western blotting: tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) (100 ng/ml)-treated Jurkat cells were used as a positive control of typical apoptosis and LBH589 (12 n)-treated K562 cells were used as a negative control. (d) Activation of RAIDD by LBH589. After cells were treated with either vehicle or 50 n of LBH589 for 24 h, real-time quantitative RT-PCR for RAIDD was performed. The fold increase in each cell line was obtained by setting the value for the expression without LBH589 as 1.0.
Analysis of caspase-2-related apoptotic pathway
CASP2 and RIPK1 domain containing adaptor with death domain (also known as RAIDD), RIP, caspase-9 and cytochrome- are involved in caspase-2-mediated apoptosis and were upregulated in their expression after LBH589 treatment (Table 1). The apoptosis induced by tumor necrosis factor-R1 requires the adaptor proteins TRADD, RIP and RAIDD in addition to caspase-2.42 Recent reports suggest that caspase-2 can cause cytochrome-'s release from mitochondria directly or indirectly.43, 44 Additionally, PIDD can interact with RAIDD and caspase-2, forming a PIDDosome and inducing apoptosis.45 We investigated the expression profiles of caspase-2-related proteins by western blotting (Figures 6b and c). Caspase-2 was clearly activated by the treatment with LBH589 and cleaved forms were observed. Similar results were observed in TRAIL-treated Jurkat cells (Figure 6c). PIDD (90kDa) is constitutively processed into 51 and 37 kDa forms and cleaved PIDD (37 kDa) is essential for the activation of caspase-2.46 Furthermore, upregulation of the expression of general forms of PIDD was observed during the activation of caspase-2 induced by genotoxic agents.46 ATLL cell lines and Jurkat cells expressed various forms of PIDD, levels of which were not increased by LBH589 or TRAIL (Figures 6b and c). RIP was increased and TRADD was decreased in KK1 cells but neither was changed in ST1 or HuT102 cells. Of note, the activation of RAIDD was a common characteristic in these cells. Furthermore, the activation of RAIDD was also observed at the mRNA level in most of the ATLL-related cell lines (Figure 6d). In KK1 and ST1 cells, the activation of RAIDD mRNA expression was faint whereas that of protein production was obvious. We analyzed the 20s proteasome's activities to investigate whether RAIDD regulation by LBH589 in these cells is due to proteasome inhibition (Supplementary Figure 1C). Activities of 20S proteasome were dramatically impaired by MG132 in all cells examined and were not changed by LBH589 in KK1 and ST1 cells. The expression of RAIDD mRNA was not induced by the various chemicals used except LBH589 (Figure 6d, Supplementary Figure 1C, and data not shown). These results suggest that LBH589 caused the activation of caspase-2 and RAIDD without activating p53 and PIDD. We further observed the activation of RAIDD and caspase-2 in addition to acetylated histones in primary ATLL cells (Figure 6e).
LBH589 induces apoptosis in ATLL cells via caspase-2's activation
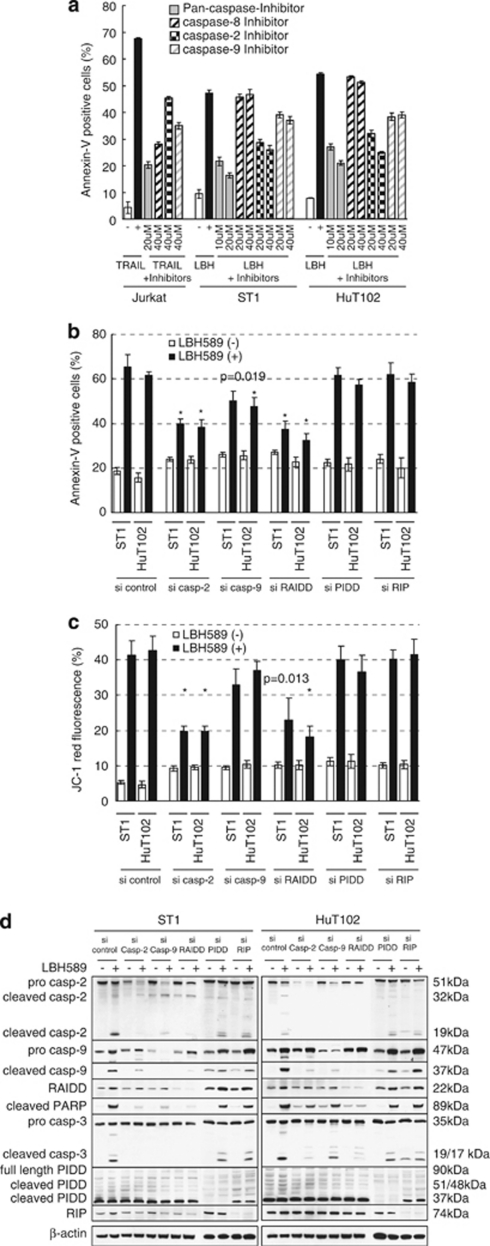
To further investigate the role of caspase-2-mediated apoptosis in LBH589-induced cell death, we first performed inhibition assays of caspases using specific inhibitors. The proportion of Annexin-V-positive cells among TRAIL-treated Jurkat cells was reduced from 67 to 20% by the pan-caspase inhibitor Z-VAD (Figure 7a). LBH589-induced cell death was also attenuated by Z-VAD with the percentage of Annexin-V-positive cells decreasing from 47 and 54% to 16 and 21% in ST1 and HuT102 cells, respectively. The caspase-8 inhibitor was not effective against LBH589-induced cell death, consistent with the finding that caspase-8 was not activated (Figure 4d). Importantly, the caspase-2 inhibitor Z-VDVAD had a similar effect to Z-VAD in LBH589-treated Jurkat cells, while it had a weak effect in TRAIL-treated cells (46%). Furthermore, western blotting revealed that the cleavage of caspase-3 by LBH589 was inhibited by Z-VDVAD as well as Z-VAD (not shown). These results suggest that LBH589 causes apoptosis in ATLL-related cell lines in a caspase-dependent manner and activation of caspase-2 is essential.
Figure 7.
RAIDD has a critical role in initiating LBH589-induced apoptosis. (a) Inhibition assay of caspases. Cells were treated with 25 n of LBH589 or 100 ng/ml of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) with indicated concentrations of caspase-inhibitors for 24 h. Results were evaluated by Annexin-V/PI staining. Experiments were performed in triplicate and results were expressed as the mean±s.d. (b and c) Effects of siRNA against caspase-2, caspase-9, RAIDD, PIDD and RIP. At 24 h after transfection, cells were incubated for 24 h with either vehicle or 50 n of LBH589. Results were evaluated using Annexin-V/PI staining or the JC-1 dye and analyzed with flow cytometric analysis. Results were expressed as the mean±s.d. for three independent experiments and were also analyzed using Student's t-test. *P<0.01 compared with si-control. (d) Western blotting. At 24 h after transfection, cells were incubated for 24 h with either vehicle or 50 n of LBH589 and western blotting was performed.
RAIDD as well as caspase-2 has a critical role in LBH589-induced apoptosis
Next, we performed siRNA experiments targeting caspase-2, caspase-9, RAIDD, PIDD and RIP to disclose the key triggers of apoptosis. Annexin-V/PI assays revealed that siRNA of RAIDD significantly repressed LBH-induced cell death, as did si-caspase-2 (Figure 7b). The proportion of Annexin-V-positive cells was ∼65% in si-control cells and 37% in ST1 si-RAIDD cells. Similar results were obtained with HuT102 cells using siRNA set #1 (Figure 7b), set #2, and set #3 (Supplementary Figure 2C and E). Si-caspase-9 also inhibited LBH589-induced cell death while si-PIDD and si-RIP caused no change (Figure 7b). Similar inhibitory effects by si-RAIDD and si-caspase-2 were observed in JC-1 experiments (Figure 7c). The effect of si-caspase-9 was less extensive than that of si-PIDD or si-RIP (Figure 7c). These results suggest that RAIDD as well as caspase-2 has a critical role in LBH589-induced apoptosis and, importantly, they have active roles upstream of the mitochondria.
RAIDD has an initiating role in LBH589-induced apoptosis
Western blotting revealed that si-caspase-2 suppressed the cleavage of caspase-9, caspase-3 and PARP as well as the expression of caspase-2 itself. However, it could not suppress the activation of RAIDD (Figure 7d). Si-caspase-9 suppressed the cleavage of caspase-3 and PARP as well as the expression of caspase-9 itself, but could not suppress the activation of caspase-2 or RAIDD. Of note, si-RAIDD most effectively suppressed the cleavage of caspase-2, caspase-9, caspase-3 and PARP as well as the upregulation of RAIDD expression. Si-PIDD and si-RIP did not alter the expression profiles of caspases and RAIDD. These changes were basically similar in ST1 and HuT102 cells. An additional siRNA experiment was performed using set #2 and set #3 siRNAs and results were similar to Figure 7d (Supplementary Figure 2D and F). The results indicate that the activation of RAIDD occurs upstream of the activation of caspase-2 and has an initiating role in LBH589-induced apoptosis.
Discussion
DACi achieved significant biological effects in preclinical models of cancer including hematological malignancies which has led to clinical trials.4, 47, 48, 49 Then, it has become important to re-evaluate the mechanisms of tumor cell death caused by DACi. These processes will improve the design of further clinical trials and help in developing novel DACi. In this study, we showed that a newly developed DACi, LBH589, effectively induces ATLL cell death both in vitro and in vivo by a unique mechanism that has not been reported. Our findings uncovered a new role of caspase-2's activation in the induction of apoptosis.
It has been recognized that caspase-2-mediated apoptosis is initiated by death receptors.42, 50 However, recent studies revealed the ability of caspase-2 to engage the intrinsic pathway in response to DNA damage.43, 51, 52, 53 One possibility is that caspase-2 acts indirectly on the mitochondria, by cleaving the pro-apoptotic protein BID or by activating Bax and inducing the release of cytochrome-.44 Another alternative is that caspase-2 directly permeabilizes the mitochondrial membrane and stimulates the release of cytochrome- independently of the Bcl-2 family including Bax and Bcl-2.54 In addition, a recent report indicated that caspase-2 became activated in the so called PIDDosome, a complex of PIDD, caspase-2 and RAIDD, in a p53-dependent manner.45 In our study, si-caspase-2 and si-RAIDD inhibited LBH589-induced apoptosis as well as permeabilization of the mitochondrial membrane. This mechanism is similar to the PIDDosome, however, LBH589-induced apoptosis was independent of p53 and PIDD was not activated. In addition, si-PIDD or si-RIP did not alter LBH589-induced apoptosis. Si-caspase-2 could not suppress the activation of RAIDD while si-RAIDD effectively suppressed the cleavage of caspase-2. These results indicated that LBH589 caused the activation of caspase-2 followed by RAIDD independent of p53 and PIDD. Regarding this point, a previous report suggested that caspase-2's activation occurred in a p53-independent manner.55 Furthermore, two recent studies using PIDD-deficient mice demonstrated the PIDDosome-independent activation of caspase-2.56, 57 From our results, si-caspase-2 attenuated the LBH589-induced permeabilization of the mitochondrial membrane and the mRNA expression of cytochrome- was highly upregulated by LBH589 probably due to cytochrome-'s release. In contrast, most of the Bcl-2 family proteins were not altered by LBH589. In this regard, Robertson et al. suggested that caspase-2 can directly stimulate the intrinsic pathway independent of the Bcl-2 family.43 More recently, Sidi et al. demonstrated a caspase-2-dependent apoptotic program that bypasses a deficiency of p53 and excess of Bcl-2.58 These reports are partly consistent with our own observations. However, our scenario does not work without the activation of RAIDD. Therefore, our results suggest that ATLL cells make use of a unique RAIDD-caspase-2-induced intrinsic pathway, which has not been reported previously. Additionally, as caspase-2-mediated apoptosis requires caspase-9,51 upregulation of caspase-9 expression by LBH589 is advantageous to the induction of apoptosis. Although a number of independent studies strongly support a role for the intrinsic pathway in DACi-induced apoptosis, the mechanism remains to be fully elucidated.1 Our findings may provide some explanation.
Although there is a consensus that the p53 pathway is inactivated by Tax in ATLL cells,59 we recently found that ATLL-related cell lines expressing the wild-type p53 harbor an intact p53 pathway.31 Some previous findings highlighted the importance of p53 in the effect of DACi, however, other studies demonstrated that the expression of wild-type p53 is not necessary for DACi-induced apoptosis.2, 60, 61 Meanwhile, it has been reported that p21, downstream of p53, is activated by DACi independent of p53.62, 63 Indeed, the expression of p21 was increased by LBH589 in ATLL-related cell lines irrespective of their p53 status. In the present study, we clearly showed that LBH589 does not activate p53 in ATLL-related cell lines with the wild-type p53, and LBH589 was also effective against p53-mutant cells via the induction of a p53-like pro-apoptotic response. A similar phenomenon was reported previously in that the DACi FR901228 caused a p53-like pro-apoptotic response in p53 mutant cells and the degradation of the mutant p53 protein.64 We further confirmed that LBH589 caused the degradation of p53 protein in p53 wild-type cells as well as p53-mutant cells. These results suggest that some types of DACi including LBH589 act upstream of the p53 pathway and cause the degradation of even mutated p53 protein via an unknown mechanism.
We also explored the ATLL-specific mechanisms in LBH589-induced cell death. Notably, CCR4 and IL-2R, well-known molecular targets in ATLL therapy, were repressed by the LBH589 treatment.65, 66 We speculated that Tax participates in the suppression of IL-2R, however, cells with high Tax mRNA levels actually showed an increase in Tax mRNA expression after LBH589 treatment. These results indicate that LBH589 can cause apoptosis even in Tax-expressing ATLL cells. In our study, mRNA levels of Tax and HBZ in ATLL-related cell lines were not inversely correlated. In addition, after treatment with LBH589, mRNA levels of Tax and HBZ changed in parallel. These results are not consistent with the idea that HBZ can inhibit Tax's activation.19 Meanwhile, the expression of HBZ-SI was suppressed by LBH589 in most of the ATLL-related cell lines. Although the precise function of HBZ-SI is still under investigation,24 a previous study found that inhibition of HBZ-SI by shRNA resulted in cell growth inhibition in ATLL cells.23 Thus, HBZ-SI could potentially be a molecular target in ATLL therapy and the decrease in HBZ-SI caused by LBH589 treatment may also contribute to the apoptosis of ATLL cells. Consequently, we demonstrated that LBH589 is a promising drug against ATLL, and also identified a novel intrinsic pathway in LBH589-induced apoptosis.
Acknowledgments
We would like to express our gratitude to Dr Mesnard JM, Institut de Biologie, Laboratoire Centre d'étude d'agents Pathogènes et Biotechnologies pour la Santé Montpellier, France. Grant support note: This study was supported in part by a Grant-in-aid for Scientific Research (20590580) from the Japan Society for the Promotion of Science and a grant from Novartis.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Leukemia website (http://www.nature.com/leu)
Supplementary Material
References
- Baylin SB, Ohm JE. Epigenetic gene silencing in cancer—a mechanism for early oncogenic pathway addiction. Nat Rev Cancer. 2006;2:107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006;9:769–984. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer. 2006;1:38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- Xu WS, Parmigiani RB, Marks PA. Histone deacetylase inhibitors: molecular mechanisms of action. Oncogene. 2007;37:5541–5552. doi: 10.1038/sj.onc.1210620. [DOI] [PubMed] [Google Scholar]
- Peart MJ, Tainton KM, Ruefli AA, Dear AE, Sedelies KA, O'Reilly LA, et al. Novel mechanisms of apoptosis induced by histone deacetylase inhibitors. Cancer Res. 2003;63:4460–4471. [PubMed] [Google Scholar]
- Henderson C, Mizzau M, Paroni G, Maestro R, Schneider C, Brancolini C. Role of caspases, Bid, and p53 in the apoptotic response triggered by histone deacetylase inhibitors trichostatin-A (TSA) and suberoylanilide hydroxamic acid (SAHA) J Biol Chem. 2003;14:12579–12589. doi: 10.1074/jbc.M213093200. [DOI] [PubMed] [Google Scholar]
- Mitsiades N, Mitsiades CS, Richardson PG, McMullan C, Poulaki V, Fanourakis G, et al. Molecular sequelae of histone deacetylase inhibition in human malignant B cells. Blood. 2003;101:4055–4062. doi: 10.1182/blood-2002-11-3514. [DOI] [PubMed] [Google Scholar]
- Maiso P, Carvajal-Vergara X, Ocio EM, Lopez-Perez R, Mateo G, Gutierrez N, et al. The histone deacetylase inhibitor LBH589 is a potent antimyeloma agent that overcomes drug resistance. Cancer Res. 2006;66:5781–5789. doi: 10.1158/0008-5472.CAN-05-4186. [DOI] [PubMed] [Google Scholar]
- Fulda S, Debatin KM. Extrinsic versus intrinsic apoptosis pathways in anticancer chemotherapy. Oncogene. 2006;25:4798–4811. doi: 10.1038/sj.onc.1209608. [DOI] [PubMed] [Google Scholar]
- Bao Q, Shi Y. Apoptosome: a platform for the activation of initiator caspases. Cell Death Differ. 2007;14:56–65. doi: 10.1038/sj.cdd.4402028. [DOI] [PubMed] [Google Scholar]
- Nimmanapalli R, Fuino L, Bali P, Gasparetto M, Glozak M, Tao J, et al. Histone deacetylase inhibitor LAQ824 both lowers expression and promotes proteasomal degradation of Bcr-Abl and induces apoptosis of imatinib mesylate-sensitive or -refractory chronic myelogenous leukemia-blast crisis cells. Cancer Res. 2003;63:5126–5135. [PubMed] [Google Scholar]
- Bali P, Pranpat M, Bradner J, Balasis M, Fiskus W, Guo F, et al. Inhibition of histone deacetylase 6 acetylates and disrupts the chaperone function of heat shock protein 90: a novel basis for antileukemia activity of histone deacetylase inhibitors. J Biol Chem. 2005;280:26729–26734. doi: 10.1074/jbc.C500186200. [DOI] [PubMed] [Google Scholar]
- Kovacs JJ, Murphy PJ, Gaillard S, Zhao X, Wu JT, Nicchitta CV, et al. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol Cell. 2005;18:601–607. doi: 10.1016/j.molcel.2005.04.021. [DOI] [PubMed] [Google Scholar]
- George P, Bali P, Annavarapu S, Scuto A, Fiskus W, Guo F, et al. Combination of the histone deacetylase inhibitor LBH589 and the hsp90 inhibitor 17-AAG is highly active against human CML-BC cells and AML cells with activating mutation of FLT-3. Blood. 2005;105:1768–1776. doi: 10.1182/blood-2004-09-3413. [DOI] [PubMed] [Google Scholar]
- Fiskus W, Pranpat M, Bali P, Balasis M, Kumaraswamy S, Boyapalle S, et al. Combined effects of novel tyrosine kinase inhibitor AMN107 and histone deacetylase inhibitor LBH589 against Bcr-Abl-expressing human leukemia cells. Blood. 2006;108:645–652. doi: 10.1182/blood-2005-11-4639. [DOI] [PubMed] [Google Scholar]
- Yamada Y, Tomonaga M. The current status of therapy for adult T-cell leukaemia-lymphoma in Japan. Leuk Lymphoma. 2003;44:611–618. doi: 10.1080/1042819021000055039. [DOI] [PubMed] [Google Scholar]
- Yoshida M. Discovery of HTLV-1, the first human retrovirus, its unique regulatory mechanisms, and insights into pathogenesis. Oncogene. 2005;24:5931–5937. doi: 10.1038/sj.onc.1208981. [DOI] [PubMed] [Google Scholar]
- Marriott SJ, Semmes OJ. Impact of HTLV-I Tax on cell cycle progression and the cellular DNA damage repair response. Oncogene. 2005;24:5986–5995. doi: 10.1038/sj.onc.1208976. [DOI] [PubMed] [Google Scholar]
- Gaudray G, Gachon F, Basbous J, Biard-Piechaczyk M, Devaux C, Mesnard JM. The complementary strand of the human T-cell leukemia virus type 1 RNA genome encodes a bZIP transcription factor that down-regulates viral transcription. J Virol. 2002;76:12813–12822. doi: 10.1128/JVI.76.24.12813-12822.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold J, Yamamoto B, Li M, Phipps AJ, Younis I, Lairmore MD, et al. Enhancement of infectivity and persistence in vivo by HBZ, a natural antisense coded protein of HTLV-1. Blood. 2006;107:3976–3982. doi: 10.1182/blood-2005-11-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh MH, Landry S, Audet B, Arpin-Andre C, Hivin P, Pare ME, et al. HTLV-I antisense transcripts initiating in the 3′LTR are alternatively spliced and polyadenylated. Retrovirology. 2006;3:15. doi: 10.1186/1742-4690-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata K, Hayashibara T, Sugahara K, Uemura A, Yamaguchi T, Harasawa H, et al. A novel alternative splicing isoform of human T-cell leukemia virus type 1 bZIP factor (HBZ-SI) targets distinct subnuclear localization. J Virol. 2006;80:2495–2505. doi: 10.1128/JVI.80.5.2495-2505.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satou Y, Yasunaga J, Yoshida M, Matsuoka M. HTLV-I basic leucine zipper factor gene mRNA supports proliferation of adult T cell leukemia cells. Proc Natl Acad Sci USA. 2006;103:720–725. doi: 10.1073/pnas.0507631103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesnard JM, Barbeau B, Devaux C. HBZ, a new important player in the mystery of adult T-cell leukemia. Blood. 2006;108:3979–3982. doi: 10.1182/blood-2006-03-007732. [DOI] [PubMed] [Google Scholar]
- Yamada Y, Tomonaga M, Fukuda H, Hanada S, Utsunomiya A, Tara M, et al. A new G-CSF-supported combination chemotherapy, LSG15, for adult T-cell leukaemia-lymphoma: Japan Clinical Oncology Group Study 9303. Br J Haematol. 2001;113:375–382. doi: 10.1046/j.1365-2141.2001.02737.x. [DOI] [PubMed] [Google Scholar]
- Tsukasaki K, Utsunomiya A, Fukuda H, Shibata T, Fukushima T, Takatsuka Y, et al. VCAP-AMP-VECP compared with biweekly CHOP for adult T-cell leukemia-lymphoma: Japan Clinical Oncology Group Study JCOG9801. J Clin Oncol. 2007;25:5458–5464. doi: 10.1200/JCO.2007.11.9958. [DOI] [PubMed] [Google Scholar]
- Maeda T, Yamada Y, Moriuchi R, Sugahara K, Tsuruda K, Joh T, et al. Fas gene mutation in the progression of adult T cell leukemia. J Exp Med. 1999;189:1063–1071. doi: 10.1084/jem.189.7.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa H, Yamada Y, Harasawa H, Tsuji T, Murata K, Sugahara K, et al. Sensitivity of adult T-cell leukaemia lymphoma cells to tumour necrosis factor-related apoptosis-inducing ligand. Br J Haematol. 2005;128:253–265. doi: 10.1111/j.1365-2141.2004.05289.x. [DOI] [PubMed] [Google Scholar]
- Yoshida M, Miyoshi I, Hinuma Y. Isolation and characterization of retrovirus from cell lines of human adult T-cell leukemia and its implication in the disease. Proc Natl Acad Sci USA. 1982;79:2031–2035. doi: 10.1073/pnas.79.6.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posner LE, Robert-Guroff M, Kalyanaraman VS, Poiesz BJ, Ruscetti FW, Fossieck B, et al. Natural antibodies to the human T cell lymphoma virus in patients with cutaneous T cell lymphomas. J Exp Med. 1981;154:333–346. doi: 10.1084/jem.154.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa H, Yamada Y, Iha H, Tsukasaki K, Nagai K, Atogami S, et al. Activation of p53 by Nutlin-3a, an antagonist of MDM2, induces apoptosis and cellular senescence in adult T-cell leukemia cells. Leukemia. 2009;23:2090–2101. doi: 10.1038/leu.2009.171. [DOI] [PubMed] [Google Scholar]
- Hasegawa H, Yamada Y, Komiyama K, Hayashi M, Ishibashi M, Sunazuka T, et al. A novel natural compound, a cycloanthranilylproline derivative (Fuligocandin B), sensitizes leukemia cells to apoptosis induced by tumor necrosis factor related apoptosis-inducing ligand (TRAIL) through 15-deoxy-Delta 12, 14 prostaglandin J2 production. Blood. 2007;110:1664–1674. doi: 10.1182/blood-2007-01-068981. [DOI] [PubMed] [Google Scholar]
- Usui T, Yanagihara K, Tsukasaki K, Murata K, Hasegawa H, Yamada Y, et al. Characteristic expression of HTLV-1 basic zipper factor (HBZ) transcripts in HTLV-1 provirus-positive cells. Retrovirology. 2008;5:34. doi: 10.1186/1742-4690-5-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy S, Nakamura N, Vazquez F, Batt DB, Perera S, Roberts TM, et al. Regulation of G1 progression by the PTEN tumor suppressor protein is linked to inhibition of the phosphatidylinositol 3-kinase/Akt pathway. Proc Natl Acad Sci USA. 1999;96:2110–2115. doi: 10.1073/pnas.96.5.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiskus W, Ren Y, Mohapatra A, Bali P, Mandawat A, Rao R, et al. Hydroxamic acid analogue histone deacetylase inhibitors attenuate estrogen receptor-alpha levels and transcriptional activity: a result of hyperacetylation and inhibition of chaperone function of heat shock protein 90. Clin Cancer Res. 2007;13:4882–4890. doi: 10.1158/1078-0432.CCR-06-3093. [DOI] [PubMed] [Google Scholar]
- Cardone MH, Roy N, Stennicke HR, Salvesen GS, Franke TF, Stanbridge E, et al. Regulation of cell death protease caspase-9 by phosphorylation. Science. 1998;282:1318–1321. doi: 10.1126/science.282.5392.1318. [DOI] [PubMed] [Google Scholar]
- Yoshie O, Fujisawa R, Nakayama T, Harasawa H, Tago H, Izawa D, et al. Frequent expression of CCR4 in adult T-cell leukemia and human T-cell leukemia virus type 1-transformed T cells. Blood. 2002;99:1505–1511. doi: 10.1182/blood.v99.5.1505. [DOI] [PubMed] [Google Scholar]
- Mori N, Fujii M, Ikeda S, Yamada Y, Tomonaga M, Ballard DW, et al. Constitutive activation of NF-kappaB in primary adult T-cell leukemia cells. Blood. 1999;93:2360–2368. [PubMed] [Google Scholar]
- Matsuoka M, Jeang KT. Human T-cell leukaemia virus type 1 (HTLV-1) infectivity and cellular transformation. Nat Rev Cancer. 2007;4:270–280. doi: 10.1038/nrc2111. [DOI] [PubMed] [Google Scholar]
- Mori N, Matsuda T, Tadano M, Kinjo T, Yamada Y, Tsukasaki K, et al. Apoptosis induced by the histone deacetylase inhibitor FR901228 in human T-cell leukemia virus type 1-infected T-cell lines and primary adult T-cell leukemia cells. J Virol. 2004;78:4582–4590. doi: 10.1128/JVI.78.9.4582-4590.2004. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Nishioka C, Ikezoe T, Yang J, Komatsu N, Bandobashi K, Taniguchi A, et al. Histone deacetylase inhibitors induce growth arrest and apoptosis of HTLV-1-infected T-cells via blockade of signaling by nuclear factor kappaB. Leuk Res. 2008;32:287–296. doi: 10.1016/j.leukres.2007.05.026. [DOI] [PubMed] [Google Scholar]
- Duan H, Dixit VM. RAIDD is a new ‘death' adaptor molecule. Nature. 1997;385:86–89. doi: 10.1038/385086a0. [DOI] [PubMed] [Google Scholar]
- Robertson JD, Enoksson M, Suomela M, Zhivotovsky B, Orrenius S. Caspase-2 acts upstream of mitochondria to promote cytochrome c release during etoposide-induced apoptosis. J Biol Chem. 2002;277:29803–29809. doi: 10.1074/jbc.M204185200. [DOI] [PubMed] [Google Scholar]
- Guo Y, Srinivasula SM, Druilhe A, Fernandes-Alnemri T, Alnemri ES. Caspase-2 induces apoptosis by releasing proapoptotic proteins from mitochondria. J Biol Chem. 2002;277:13430–13437. doi: 10.1074/jbc.M108029200. [DOI] [PubMed] [Google Scholar]
- Tinel A, Tschopp J. The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science. 2004;304:843–846. doi: 10.1126/science.1095432. [DOI] [PubMed] [Google Scholar]
- Tinel A, Janssens S, Lippens S, Cuenin S, Logette E, Jaccard B, et al. Autoproteolysis of PIDD marks the bifurcation between pro-death caspase-2 and pro-survival NF-kappaB pathway. EMBO J. 2007;26:197–208. doi: 10.1038/sj.emboj.7601473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosato RR, Grant S. Histone deacetylase inhibitors in cancer therapy. Cancer Biol Ther. 2003;1:30–37. doi: 10.4161/cbt.190. [DOI] [PubMed] [Google Scholar]
- Rasheed W, Bishton M, Johnstone RW, Prince HM. Histone deacetylase inhibitors in lymphoma and solid malignancies. Expert Rev Anticancer Ther. 2008;3:413–432. doi: 10.1586/14737140.8.3.413. [DOI] [PubMed] [Google Scholar]
- Ellis L, Pan Y, Smyth GK, George DJ, McCormack C, Williams-Truax R, et al. Histone deacetylase inhibitor panobinostat induces clinical responses with associated alterations in gene expression profiles in cutaneous T-cell lymphoma. Clin Cancer Res. 2008;14:4500–4510. doi: 10.1158/1078-0432.CCR-07-4262. [DOI] [PubMed] [Google Scholar]
- Ahmad M, Srinivasula SM, Wang L, Talanian RV, Litwack G, Fernandes-Alnemri T, et al. CRADD, a novel human apoptotic adaptor molecule for caspase-2, and FasL/tumor necrosis factor receptor-interacting protein RIP. Cancer Res. 1997;57:615–619. [PubMed] [Google Scholar]
- Lassus P, Opitz-Araya X, Lazebnik Y. Requirement for caspase-2 in stress-induced apoptosis before mitochondrial permeabilization. Science. 2002;297:1352–1354. doi: 10.1126/science.1074721. [DOI] [PubMed] [Google Scholar]
- Zhivotovsky B, Orrenius S. Caspase-2 function in response to DNA damage. Biochem Biophys Res Commun. 2005;331:859–867. doi: 10.1016/j.bbrc.2005.03.191. [DOI] [PubMed] [Google Scholar]
- Mhaidat NM, Wang Y, Kiejda KA, Zhang XD, Hersey P. Docetaxel-induced apoptosis in melanoma cells is dependent on activation of caspase-2. Mol Cancer Ther. 2007;6:752–761. doi: 10.1158/1535-7163.MCT-06-0564. [DOI] [PubMed] [Google Scholar]
- Robertson JD, Gogvadze V, Kropotov A, Vakifahmetoglu H, Zhivotovsky B, Orrenius S. Processed caspase-2 can induce mitochondria-mediated apoptosis independently of its enzymatic activity. EMBO Rep. 2004;6:643–648. doi: 10.1038/sj.embor.7400153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castedo M, Perfettini JL, Roumier T, Valent A, Raslova H, Yakushijin K, et al. Mitotic catastrophe constitutes a special case of apoptosis whose suppression entails aneuploidy. Oncogene. 2004;23:4362–4370. doi: 10.1038/sj.onc.1207572. [DOI] [PubMed] [Google Scholar]
- Manzl C, Krumschnabel G, Bock F, Sohm B, Labi V, Baumgartner F, et al. Caspase-2 activation in the absence of PIDDosome formation. J Cell Biol. 2009;185:291–303. doi: 10.1083/jcb.200811105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim IR, Murakami K, Chen NJ, Saibil SD, Matysiak-Zablocki E, Elford AR, et al. DNA damage- and stress-induced apoptosis occurs independently of PIDD. Apoptosis. 2009;14:1039–1049. doi: 10.1007/s10495-009-0375-1. [DOI] [PubMed] [Google Scholar]
- Sidi S, Sanda T, Kennedy RD, Hagen AT, Jette CA, Hoffmans R, et al. Chk1 suppresses a caspase-2 apoptotic response to DNA damage that bypasses p53, Bcl-2, and caspase-3. Cell. 2008;133:864–877. doi: 10.1016/j.cell.2008.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pise-Masison CA, Jeong SJ, Brady JN. Human T cell leukemia virus type 1: the role of Tax in leukemogenesis. Arch Immunol Ther Exp (Warsz) 2005;53:283–296. [PubMed] [Google Scholar]
- Ruefli AA, Ausserlechner MJ, Bernhard D, Sutton VR, Tainton KM, Kofler R, et al. The histone deacetylase inhibitor and chemotherapeutic agent suberoylanilide hydroxamic acid (SAHA) induces a cell-death pathway characterized by cleavage of Bid and production of reactive oxygen species. Proc Natl Acad Sci USA. 2001;98:10833–10838. doi: 10.1073/pnas.191208598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insinga A, Monestiroli S, Ronzoni S, Gelmetti V, Marchesi F, Viale A, et al. Inhibitors of histone deacetylases induce tumor-selective apoptosis through activation of the death receptor pathway. Nature Med. 2005;11:71–76. doi: 10.1038/nm1160. [DOI] [PubMed] [Google Scholar]
- Richon VM, Sandhoff TW, Rifkind RA, Marks PA. Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acetylation. Proc Natl Acad Sci USA. 2000;97:10014–10019. doi: 10.1073/pnas.180316197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui CY, Ngo L, Xu WS, Richon VM, Marks PA. Histone deacetylase (HDAC) inhibitor activation of p21WAF1 involves changes in promoter-associated proteins, including HDAC1. Proc Natl Acad Sci USA. 2004;101:1241–1246. doi: 10.1073/pnas.0307708100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagosklonny MV, Trostel S, Kayastha G, Demidenko ZN, Vassilev LT, Romanova LY, et al. Depletion of mutant p53 and cytotoxicity of histone deacetylase inhibitors. Cancer Res. 2005;65:7386–7392. doi: 10.1158/0008-5472.CAN-04-3433. [DOI] [PubMed] [Google Scholar]
- Ishida T, Ueda R. CCR4 as a novel molecular target for immunotherapy of cancer. Cancer Sci. 2006;11:1139–1146. doi: 10.1111/j.1349-7006.2006.00307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zhang M, Ju W, Waldmann TA. Effective treatment of a murine model of adult T-cell leukemia using depsipeptide and its combination with unmodified daclizumab directed toward CD25. Blood. 2009;113:1287–1293. doi: 10.1182/blood-2008-04-149658. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.