Summary
PPARγ and Wnt signaling are central positive and negative regulators of adipogenesis, respectively. Here we identify the groucho family member TLE3 as a transcriptional integrator of the PPARγ and Wnt pathways. TLE3 is a direct target of PPARγ that participates in a feed-forward loop during adipocyte differentiation. TLE3 enhances PPARγ activity and functions synergistically with PPARγ on its target promoters to stimulate adipogenesis. At the same time, induction of TLE3 during differentiation provides a mechanism for termination of Wnt signaling. TLE3 antagonizes TCF4 activation by β-catenin in preadipocytes, thereby inhibiting Wnt target gene expression and reversing β-catenin-dependent repression of adipocyte gene expression. Transgenic expression of TLE3 in adipose tissue in vivo mimics the effects of PPARγ agonist and ameliorates high fat diet-induced insulin resistance. Our data suggest that TLE3 acts as a dual function switch, driving the formation of both active and repressive transcriptional complexes that facilitate the adipogenic program.
Introduction
Adipocytes are specialized cells that store excess energy in the form of triglycerides and also serve an endocrine function, secreting adipokines that influence systemic energy homeostasis (Halaas et al., 1995; Steppan et al., 2001; Yamauchi et al., 2001). The formation of adipocytes is dependent on peroxisome proliferator-activated receptor γ (PPARγ) and CCAAT/enhancer binding proteins (CEBPs), transcription factors that coordinately regulate genes involved in lipid metabolism (Freytag et al., 1994; Tontonoz et al., 1994b). Ectopic expression of PPARγ programs fibroblasts to differentiate into adipocytes (Tontonoz et al., 1994c). Many of the genes characteristic of the differentiated adipocyte are direct targets of PPARγ and/or C/EBPα (Christy et al., 1989; Dalen et al., 2004; Schoonjans et al., 1996; Tontonoz et al., 1995; Tontonoz et al., 1994b). PPARγ is also the therapeutic target of the thiazolidinedione anti-diabetic drugs that promote lipid storage and adipokine production in adipose tissue (Lehmann et al., 1995).
Cell-specific gene regulation is driven by DNA-binding factors working in concert with cofactors (Roeder, 2005). Several cofactors have been identified that interact with PPARγ and facilitate its action (Cho et al., 2009; Ge et al., 2002; Gelman et al., 1999; Grontved et al.; Qi et al., 2003). Interestingly, however, few if any of these factors are regulated components of the differentiation program, i.e. their expression does not change during differentiation. Rather, they act as constitutive factors to permit PPARγ-dependent transcription. In line with this constitutive role, increasing the expression of most PPARγ coactivators above the basal state does not enhance adipogenesis. Expression of PPARγ coactivator-1α (PGC-1α) is highly regulated in brown adipose tissue (BAT) and promotes the expression of genes important for thermogenesis (Puigserver et al., 1998). However, PGC-1α is not believed to play an important role in the development of white adipose tissue (WAT), and therefore the question of whether coactivators may be regulated components of the white adipose tissue differentiation program remains to be addressed.
The Wnt signaling pathway as a major physiological inhibitor of adipogenesis that is responsible for maintaining preadipocytes in an undifferentiated state (Ross et al., 2000). Wnts are secreted glycoproteins that signal through frizzled receptors leading to the inhibition of the Disheveled/axin/GSK3β complex, thereby preventing the targeted degradation of β-catenin (MacDonald et al., 2009). Accumulation of nuclear β-catenin activates TCF/LEF transcription factors and increases the expression of Wnt target genes (Molenaar et al., 1996). A number of studies have shown that Wnt opposes the actions of PPARγ in adipogenesis (Bennett et al., 2002; Liu and Farmer, 2004). Blocking TCF signaling, for example by ectopic expression of a dominant negative or conditional deletion of β-catenin in mesenchyme, is sufficient to promote differentiation (Arango et al., 2005; Ross et al., 2000). It has also been suggested that the Wnt signaling pathway is down regulated through the action of PPARγ (Moldes et al., 2003). However, the molecular mechanisms by which Wnt blocks adipogenesis, as well as those that serve to integrate the PPARγ and Wnt signaling pathways, remain to be elucidated. Previously, we developed a high-throughput phenotypic screening platform for the identification of modulators of adipogenesis (Waki et al., 2007). We utilized this approach to identify small molecules that drive differentiation through the induction of PPARγ expression (Park et al., 2010; Waki et al, 2007). Here we report the adaptation of this strategy for cDNA library screening and the identification of the groucho family member transducin-like enhancer of split 3 (TLE3) as an adipogenic factor. Despite the fact that TLE proteins have been studied primarily as transcriptional repressors, we find that TLE3 is a potent facilitator of PPARγ activity on its target promoters. We further uncover a mechanism for Wnt-dependent inhibition of adipogenesis and demonstrate that TLE3 antagonizes the Wnt pathway during differentiation. These studies identify TLE3 as a dual function modulator of adipogenesis that augments PPARγ action and inhibits Wnt signaling.
Results
A high-throughput screen for cDNA modulators of adipogenesis
We previously validated a phenotype-based high-throughput screen for chemical modulators of adipogenesis (Waki et al., 2007). We modified this approach to screen genome-size cDNA libraries in 384-well format and used it to identify candidate regulators of adipocyte differentiation (Fig. S1A). 10T1/2 cells were retro-transfected simultaneously with a luciferase reporter driven by the −5.4 kb aP2 promoter and a collection of 18,292 individually-spotted mammalian cDNA expression vectors. The day after transfection, cells were treated with insulin and a PPARγ agonist (rosiglitazone) to induce adipogenic differentiation; luciferase activity was evaluated four days later (Fig. S1B). The screen was run in duplicate; each plate contained cDNAs encoding PPARγ and CEBPα as positive controls. Relative intensities were normalized to their respective plate median values, and mean values and standard deviations calculated for each well from the replicate screens to identify hits. For reconfirmation, a set of 96 cDNAs encoding putative adipogenesis regulators was chosen and re-assayed, and luciferase values were normalized to empty vector controls (Fig. S1C). A number of cDNAs were identified as activators of aP2-driven luciferase activity in our screen. PPARγ emerged as the most potent activator, and several additional known adipogenic factors were also represented, including CEBPα, CEBPδ, early B-cell factor 1 (EBF1), and mitogen activated protein kinase kinase 6 (MAPKK6). Select cDNAs were subsequently evaluated for adipogenic potential by means of stable retroviral transduction of 10T1/2 cells (Fig. S1D). TLE3 was chosen for further analysis, as this factor had not previously been linked with adipocyte biology.
TLE3 expression is regulated during adipocyte differentiation
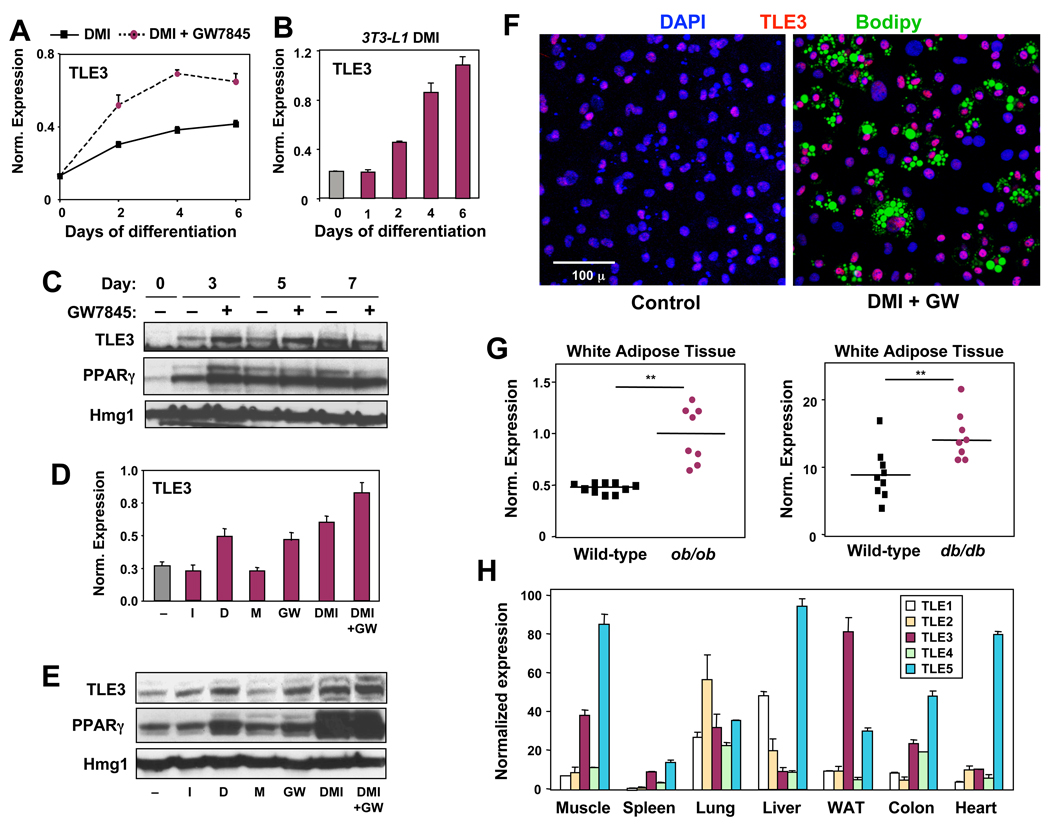
We reasoned that if TLE3 was a regulated component of the differentiation program, then its expression should change over the course of differentiation. Indeed, analysis of a time course of 10T1/2 and 3T3-L1 differentiation revealed that TLE3 mRNA expression rose during differentiation and was further enhanced by treatment of the cells with PPARγ agonist (Fig. 1A,B). A strong increase in TLE3 protein expression was also observed, and again treatment with GW7845 increased its levels (Fig. 1C). To determine which component of the differentiation cocktail was primarily responsible for TLE3 induction, 10T1/2 cells were stimulated for 2 days with insulin (I), dexamethasone (D), methylisobutyl xanthine (M), and/or GW7845. TLE3 mRNA and protein expression were found to be responsive to both dexamethasone and PPARγ ligand (Fig. 1D,E).
Fig 1. Regulation of TLE3 expression during adipocyte differentiation.
(A) Realtime PCR analysis of TLE3 mRNA expression during differentiation of 10T1/2 cells treated with differentiation cocktail (DMI = 1 µM dexamethasone, 0.5 mM IBMX, 5 µg/ml insulin) or DMI and GW7845 (20 nM). mRNA expression in this and all subsequent figures was normalized to 36B4 control. (B) TLE3 mRNA expression during differentiation of 3T3-L1 preadipocyte differentiation. Cells were treated as in A. (C) Immunoblot analysis of TLE3 protein expression in 10T1/2 cells treated with DMI plus DMSO (−) or DMI plus GW7845 (20 nM). (D) Realtime PCR analysis of TLE3 mRNA expression in 10T1/2 cells treated for 2 d with individual components of the differentiation cocktail. D, 1 µM dexamethasone; M, 0.5 mM IBMX; I, 5 µg/ml insulin; GW, 20 nM GW7845. (E) Immunoblot analysis of total cell lysates from cells treated as in (D). (F) TLE3 expression visualized by fluorescent confocal microscopy in undifferentiated (control) and differentiating (DMI + 20 nM GW for 4 d) 10T1/2 cells. TLE3 (red) colocalizes with DAPI (blue) staining nuclei, with highest expression observed in bodipy-staining (green) adipocytes. (G) Realtime PCR analysis of TLE3 mRNA expression in epididymal white adipose tissue from ob/ob and db/db mice. N = 8–10 per group, ** P<0.01. (H) Realtime PCR analysis of the relative tissue distribution of mRNAs encoding murine TLE (1–5) family members. Error bars represent mean +/− S.D. See also Figure S1.
We also employed confocal immunofluorescence microscopy to visualize TLE3 expression. In 10T1/2 cells, TLE3 expression colocalized with DAPI staining, consistent with nuclear localization (Fig. 1F). Furthermore, the level of TLE3 protein increased robustly in cells induced to differentiate (DMI+GW), and lipid-laden mature adipocytes were consistently TLE3-positive (Fig. 1F). In vivo, TLE3 protein expression was readily detected in WAT and BAT, but not in adjacent skeletal muscle (Fig. S1E). Interestingly, immunoblot analysis revealed more prominent expression of TLE3 in WAT compared to BAT (Supplemental Figure 2A). In fractionated mouse WAT, TLE3 was more abundant in adipocytes compared to the stromal-vascular fraction (Fig. S2B).
We also investigated whether TLE3 levels were altered in murine models of obesity. We found that ob/ob and db/db mice expressed more TLE3 mRNA in WAT compared to WT controls (Fig. 1G and Fig. S2C). Examination of the tissue distribution of the mammalian TLE family of proteins revealed that, although TLE3 was expressed in a number of tissues, its expression was particularly prominent in WAT (Fig. 1H).
TLE3 is a direct target of PPARγ
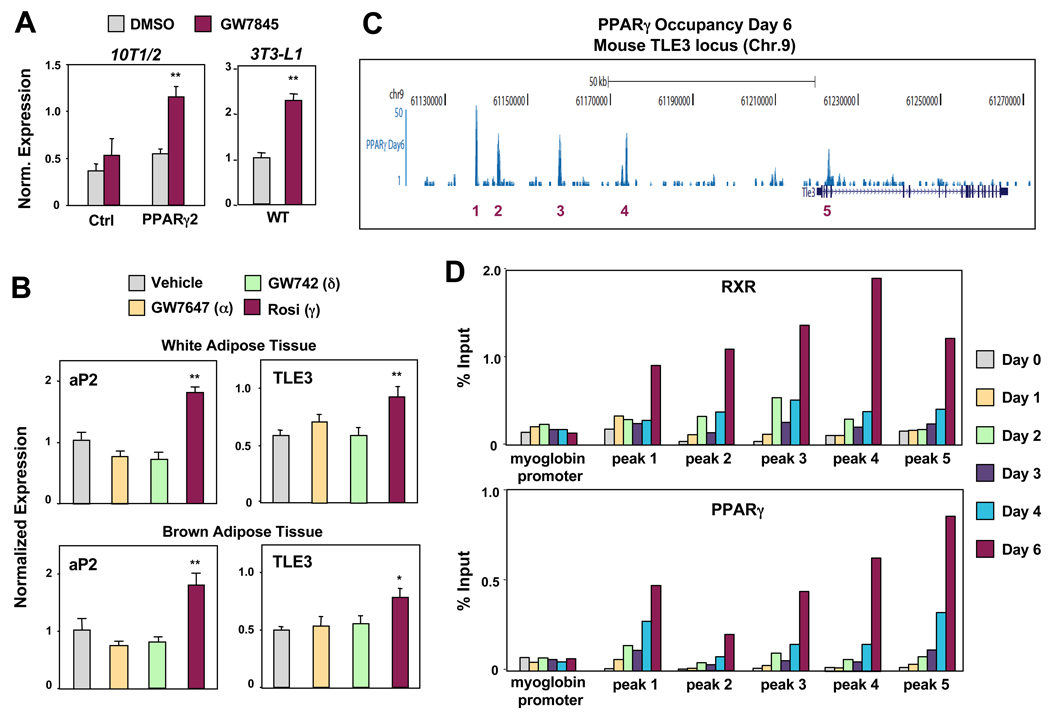
The observation that TLE3 expression increased during differentiation and was responsive to PPARγ agonist administration led us to explore whether TLE3 might be a direct target of PPARγ. We confirmed that short-term treatment of PPARγ-expressing 10T1/2 cells with PPARγ agonist induced TLE3 mRNA (Fig. 2A). A similar induction by GW7845 was seen in 3T3-L1 cells. TLE3 expression was also responsive to PPARγ administration in vivo. Treatment of mice with PPARγ (rosiglitazone), but not PPARα (GW7647) or PPARδ (GW742) agonists induced TLE3 mRNA in WAT and BAT (Fig. 2B).
Fig 2. TLE3 is a PPARγ target gene.
(A) Realtime PCR analysis of TLE mRNA expression in PPARγ2-expressing 10T1/2 preadipocytes treated with 100 nM GW7845 for 2 d (left) and 3T3-L1 preadipocytes treated with DMI + 20 nM GW for 2 d (right). (B) Induction of TLE3 and aP2 mRNA by PPARγ agonist in white and brown adipose tissue in vivo. Mice were gavaged twice daily for 2 d with vehicle, PPARα agonist (10 mg/kg GW7647), PPARδ (10 mg/kg GW742) agonist, or PPARγ agonist (30 mg/kg rosiglitazone). Male mice, n = 10 per group, * P<0.05, ** P<0.01. (C) High-resolution ChIP-Seq analysis of PPARγ bindings sites within the mouse TLE3 locus from 3T3-L1 cells differentiated for 6 d with DMI. These data are from the deep sequencing study of Nielsen et al. (2008). (D) Differentiation dependent PPARγ/RXR occupancy in the vicinity of the TLE3 gene. ChIP of PPARγ and RXR in 3T3-L1 cells was followed by qPCR analysis using primers flanking individual PPARy bindings sites in the TLE3 gene region at the indicated time points. A region of the myoglobin promoter served as a negative control. Error bars represent mean +/− S.D. See also Fig. S2.
To address whether PPARγ bound directly to the TLE3 promoter, we employed chromatin immunoprecipitation (ChIP) assays combined with deep sequencing (Nielsen et al., 2008). Through analysis of the global PPARγ DNA binding data of Nielsen et al. we identified several putative PPARγ binding regions in the mouse TLE3 locus (Fig. 2C). Four of these regions were located more than 50 kb upstream of the transcriptional start site (peaks 1–4), while one was located in an intronic region (peak 5). Sequence analysis revealed that DR-1 sequences were associated with each of these peaks, increasing our confidence that these were likely to be bona fide PPARγ binding sites. To confirm this, we performed ChIP-PCR analysis over the time course of 3T3-L1 adipocyte differentiation. Both RXR and PPARγ bound to the 5 putative binding sites in the TLE3 genomic region in a differentiation-dependent manner (Fig. 2D). No binding was observed with a control region from the myoglobin promoter. These results indicated that TLE3 was a direct PPARγ target gene and suggested that induction of TLE3 by PPARγ might contribute to a positive feedback loop to promote adipogenesis.
TLE3 is an adipogenic factor that acts synergistically with PPARγ
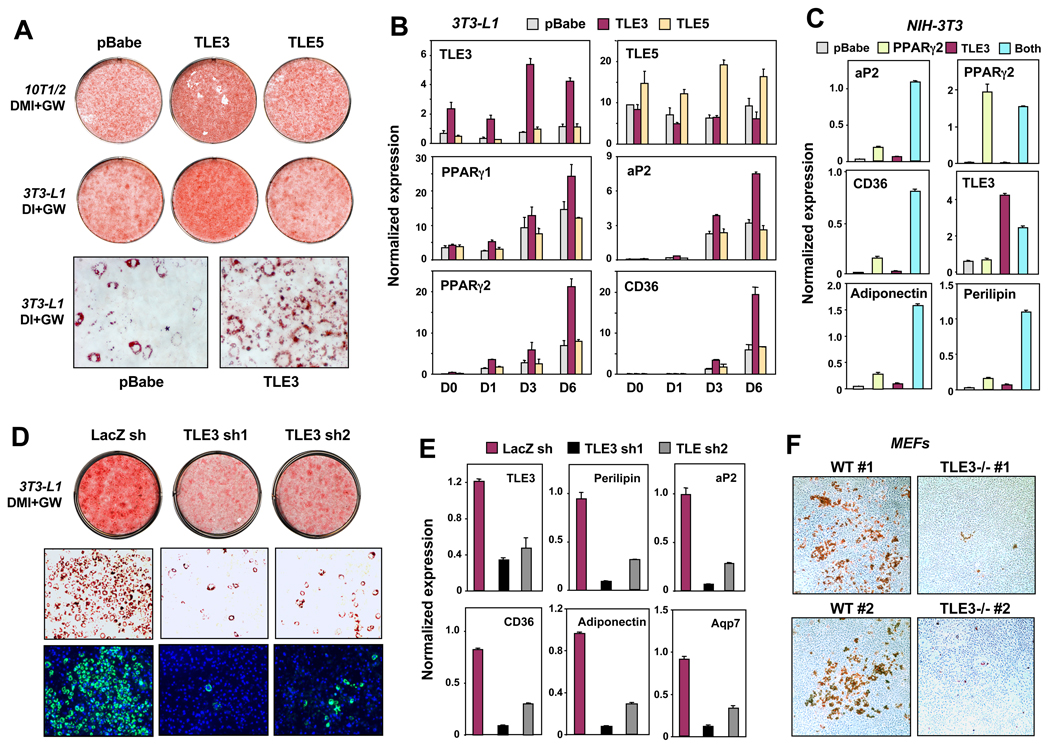
To further validate the adipogenic action of TLE3, we generated stable cell lines. Importantly, these lines expressed TLE3 at only moderately elevated levels, consistent with the degree of TLE3 regulation during adipogenesis. Retroviral TLE3 expression strongly promoted adipocyte differentiation in both 10T1/2 and 3T3-L1 cells, as assessed by oil red O staining (Fig. 3A). We also assayed the activity of TLE5 which lacks a WD40 domain and is postulated to act as a dominant negative (Chen and Courey, 2000). TLE5 expression did not affect differentiation, suggesting that a functional WD40 domain is required for the adipogenic effect. Prior studies have identified loss of function groucho point mutations (Jennings et al., 2006). Introduction of a loss-of-function point mutation in the WD40 domain (V708D) blocked the ability of TLE3 to stimulate adipogenesis (Supplemental Fig. 3B). Gene expression analysis confirmed increased expression of PPARγ and many of its downstream target genes in cells stably expressing TLE3 (Fig. 3B and Fig. S3A,B).
Fig 3. TLE3 is a transcriptional modulator of adipogenesis.
(A) Analysis of differentiation by oil red-O (ORO) staining of retrovirally-derived stable 10T1/2 and 3T3-L1 cell lines expressing vector, TLE3 or TLE5. 10T1/2 and 3T3-L1 cells were stimulated to differentiate with DMI + 20 nM GW for 7 d and 10 d, respectively. Top: plate view of ORO stained cultures; bottom: microscopic view. (B) Realtime PCR analysis of adipogenic gene expression in 3T3-L1 cells transduced with TLE3 or TLE5. (C) NIH-3T3 cells stably expressing TLE3, PPARγ or both from retroviral vectors were stimulated to differentiate with dexamethasone (2 µM), insulin (5 µg/ml) and GW7845 (20 nM) for 10 d. (D) Adipogenic potential of 3T3-L1 cells expressing lentivirally-delivered shRNAs targeting TLE3 or LacZ shRNA control. Infected 3T3-L1 cells were stimulated to differentiate with DMI + 10 nM GW for 7d. Top: plate view of ORO stained cultures; Middle: microscopic view; bottom: microscopic view of Bodipy (lipid) and DAPI (nuclei) stained cells. (E) Expression of PPARγ target genes in 3T3-L1 cells expressing TLE3 shRNAs as determined by realtime PCR. Cells were treated with DMI+GW (10 nM) for 7d. (F) Adipogenic potential of individually-derived primary WT or TLE3 null mouse embryonic fibroblasts (MEFs). Cells were stained with ORO after stimulation with DMI + 1 µM rosiglitazone for 6 d, followed by 6 d with rosiglitazone and insulin. Error bars represent mean +/− S.D. See also Fig. S3.
We turned to the NIH-3T3 fibroblast system to investigate potential combinatorial effects of TLE3 and PPARγ. NIH-3T3 cells are unable to differentiate into adipocytes because they lack PPARγ. Expression of TLE3 in this context had little if any effect on the expression of PPARγ target genes (Fig. 3C and Fig. S3C). Consistent with prior work (Tontonoz et al., 1994c), introduction of PPARγ conferred the ability to accumulate lipid and express adipogenic genes (Fig. 3C and data not shown). The combination of PPARγ and TLE3 was highly synergistic, both in terms of target gene expression and morphological differentiation (Fig. 3C and data not shown). These data demonstrate that the effects of TLE3 on adipogenic gene expression are highly dependent on PPARγ expression.
In order to address whether endogenous TLE3 activity contributes to adipogenesis, we used retroviruses encoding inhibitory shRNAs to knockdown TLE3 expression. TLE3 knockdown was confirmed by immunoblotting (Fig. S3D). Stable 3T3-L1 cell lines expressing two different shRNA sequences targeting TLE3 exhibited reduced differentiation capacity compared to those expressing a control shRNA (Fig. 3D). In agreement with the morphological differentiation, the expression of adipocyte-selective genes was also impaired in TLE3 knockdown cells (Fig. 3E and Fig. S3E).
As a complement to the knockdown studies we generated mice lacking TLE3 using a Genetrap embryonic stem cell line obtained from The Sanger Centre (XP0165), in which the insertion of the trapping vector resulted in a null allele. Unfortunately, homozygous deletion of TLE3 results in embryonic lethality due to multiple developmental defects (data not shown). However, we succeeded in deriving primary MEFs from TLE3−/− mice. Multiple preparations of primary embryonic fibroblasts derived from TLE3−/− mice showed reduced capacity for adipogenesis compared to WT controls (Fig. 3F). Adipogenic gene expression was correspondingly reduced in MEFs lacking TLE3 (Fig. S3F). Together, these data demonstrate that TLE3 is a physiologic component of the adipogenic program that works cooperatively with PPARγ to promote differentiation.
TLE3 facilitates PPARγ-dependent gene activation
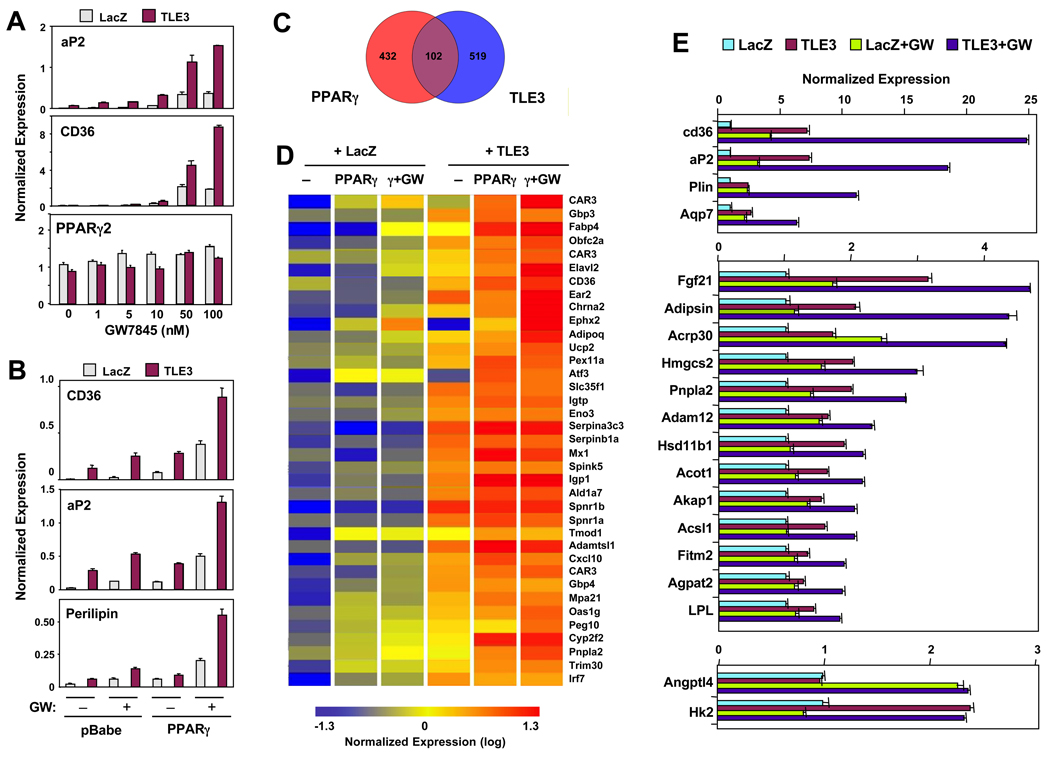
Next we addressed the mechanism of TLE3 action in adipogenesis. As chronic (stable) overexpression of TLE3 was able to induce expression of PPARγ, aP2 and CD36 in undifferentiated 10T1/2 cells, it was possible that TLE3 was functioning primarily by inducing PPARγ expression, leading to secondary effects on downstream target genes. To address whether acute expression of TLE3 could also induce adipogenic gene expression, we transduced 10T1/2 cells expressing the coxsackie adenovirus receptor (CAR) with an adenoviral vector encoding TLE3. Surprisingly, TLE3-tranduced cells showed increased expression of aP2 and CD36 and enhanced response to PPARγ agonist, despite expressing similar endogenous levels of both PPARγ1 and PPARγ2 (Fig. 4A and data not shown). These effects occurred in the absence of morphological differentiation. Many PPARγ target genes are also regulated by C/EBPs; however, the expression of C/EBPs was also not altered by TLE3 (Fig. S4A). The ability of TLE3 to regulate target genes without affecting PPARγ expression suggested that TLE3 might be enhancing PPARγ activity. We further explored this possibility by acutely expressing TLE3 in the presence of stably expressed PPARγ. As shown in Figure 4B, TLE3 strongly promoted the ability of PPARγ to induce its target genes in response to agonist. These data suggest that the primary effect of TLE3 is on PPARγ activity and that the elevated level of PPARγ observed in cells stably expressing TLE3 is secondary to increased adipocyte differentiation.
Fig 4. Overlapping transcriptional profiles of PPARγ and TLE3 regulated genes.
(A) Confluent 10T1/2 cells expressing the coxsackie adenovirus receptor (CAR) were infected with LacZ-or TLE3-expressing adenoviruses and simultaneously treated with GW7845 at the indicated concentrations for 48 h. Gene expression was determined by realtime PCR. (B) Effects of TLE3 expression on adipogenic genes are dependent on the level PPARγ expression. Postconfluent 10T1/2 CAR cells stably expressing vector (pBabe) or PPARγ2 were infected overnight with LacZ or TLE3 expressing adenovirus. 48 h post infection cells were treated with DMSO or 10 nM GW7845. (C) Venn diagram of overlapping transcriptional programs of PPARγ and TLE3 in 10T1/2 cells. (D) Heatmap representation of selected PPARγ and TLE3-responsive genes (> 1.4 change) identified by analysis of Affymetrix arrays. (E) Validation of gene expression changes from microarray analysis by realtime PCR. PPARγexpressing 10T1/2 cells infected with LacZ or TLE3 adenovirus were treated with DMSO or 10 nM GW7845 for 24 h. Error bars represent mean +/− S.D. See also Fig. S4.
We next employed transcriptional profiling to compare the PPARγ and TLE3 transcriptomes. We acutely expressed TLE3 and PPARγ individually and in combination in 10T1/2 cells by viral transduction. Gene expression was analyzed after 48 h using Affymetrix Mouse Gene 1.0 ST Arrays. Despite previous characterization of TLEs as repressors, a greater number of transcripts were induced in response to TLE3 expression than suppressed (Fig S4B). Furthermore, the transcriptional programs engaged by TLE3 and PPARγ were highly overlapping, as indicated by the Venn diagram in Fig. 4C. Approximately 25% of PPARγ-responsive genes overlapped with those regulated by TLE3, indicating a significant degree of specificity for the PPARγ transcriptional program. Cluster analysis of the entire set of overlapping genes is presented in Supplemental Fig. 4C and Table S1 and S2. A more limited set of genes whose expression was altered more than 1.4 fold is presented in Fig. 4D. Importantly, the common regulated genes identified in this analysis included many established PPARγ targets, including aP2, CD36, and adiponectin.(Fig. 4D). Furthermore, it is clear from the heat map of Fig. 4D that PPARγ and TLE3 have additive or synergistic effects on the expression of a large battery of genes. In agreement with the additive effects of these factors in adipogenesis (Fig. 3C), maximal expression of this gene set was achieved in the presence of TLE3, PPARγ, and GW78545 (Fig. 4D). Our global transcriptional analysis did not reveal a significant effect of TLE3 expression on the programs of other nuclear receptors, including LXR, FXR or RAR (data not shown).
We validated our array results for a number of direct PPARγ targets by realtime PCR analysis (Fig. 4E). The results confirmed that the vast majority of established direct PPARγ target genes in adipocytes were additively responsive to TLE3 and PPARγ. For some genes, such as aP2 and perilipin, the response was synergistic. A minority of putative PPARγ target genes was responsive to PPARγ but not TLE3 (e.g. Angptl4) or TLE3 but not PPARγ (e.g. HK2) (Fig. 4E).
Differentiation-dependent recruitment of TLE3 to PPREs
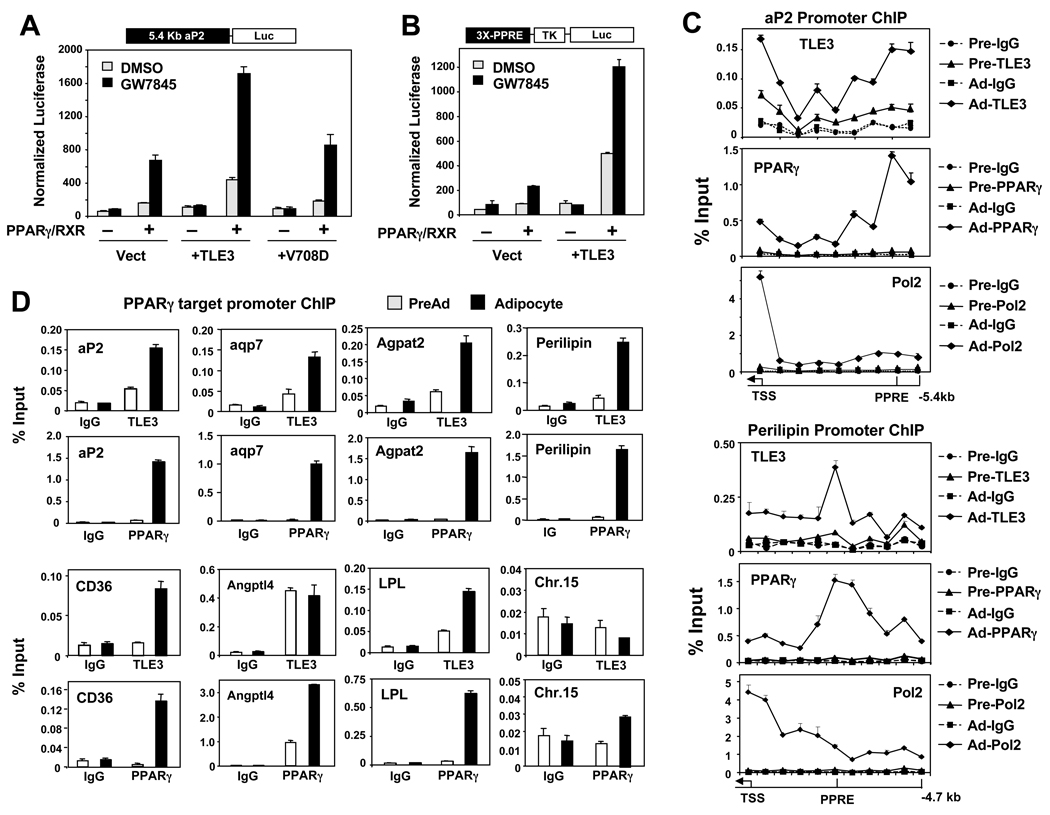
The ability of TLE3 to selectively promote PPAR γ-dependent target gene expression suggested that TLE might function as a positive transcriptional cofactor for PPARγ. To test this idea, we performed transfection assays with a −5.4kb aP2-luciferease reporter. 10T1/2 cells transfected with PPARγ and RXR expression vectors showed robust luciferase activity in the presence of GW7845 (Fig. 5A). TLE3 had no effect in the absence of PPARγ and RXR, but enhanced reporter activity when these nuclear receptors were present. Introduction of the point mutation in the WD40 domain of TLE3 (V708D) that prevented TLE from inducing adipogenesis also prevented coactivation of PPARγ (Fig. 5A). TLE3 showed similar ability to enhance PPARγ-dependent transcription when a reporter driven by isolated PPAR response elements (PPREs) was used (Fig. 5B). This result strongly suggested that TLE3 was acting to increase aP2 promoter transcription by increasing PPARγ activity on its cognate response element, rather than by acting through other binding sites. Preliminary studies indicated that TLE3 also promoted the action of PPARα and PPARδ in transfection assays (Fig. S5A).
Fig 5. TLE3 coactivates PPARγ-dependent gene expression.
(A) Analysis of −5.4kb aP2 enhancer activation by coexpression of PPARγ/RXR and TLE3 or TLE3 carrying a mutation in the WD40 (V708D) domain in undifferentiated 10T1/2 cells treated with DMSO or 100 nM GW7845. (B) TLE3 enhances PPARγ/RXR activation of a luciferase reporter driven by minimal PPAR responsive elements (3X-PPRE). Cells were treated as in (A). (C) Differentiation-dependent recruitment of TLE3 to the endogenous aP2 and perilipin promoters in 10T1/2 cells. ChIP assays were carried out using TLE3, PPARγ, RNA Pol2, and control IgG in preadipocytes (Pre) and adipocytes (day 6 DMI + 20nM GW; Ad). Individual regions of the aP2 and perilipin upstream regions from the (TSS) to 5.4 kb upstream and 4.7 kb upstream were amplified by realtime PCR, respectively. (D) ChIP analysis of TLE3 recruitment to the PPREs of PPARγ target gene promoters in differentiated adipocytes. A non-specific region in Chr. 15 was used as a control. ChIP signals were quantified by realtime PCR. Error bars represent mean +/− S.D. See also Fig. S5.
To provide additional evidence for the ability of TLE3 to enhance PPARγ activity on PPREs, we performed ChIP assays in 10T1/2 cells. We initially analyzed PPARγ and TLE3 occupancy along the 5’-flanking region of the aP2 gene extending from −5.4 kb to the transcriptional start site. Strong differentiation-dependent binding of PPARγ was detected in the region corresponding to the previously characterized PPREs in the aP2 enhancer (Tontonoz et al., 1994a). Remarkably, TLE3 was also found to occupy the aP2 promoter/enhancer in a differentiation-dependent manner (Fig. 5C,D). A strong peak was detected in the region corresponding to the PPREs, coincident with PPARγ binding, and an additional peak was noted closer to the proximal promoter. As expected, Pol2 occupancy was detected at the transcriptional start site in adipocytes but not preadipocytes. Analysis of the 5’-flanking region of the perilipin gene revealed similar differentiation-dependent co-occupancy of PPARγ and TLE3 in the region of the PPRE (Fig. 5C).
We next addressed whether TLE3 was localized with PPARγ on the regulatory regions of other adipocyte PPARγ target genes. Indeed, regulatory regions containing previously validated PPREs from the CD36, Aqp7, Agpat2, LPL, and perilipin genes were all co-occupied by PPARγ and TLE3 in a differentiation-dependent manner (Fig. 5D). By contrast, there was no enrichment of PPARγ or TLE3 on control regions not containing PPREs, such as a sequence on Chr. 15 (Fig. 5D). Interestingly, we did not observe differentiation-dependent changes in TLE3 occupancy on the Angptl4 promoter. This finding was consistent with our prior observation that TLE3 expression did not enhance Angptl4 expression (Fig. 4E).
We also investigated whether TLE3 could be localized to PPARγ-containing complexes in adipocytes. We were unable to detect a direct interaction between TLE3 and PPARγ using standard in vitro pull-down assays (data not shown). However, two different lines of evidence support the hypothesis that these proteins are present in a transcriptional complex. First, TLE3 could be identified in complexes immunoprecipitated from 10T1/2 adipocytes with a PPARγ antibody (Fig. S5B). Second, TLE3 was independently identified as a PPARγ-associated protein in mass spectrometry analysis of PPARγ-containing fractions purified from 3T3-L1 adipocytes (Mandrup and colleagues, manuscript in preparation). Together, these data indicate that TLE3 is present in PPARγ-containing differentiation-dependent transcriptional complexes and facilitates adipocyte differentiation by enhancing the activity of PPARγ on its target promoters.
TLE3 counteracts Wnt signaling in preadipocytes
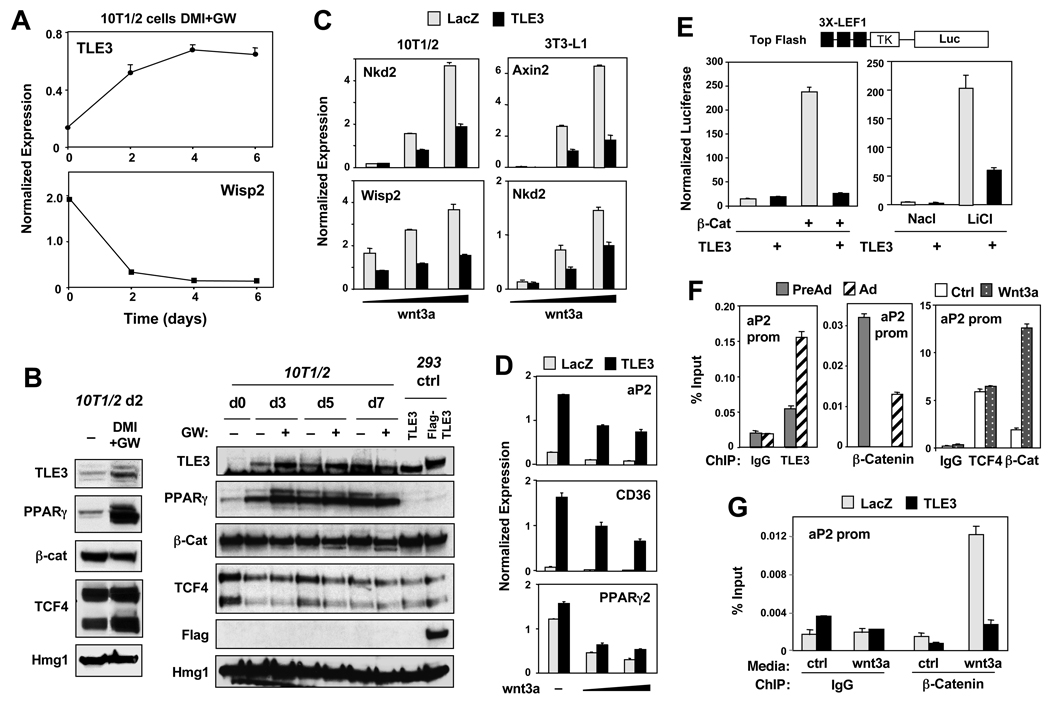
Studies in drosophila have characterized the TLE ortholog groucho as a repressor that binds TCF and inhibits Wnt signaling (Cavallo et al., 1998). We hypothesized that increased TLE3 expression during differentiation might provide a mechanism for counteracting Wnt signaling in this context. In support of this idea, we found that TLE3 could be identified in complexes immunoprecipitated from 3T3-L1 adipocytes with a TCF4 antibody (Fig. S5B). Furthermore, the expression of Wisp2, a Wnt-responsive gene highly expressed in preadipocytes, was strongly downregulated during adipogenesis, coincident with upregulation of TLE3 (Fig. 6A). We observed substantial increases in TLE3 protein levels 2 days following stimulation of preadipocytes with differentiation cocktail, and this correlated with the time course of Wnt target gene decline. In fact, TLE3 protein expression was more highly regulated during differentiation compared to the expression of either TCF4 or β-catenin (Fig. 6B).
Fig 6. TLE3 counteracts Wnt repression of adipocyte promoters through corepression of TCF4.
(A) Realtime PCR analysis of mRNA expression in 10T1/2 cells induced to differentiate with DMI and GW7845. (B) Immunoblot analysis of 10T1/2 cells stimulated to differentiate with DMI −/20 nM GW as indicated. (C) Undifferentiated 10T1/2 and 3T3-L1 cells stably expressing CAR were infected with LacZ or TLE3 adenovirus overnight. Cells were treated 2 d post infection with control or Wnt3a conditioned media for 24 h. Gene expression was determined by realtime PCR. (D) 10T1/2 cells were transduced with control or TLE3 adenovirus vectors and treated for 24 h with Wnt-conditioned media. Gene expression was determined by realtime PCR. (E) Effect of TLE3 transfection on activation of the Wnt-responsive Top-Flash reporter by β-catenin (left) and GSK-3β inhibitor LiCl (25 mM for 24 h) (Right) in 293T cells. (F) Reciprocal occupancy of β-catenin and TLE3 on the aP2 promoter. ChIP assays of aP2 promoter binding were carried out for TLE3, TCF4 and β-catenin in pre-adipocytes (Pre-Ad) and adipocytes (Ad). (G) 10T/12 cells stably expressing CAR were infected overnight with LacZ or TLE3 adenovirus. 48 h post-infection cells were treated with control or Wtn3a conditioned media for 24 h. ChIP assays were carried out using control IgG and β-catenin antibodies. Note, the data for TLE3 in A and B are the same as shown in Figure 1 and are included here for comparison. Error bars represent mean +/− S.D. See also Fig. S6.
This led us to propose that TLE3 may antagonize β-catenin binding to TCF during differentiation and thereby inhibit TCF action. To test this possibility, we expressed TLE3 in preadipocytes and then challenged cells with control or Wnt-conditioned media. TLE3 blunted the induction of both Nkd2 and Wisp2 upon Wnt activation in 10T1/2 cells (Fig. 6C). A similar result was observed in undifferentiated 3T3-L1 cells for Axin2 and Nkd2 (Fig. 6C). We also found that TLE3 expression could counteract the repressive action of Wnt3a on adipogenic genes, including aP2, CD36, and to a lesser degree, PPARγ (Fig. 6D).
To test whether TLE3 could directly inhibit Wnt-dependent transcription, we performed transient transfection assays using the Wnt-responsive TOP-FLASH reporter (3X LEF1). Cotransfection of a TLE3 expression vector antagonized the ability of β-catenin to activate the TOP-FLASH reporter. Similarly, TLE3 expression blocked activation of the TOP-FLASH reporter in cells stimulated with LiCl, a potent inhibitor of GSK3β and activator of TCF-dependent transcription (Fig. 6E).
Although the ability of β-catenin and TCF to inhibit adipogenesis is well-documented, the mechanisms involved are poorly understood. Recent studies in other systems have shown that β-catenin can act as a repressor of transcription through a TCF/Lef1-dependent mechanism (Blauwkamp et al., 2008). We hypothesized that TCF-β-catenin complexes might be exerting repressive effects directly at the promoters of adipogenic genes. Remarkably, we found using ChIP that TCF and β-catenin colocalized along the −5.4 kb aP2 promoter/enhancer in undifferentiated cells (Fig. S6 and 6F). This occupancy decreased upon differentiation into adipocytes, consistent with Wnt signaling diminishing over the course of differentiation (Fig. 6F). Interestingly, treatment of preadipocytes with Wnt3a-conditioned media increased β-catenin occupancy, but TCF4 occupancy remained constant. Thus, recruitment of β-catenin was associated with repression of the aP2 promoter in undifferentiated cells. Furthermore, the presence of β-catenin was inversely correlated with the presence of TLE3 (Fig. 6F).
To address the effect of TLE3 on recruitment of β-catenin, we used adenoviral vectors to express TLE3 in undifferentiated cells. ChIP assays demonstrated that TLE3 strongly inhibited β-catenin occupancy of the aP2 promoter, both in the presence and absence of exogenous Wnt (Fig. 6G). Together, our observations suggest that increased TLE 3 expression during adipogenesis promotes differentiation by two related mechanisms. First, it binds together with PPARγ to adipocyte gene promoters and facilitates PPARγ-dependent activation. Second, TLE3 forms complexes with TCFs that fail to recruit β-catenin, thus relieving the repression of adipocyte promoters.
Expression of TLE3 in adipose tissue mimics PPARγ activation
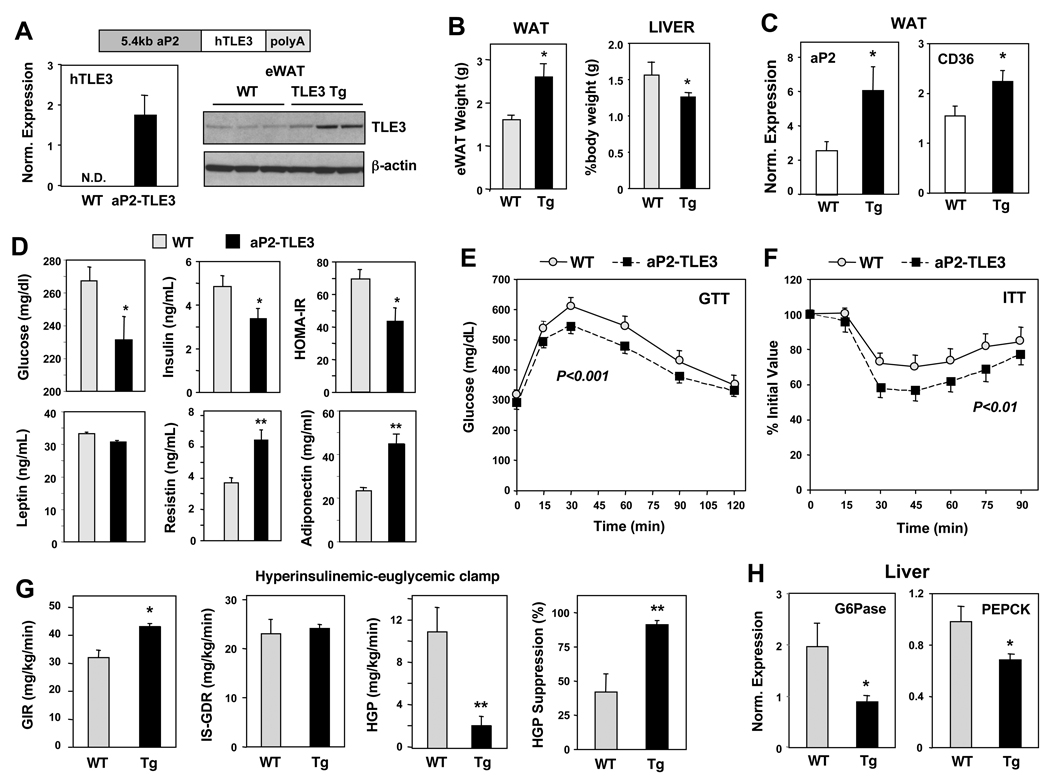
Because our studies indicated that TLE3 selectively promoted the PPARγ transcriptional program in cultured cells, we investigated its activity in adipose tissue in vivo. We generated transgenic mice expressing human TLE3 in adipose tissue using the −5.4 kb aP2 promoter/enhancer (Fig. 7A). Adipose-selective expression of transgenic TLE3 was confirmed using qPCR primers specific for human TLE3 (Fig. 7A). Immunoblot analysis revealed modestly increased TLE3 protein levels in WAT and BAT from aP2-TLE3 mice (Fig. 7A and Fig. S7A). DEXA analysis revealed a trend toward increased fat mass, but no differences in total body weight, bone mineral density or lean body mass between WT and aP2-TLE3 mice (Table S3). However, aP2-TLE3 mice fed a high-fat diet for 10 weeks exhibited increased epidydmal WAT mass when compared to WT littermates (Fig. 7B). By contrast, liver mass was slightly reduced when corrected for body weight. Furthermore, realtime PCR analysis demonstrated that expression of the PPARγ target genes aP2 and CD36 was modestly elevated in WAT from aP2-TLE3 mice (Fig. 7C). Notably, the magnitude of these changes was consistent with the expected effect of synthetic PPARγ agonist (Fig. 2B). Plasma levels of the PPARγ-responsive adipokines adiponectin and resistin were also found to be elevated in aP2-TLE3 mice (Fig. 7D). However, plasma leptin levels were not different between groups. The lack of a change in leptin levels despite increase fat mass is consistent with prior reports that PPARγ ligands suppress leptin expression (De Vos et al., 1996).
Fig 7. Transgenic expression of TLE3 in adipose tissue ameliorates insulin resistance.
(A) Expression of TLE3 in epididymal WAT of aP2-TLE3 transgenic mice. Left: realtime PCR analysis of human TLE3 transcript; right: immunoblot analysis of TLE3 and β-actin. Chow-fed C57BL6 male mice, N = 3 per group. (B) Weight of eWAT and liver % of body mass in high-fat fed WT and aP2-TLE3 (Tg) transgenic mice. 14 weeks HF-diet, N = 6 per group, * P<0.05. (C) Realtime PCR analysis of aP2 and CD36 expression in eWAT from 14 week HF-fed mice. N=5–6 per group, * P<0.05. (D) Plasma glucose, insulin, leptin, resistin, and adiponectin levels from mice fed high-fat diet for 11-weeks. Homeostatic model assessment-insulin resistance (HOMA-IR) index was computed from glucose and insulin values. 6 h fasting, N = 5–6 per group, * P<0.05. (E and F) Glucose tolerance test (GTT) and insulin tolerance test (ITT) wa performed after 6-weeks and 9-weeks of HF-diet, respectively. N = 5–6 per group. P values were determined by 2-way ANOVA followed by Bonferroni post-hoc test. (G) Improved insulin sensitivity and reduced hepatic glucose production in aP2-TLE3 transgenic mice. Glucose infusion rate (GIR), insulin-stimulated glucose disposal (IS-GDR) and hepatic glucose production (HGP) was determined by hyperinsulinemic euglycemic clamp studies of 14-week HF-fed mice WT and aP2-TLE3 mice. N =5–6 per group, * P<0.05, ** P<0.01. (H) Reduced expression of the gluconeogenic enzymes glucose-6-phosphatase (G-6-Pase) and PEPCK determined by realtime PCR analysis of livers from 14-week HF fed mice. N = 5–6 per group, * P<0.05. Error bars represent mean +/− S.D. See also Fig. S7.
Elevated adiponectin levels often coincide with improved insulin sensitivity. We therefore investigated whether adipose tissue expression of TLE3 would impact glucose homeostasis in the context of diet-induced obesity. Compared to WT controls, aP2-TLE3 mice fed a high fat diet for 8 weeks showed reduced fasting (6 h) glucose, insulin, and homeostatic model assessment insulin resistance (HOMA-IR) index (Fig. 7D). All of these parameters are indicative of improved glucose homeostasis. To further confirm this, we performed glucose and insulin tolerance tests (GTT, ITT). Adipose TLE3 expression was associated with both increased glucose tolerance and increased insulin sensitivity (Fig. 7F). A trend toward improved glucose homeostasis was observed in a second line of transgenic mice (aP2-TLE3B) that exhibited lower TLE3 expression (Fig. S7B). We did not observe a difference in glucose tolerance between aP2-TLE3 and WT mice fed a normal chow diet (Fig. S7C). Metabolic cage analysis revealed modest but significant reduction in energy expenditure in Tg compared to WT mice (Fig. S7D). Thus, although the aP2-TLE3 transgene is also expressed in BAT, these data suggest that the improvement in glucose homeostasis is unlikely to be due to enhanced fatty acid oxidation in BAT. Food intake was also reduced in aP2-TLE3 mice compared to WT.
Finally, we performed hyperinsulinemic euglycemic clamp studies in order to more directly measure insulin sensitivity and gain insight into the tissues involved in the phenotype of aP2-TLE3 mice. aP2-TLE3 mice required an increased glucose infusion rate (GIR) to maintain euglycemia, a sensitive measurement of whole-body insulin sensitivity (Fig. 7G). Insulin stimulated glucose disposal rate (IS-GDR), which primarily reflects skeletal muscle insulin sensitivity, was similar between genotypes (Fig. 7G). By contrast, the suppression of hepatic glucose production (HGP) by insulin was improved in aP2-TLE3 mice relative to controls. These findings indicate that the observed changes in GIR largely reflected improved hepatic insulin sensitivity. Histological analysis of livers revealed that aP2-TLE3 mice had reduced lipid accumulation in the liver (Fig. S7E). The expression of gluconeogenic genes was reduced in aP2-TLE3 mice in accordance with the HGP results (Fig. 7H). Interestingly, a selective effect on HGP compared to IS-GDR is consistent with the effect of low-dose TZD administration (Kubota et al., 2006). Thus, our results are in line with the expected effects of modest stimulation of the adipocyte PPARγ pathway.
Discussion
Here we outline a role for a member of the highly conserved groucho transcription factor family as a facilitator of nuclear receptor action during cell differentiation. TLE3 is a direct target for regulation by PPARγ and functions in a feed-forward loop with PPARγ to promote differentiation. Mechanistically, TLE3 functions as a coregulator of both PPARγ and Wnt signaling, driving the formation of active and repressive transcriptional complexes on the promoters of adipocyte genes. The dual ability of TLE3 to function as a coactivator for PPARγ and a corepressor for TCF provides an elegant mechanism for the integration of pro- and anti-adipogenic signals during adipocyte development.
The function of TLE proteins in adipocyte development has not previously been investigated. Our discovery of TLE3 as a coactivator for PPARγ-dependent transcription was unexpected, as groucho and mammalian TLEs have been primarily studied for their roles as corepressors (Chen and Courey, 2000). TLE proteins lack a DNA-binding domain, and therefore their ability to regulate transcription is believed to be dependent on interaction with other proteins. Several transcription factors are known to interact with TLEs, including PAX, Hes, Engrailed, and TCFs (Buscarlet and Stifani, 2007). TLEs are recruited to silence gene expression in various contexts through direct interactions with histones and histone modifying enzymes (Chen et al., 1999; Sekiya and Zaret, 2007). Our demonstration that TLE3 expression acts to positively reinforce PPARγ action has uncovered a previously unrecognized mode of action for this transcriptional cofactor.
Previous work has identified several coactivators that interact with PPARγ (Cho et al., 2009; Ge et al., 2002; Gelman et al., 1999; Grontved et al.; Louet et al., 2006; Qi et al., 2003; Takahashi et al., 2002). Unlike TLE3, however, the levels of these factors are not regulated during differentiation. They are likely required for the efficient action of PPARγ and other transcription factors, but are not utilized as developmental switches per se. PGC-1α, a coregulator whose expression is highly regulated by physiological stimuli, is critical for the in brown adipocyte thermogenic program, but is not believed to play a central role in white adipose differentiation (Puigserver et al., 1998). The high expression of TLE3 in WAT, relative to BAT, leads us to speculate that TLE3 may function as a white adipocyte counterpart to PGC-1α in brown adipocytes. Transcriptional profiling supports this, as the brown adipocyte markers, PRDM16, Cidea, Elovl3, Ucp1, and PGC-1a were not upregulated in 10T1/2 cells expressing TLE3.
TLE3 mRNA and protein expression accumulate during preadipocyte differentiation and in response to PPARγ activation. Thus, coactivation of PPARγ by TLE3 may serve as a feed-forward mechanism to enhance differentiation. Expression of TLE3 at levels present in differentiated adipocytes promotes preadipocyte differentiation, and this effect is highly dependent on PPARγ expression. Indeed, co-expression of PPARγ and TLE3 has a synergistic effect on the expression of a number of terminal adipocyte genes. Moreover, we found a high degree of overlap between the transcriptional programs regulated by TLE3 and PPARγ, indicating that TLE3 exerts a preferential effect on the PPARγ signaling pathway in this cell type. Further studies will be needed to determine whether there may be additional transcription factors other than PPARγ involved in TLE3 signaling in preadipocytes. Mechanistic studies indicate that TLE3 is recruited along with PPARγ to PPREs in adipocyte target genes in a differentiation-dependent manner. PPARγ and TLE3 can be localized to common transcription complexes by immunoprecipitation and biochemical purification, although the two proteins do not appear to interact directly.
The Wnt signaling pathway is important for the maintenance and proliferation of preadipocytes (Ross et al., 2000). Differentiation is accompanied by the suppression of Wnt signaling and the concurrent activation of PPARγ (Ross et al., 2002). Surprisingly, the mechanisms underlying this switch are poorly understood. In particular, it is unclear how the Wnt pathway is shut off. We propose that induction of TLE3 expression is a component of a developmental switch that silences Wnt signaling and allows adipogenesis to proceed. TCF4 and β-catenin are present on the aP2 promoter in preadipocytes, and this correlates with the suppression of transcription. In the course of differentiation, endogenous TLE3 is recruited to differentiation-dependent adipocyte promoters, where it can interact directly with TCF4 and compete for the binding of β-catenin. Forced expression of TLE3 in preadipocytes displaces β-catenin and relieves repression of differentiation-dependent genes. Interestingly, a similar mode of TCF action has recently been proposed to operate in the context of skin differentiation (Nguyen et al., 2006). TCF3 was found to actively repress epidermal and sebaceous gland differentiation in the stem cell compartment through repression of lipid metabolism genes such as PPARγ and CD36.
There is precedence for dual function transcriptional cofactors. It is becoming increasingly clear that the strict labels of “coactivator” and “corepressor” may not accurately reflect the complex interactions of some of these nuclear proteins. For example, in addition to coactivating TCF/Lef1,β-catenin can also act as a repressor (Blauwkamp et al., 2008). The nuclear receptor cofactor RIP140 has also been reported to perform both coactivator and corepressor functions (Debevec et al., 2007; Subramaniam et al., 1999). Furthermore, the functional roles of transcriptional coregulators may be context-specific and vary with the transcriptional machinery present in a particular cell. One possibility is that TLE3 is directing chromatin remodeling and generating a chromatin structure that facilitates PPARγ-dependent transcription.
The physiological importance of TLE3 for the adipocyte program is illustrated by the demonstration that suppression of TLE3 expression compromises preadipocyte differentiation and PPARγ target gene expression. In addition, we showed that expression of TLE3 from the adipose-selective aP2 promoter in mice mimics the effect of synthetic PPARγ agonist administration. aP2-TLE3 transgenic mice challenged with a high-fat diet were partially protected against insulin resistance. Clamp studies showed that the improvement in glucose handling in aP2-TLE3 mice was largely attributable to improved hepatic insulin sensitivity. This result is in line with previously reported effects of low-dose TZD treatment. Submaximal doses of pioglitazone have been shown to increase the glucose infusion rate and suppress HGP in the absence of major effects on IS-GDR (Kubota et al., 2006).
Improvements in hepatic insulin sensitivity may reflect redistribution of triglycerides away from liver and into adipose tissue. We found that aP2-TLE3 mice have increased adipose tissue, reduced liver mass, and reduced hepatic lipid accumulation on high-fat diet. A similar finding was reported in ob/ob animals expressing an adiponectin transgene (Kim et al., 2007). Furthermore, adiponectin has been shown to act directly on the liver to suppress gluconeogenesis (Combs et al., 2001). Therefore, our demonstration that aP2-TLE3 mice have higher plasma adiponectin levels provides a plausible mechanistic explanation for the beneficial effects of TLE3 on systemic glucose metabolism. Interestingly, despite having reduced food intake, leptin levels were not elevated in aP2-TLE3 mice, suggestive of a change in leptin sensitivity. Additional studies will be needed to explore this issue. Since TLE3 and aP2 are also expressed in macrophages, it will be interesting to address the function of TLE3 in this cell type and its potential contribution to the phenotype of the aP2-TLE3 mice. Finally, given the lethality of global TLE3-deficiency, future in vivo loss of function studies will necessitate the generation of tissue-selective conditional deletions of TLE3.
Experimental Procedures
Cell Culture
Confluent 10T1/2 and 3T3-L1 cells were stimulated to differentiate with DMEM containing 10% FBS, 1 µM dexamethasone, 0.5 mM isobutylmethylxanthine, and 5 µg/ml insulin for 2 d, followed by 5 µg/ml insulin alone. When specified, PPARγ agonists GW7845 or rosiglitazone was included. NIH-3T3 cells were differentiated by 2 d treatment dexamethasone, insulin, and GW7845, followed by insulin and GW7845 alone. Stable cells expressing TLE3 (Puro), PPARγ (Hygro), or CAR (Neo) were generated using pBabe retroviral vectors (Hummasti and Tontonoz, 2006). For MEF studies, TLE3 null animals were generated from ES gene trap line XP0165 obtained from The Wellcome Trust Sanger Institute. Heterozygous animals were bred to generate E13 embryos that were used to derive fibroblasts. Adipocyte differentiation was induced by treating confluent MEFs with DMEM containing 10% FBS, 0.5 mM isobutylmethylxanthine, 1 µM dexamethasone, 5 µg/ml insulin and 1 µM rosiglitazone for 6 d. Subsequently, cells were treated for 6 d with insulin and rosiglitazone. LacZ or TLE3 expressing adenoviruses were generated as described in (Zelcer et al., 2009). Control and Wnt3a conditioned media was prepared using L cells (ATCC) as described (Waki et al., 2007).
Cell-based cDNA screen
An arrayed library of 18,292 human and mouse full-length cDNAs from the OriGene TrueClone Collection was screened by high-throughput reverse transfection of 10T1/2 cells as described (Cho et al., 2006; Waki et al., 2007). Briefly, a 15 µl mixture of FuGENE 6 and luciferase reporter (20 ng/well) in serum-free medium was added to pre-spotted 384-well plates containing 62.5 ng of plasmid DNA per well. PPARγ, LIP, and GAL4-LXR were used as controls. After a 30 min incubation, 2,000 cells in 20 µl of DMEM supplemented with 20% FBS were added to each well. The following day, 5 µl of differentiation-induction media containing insulin (5 µg/ml) and rosiglitazone (1 µM) were added. Plates were incubated for 4 more d and luciferase activity determined. Relative intensities were normalized to their respective plate median values then normalized by log2 transformation, and mean values and standard deviations calculated for each well from the replicate screens. These values were then reverse log2 transformed, and the ratio of “afa” (derived from means) to “mfa” (derived from standard deviations) calculated to penalize wells for replicate quality. Each cDNA was ranked by afa score. A secondary screen was performed on a set of 96 cDNAs chosen from the top afa/mfa scores re-assayed in quadruplicate, and raw luciferase values were normalized to empty vector controls.
shRNA Plasmids
TLE3 shRNA constructs were designed using BLOCK-IT RNAi designer tool (Invitrogen). Sense and antisense oligos were annealed and cloned into pENTR/U6 plasmid (Invitrogen). Using the LR recombinase (Invitrogen), shRNA constructs were recombined into a gateway adapted pBabe-Puro plasmid. The following shRNA oligos were used: LacZ shRNA CACCGGGCCAGCTGTATAGACATCTCGAA AGATGTCTATACAGCTGGCCC, TLE3sh1 CACCGCACAAGCAGACAGAGATT GCCGAAGCAATCTCTGTCTGCTTGTGC, and TLE3sh2 CACCGGGCCA GCTGTATAGACATCTCGAAAGATGTCTATACAGCTGGCCC. Only sense strands are shown.
Luciferase Reporter Assay
10T1/2 cells were seeded in 24 well culture plates at 90% confluence. Cells were cotransfected with 100 ng of pGL3-aP2-luciferase, or 100 ng of PTK-3XPPRE-luciferase, and 100 ng of pCMX-PPARγ, 20 ng of pCMX-RXR, 100 ng of pCMX-TLE3, 100 ng of pCMX-TLE3(V708D) and 5 ng of renila control vector using Lipofectamine 2000 (Invitrogen). 48 h later cells were treated with DMSO or 100 nM GW7845 in DMEM with 1% FBS for 24 h. Luciferase activity was determined with STOP&GLO (Promega) and a GLOMAX luminometer (Promega). Firefly luciferase activity was normalized to Renila luciferase. Wnt-reporter activity was determined by cotransfecting 50 ng of TOP-FLASH reporter, 25 ng of constitutively active β-catenin (S37A), 500 ng of pCMX-TLE3, and 15 ng of Renila luciferase in 293T cells. Luciferase activity was determined 48 h after transfection with Fugene 6 (Roche).
Gene Expression and Microarray Analysis
Total RNA was isolated using Trizol reagents (Invitrogen) and reverse transcribed using iScript cDNA synthesis kit (Biorad). cDNA was quantified by realtime PCR using SYBR Green (Diagenode) and an ABI 7900 instrument. Gene expression levels were determined by using a standard curve. Each gene was normalized to 36B4. Primers used for realtime PCR are listed in Table S5. For microarray experiments, 10T1/2 cells stably expressing CAR, −/+ PPARγ were infected overnight with LacZ or TLE3 expressing adenovirus. 48 h after infection cells were stimulated with DMSO or 10 nM GW7845 for 24 h. RNA was pooled from 6 biological replicates and processed in the UCLA Microarray Core Facility using GeneChip Mouse Gene 1.0 ST Arrays (Affymetrix). The results were analyzed using GenespringGX (Agilent).
Nuclear Extracts
Cells washed with 1X PBS and incubated with TEN-buffer (10mM Tris-Cl pH 8, 100 mM NaCl, 1 mM EDTA pH 8). Cells allowed to swell on ice for 15 min in 10 mM HEPES, 10 mM KCl, 0.1 mM EGTA, 0.1 mM EDTA, 1 mM DTT + complete proteinase inhibitor (Roche). Mixed with 0.6% NP-40 alternative (Calbiochem) for 10 sec and centrifuged at 12,000 g. Nuclear pellet was resuspended in ice cold 20mM HEPES, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 25% glycerol, 1 mM DTT + complete proteinase inhibitor (Roche). Nuclear extracts were spun at 12,000 g for 5 min and the supernatant used for further analysis.
Protein analysis
Proteins were diluted in Nupage loading dye (Invitrogen), heated at 70°C for 20 min, and run on 4–12% Bis-Tris Gel (Invitrogen). Proteins were transferred to hybond ECL membrane (GE healthcare) and blotted using TLE3 (M-201, Santa Cruz or 11372-1-AP, Proteintech Group), PPARγ (81B8, Cell Signaling), Hmg1 (556528, BD Pharmingen), TCF4 (C48H11, Cell Signaling), or β-catenin (H-102, Santa Cruz) antibodies. For immunoprecipitation, nuclear extracts were diluted in IP buffer (20 mM Tris, 137 mM NaCl, 2 mM EDTA, 1% NP-40, 10% glycerol) and pre-cleared with ProteinA agarose beads (Santa Cruz). Extracts were mixed with IgG (PP64, Millipore), TCF4 (C48H11, Cell signaling) or PPARγ (81B8, Cell signaling) antibodies and incubated with beads. After spinning, beads were washed with IP buffer and protein eluted for immunoblotting.
Chromatin Immunoprecipitation
ChIP experiments were performed according to standard protocols (Nielsen et al., 2008). Lysed cells were sonicated using a Bioruptor (Diagenode) according to the manufacturer’s protocol, and chromatin was immunoprecipitated with antibodies against TLE3 (11372-1-AP, Proteintech Group), TCF4 (C48H11, Cell Signaling), RNA Pol2 (CTD4H8, Millipore), IgG (PP64, Millipore), β-catenin (610154, BD Transduction Laboratories), PPARγ (H-100, sc7196; Santa Cruz Biotechnologies) or RXR (D197, sc774; Santa Cruz Biotechnologies) overnight at 4°C in the presence of protein A beads (GE Healthcare). DNA enrichment was quantified by real-time PCR (MX-3000; Stratagene or ABI 7900 (ABI)) using SYBR green Master Mix (Diagenode or Sigma-Aldrich). Primers used for these studies are listed in Table S4. Occupancy was quantified using a standard curve and normalized to input DNA.
Immunofluorescence
10T1/2 cells were plated in gelatin (0.2%) treated glass bottom dishes (Mat TLK Corp.) and differentiated with DMI+20nM GW7845. At day 4 of differentiation cells were fixed with 4% PFA (1XPBS) and washed with PBS. Cells were permebialized with 0.1% triton and blocked with 3%BSA. Cells were incubated with TLE3 antibody in 3%BSA overnight, washed and incubated with Alexa Fluor 555 (A21429, Invitrogen) for 1 h. After washing, cells were stained with 1 µg/ml Bodipy 493/503 (D3922, Invitrogen) or DAPI (Invitrogen) and washed with PBS. Cells were visualized with LSM 510 confocal laser scanning microscope (Carl Zeiss).
Animal Studies
Male C57BL6 ob/ob, db/db, and WT littermates were acquired from Jackson laboratory. Mice were sacrificed at 3 months of age. Transgenic mice were generated at the UCLA transgenic core facility. The −5.4kb enhancer region of aP2 (Graves et al., 1992) was subcloned into PCR2.1 containing TLE3 cDNA followed by bovine growth hormone polyA (TLE3-PolyA). The linearized construct was gel purified (Zymol) and microinjected into C57BL6 fertilized zygotes. Founders were identified by PCR using the following primers, Forward: AGGGAGAACCAAAGTTGAGAAAT and Reverse: GTCTTCTCGTTTGCCAGCTT. At 12 weeks of age aP2-TLE3 transgenic mice (F2 generation) and their wild-type littermates were fed a 60% high-fat diet (Research Diets) for the indicated times. For glucose tolerance tests, mice were fasted for 6 h and challenged with an i.p. injection of glucose (2 g/kg). For insulin tolerance tests, mice were fasted for 3 h and given an i.p. injection of insulin (1 U/kg). Blood glucose levels were monitored using the ACCU-CHEK active glucometer (Roche). Serum adiponectin levels were determined by Elisa (B-Bridge International Inc.) and insulin, resistin, and leptin were determined by multiplex immunoassay (Milliplex Kit, Millipore) after 6 h fast. Body composition and bone mineral density was determined by DEXA analysis. Glucose infusion rate (GIR), insulin stimulated-glucose disposal rate (IS-GDR), and glucose production was determined by euglycemic hyperinsulinemic clamp as described previously (Hevener et al., 2003; Hevener et al., 2007; Steele, 1959). No differences in clamp glucose or steady state insulin concentration were observed between the two genotypes of mice. Indirect calorimetry was performed using a Columbus Instruments Comprehensive Lab Animal Monitoring System (CLAMS). Animals were placed individually in chambers for three consecutive days at ambient temperature (26.5 °C) with 12 h light/dark cycles. Animals had free access to food and water. Respiratory measurements were made in 20 min intervals after initial 12 h acclimation period. Energy expenditure was calculated from VO2 and RER using the Lusk equation, EE in cal/min=(3.815+1.232 X RER) X VO2 in ml/min (McLean and Tobin, 1987). Statistical significance for EE measurements was determined by using 2-way-ANOVA.
Supplementary Material
Acknowledgments
We thank Stephen Young and Karen Reue for helpful discussions and Loren Fong for help with histology. P.T. is an investigator of the Howard Hughes Medical Institute and was also supported by NIH grants HL090553 and DK063491. C.J.V. was supported by NIH training grant HL069766. E.S. was supported by a Career Development Award from the American Diabetes Association, The McDonald’s Center for Type 2 Diabetes and Obesity, and NIH grant DK081003.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arango NA, Szotek PP, Manganaro TF, Oliva E, Donahoe PK, Teixeira J. Conditional deletion of beta-catenin in the mesenchyme of the developing mouse uterus results in a switch to adipogenesis in the myometrium. Dev Biol. 2005;288:276–283. doi: 10.1016/j.ydbio.2005.09.045. [DOI] [PubMed] [Google Scholar]
- Bennett CN, Ross SE, Longo KA, Bajnok L, Hemati N, Johnson KW, Harrison SD, MacDougald OA. Regulation of Wnt signaling during adipogenesis. J Biol Chem. 2002;277:30998–31004. doi: 10.1074/jbc.M204527200. [DOI] [PubMed] [Google Scholar]
- Buscarlet M, Stifani S. The 'Marx' of Groucho on development and disease. Trends Cell Biol. 2007;17:353–361. doi: 10.1016/j.tcb.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Cavallo RA, Cox RT, Moline MM, Roose J, Polevoy GA, Clevers H, Peifer M, Bejsovec A. Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature. 1998;395:604–608. doi: 10.1038/26982. [DOI] [PubMed] [Google Scholar]
- Chen G, Courey AJ. Groucho/TLE family proteins and transcriptional repression. Gene. 2000;249:1–16. doi: 10.1016/s0378-1119(00)00161-x. [DOI] [PubMed] [Google Scholar]
- Chen G, Fernandez J, Mische S, Courey AJ. A functional interaction between the histone deacetylase Rpd3 and the corepressor groucho in Drosophila development. Genes Dev. 1999;13:2218–2230. doi: 10.1101/gad.13.17.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho CY, Koo SH, Wang Y, Callaway S, Hedrick S, Mak PA, Orth AP, Peters EC, Saez E, Montminy M, Schultz PG, Chanda SK. Identification of the tyrosine phosphatase PTP-MEG2 as an antagonist of hepatic insulin signaling. Cell Metab. 2006;3:367–378. doi: 10.1016/j.cmet.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Cho YW, Hong S, Jin Q, Wang L, Lee JE, Gavrilova O, Ge K. Histone methylation regulator PTIP is required for PPARgamma and C/EBPalpha expression and adipogenesis. Cell Metab. 2009;10:27–39. doi: 10.1016/j.cmet.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christy RJ, Yang VW, Ntambi JM, Geiman DE, Landschulz WH, Friedman AD, Nakabeppu Y, Kelly TJ, Lane MD. Differentiation-induced gene expression in 3T3-L1 preadipocytes: CCAAT/enhancer binding protein interacts with and activates the promoters of two adipocyte-specific genes. Genes Dev. 1989;3:1323–1335. doi: 10.1101/gad.3.9.1323. [DOI] [PubMed] [Google Scholar]
- Combs TP, Berg AH, Obici S, Scherer PE, Rossetti L. Endogenous glucose production is inhibited by the adipose-derived protein Acrp30. J Clin Invest. 2001;108:1875–1881. doi: 10.1172/JCI14120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalen KT, Schoonjans K, Ulven SM, Weedon-Fekjaer MS, Bentzen TG, Koutnikova H, Auwerx J, Nebb HI. Adipose tissue expression of the lipid droplet-associating proteins S3-12 and perilipin is controlled by peroxisome proliferator-activated receptor-gamma. Diabetes. 2004;53:1243–1252. doi: 10.2337/diabetes.53.5.1243. [DOI] [PubMed] [Google Scholar]
- De Vos P, Lefebvre AM, Miller SG, Guerre-Millo M, Wong K, Saladin R, Hamann LG, Staels B, Briggs MR, Auwerx J. Thiazolidinediones repress ob gene expression in rodents via activation of peroxisome proliferator-activated receptor gamma. J Clin Invest. 1996;98:1004–1009. doi: 10.1172/JCI118860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debevec D, Christian M, Morganstein D, Seth A, Herzog B, Parker M, White R. Receptor interacting protein 140 regulates expression of uncoupling protein 1 in adipocytes through specific peroxisome proliferator activated receptor isoforms and estrogen-related receptor alpha. Mol Endocrinol. 2007;21:1581–1592. doi: 10.1210/me.2007-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freytag SO, Paielli DL, Gilbert JD. Ectopic expression of the CCAAT/enhancer-binding protein alpha promotes the adipogenic program in a variety of mouse fibroblastic cells. Genes Dev. 1994;8:1654–1663. doi: 10.1101/gad.8.14.1654. [DOI] [PubMed] [Google Scholar]
- Ge K, Guermah M, Yuan CX, Ito M, Wallberg AE, Spiegelman BM, Roeder RG. Transcription coactivator TRAP220 is required for PPAR gamma 2-stimulated adipogenesis. Nature. 2002;417:563–567. doi: 10.1038/417563a. [DOI] [PubMed] [Google Scholar]
- Gelman L, Zhou G, Fajas L, Raspe E, Fruchart JC, Auwerx J. p300 interacts with the N- and C-terminal part of PPARgamma2 in a ligand-independent and -dependent manner, respectively. J Biol Chem. 1999;274:7681–7688. doi: 10.1074/jbc.274.12.7681. [DOI] [PubMed] [Google Scholar]
- Graves RA, Tontonoz P, Platt KA, Ross SR, Spiegelman BM. Identification of a fat cell enhancer: analysis of requirements for adipose tissue-specific gene expression. J Cell Biochem. 1992;49:219–224. doi: 10.1002/jcb.240490303. [DOI] [PubMed] [Google Scholar]
- Grontved L, Madsen MS, Boergesen M, Roeder RG, Mandrup S. MED14 tethers mediator to the N-terminal domain of peroxisome proliferator-activated receptor gamma and is required for full transcriptional activity and adipogenesis. Mol Cell Biol. 30:2155–2169. doi: 10.1128/MCB.01238-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halaas JL, Gajiwala KS, Maffei M, Cohen SL, Chait BT, Rabinowitz D, Lallone RL, Burley SK, Friedman JM. Weight-reducing effects of the plasma protein encoded by the obese gene. Science. 1995;269:543–546. doi: 10.1126/science.7624777. [DOI] [PubMed] [Google Scholar]
- Hevener AL, He W, Barak Y, Le J, Bandyopadhyay G, Olson P, Wilkes J, Evans RM, Olefsky J. Muscle-specific Pparg deletion causes insulin resistance. Nat Med. 2003;9:1491–1497. doi: 10.1038/nm956. [DOI] [PubMed] [Google Scholar]
- Hevener AL, Olefsky JM, Reichart D, Nguyen MT, Bandyopadyhay G, Leung HY, Watt MJ, Benner C, Febbraio MA, Nguyen AK, Folian B, Subramaniam S, Gonzalez FJ, Glass CK, Ricote M. Macrophage PPAR gamma is required for normal skeletal muscle and hepatic insulin sensitivity and full antidiabetic effects of thiazolidinediones. J Clin Invest. 2007;117:1658–1669. doi: 10.1172/JCI31561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummasti S, Tontonoz P. The peroxisome proliferator-activated receptor N-terminal domain controls isotype-selective gene expression and adipogenesis. Mol Endocrinol. 2006;20:1261–1275. doi: 10.1210/me.2006-0025. [DOI] [PubMed] [Google Scholar]
- Jennings BH, Pickles LM, Wainwright SM, Roe SM, Pearl LH, Ish-Horowicz D. Molecular recognition of transcriptional repressor motifs by the WD domain of the Groucho/TLE corepressor. Mol Cell. 2006;22:645–655. doi: 10.1016/j.molcel.2006.04.024. [DOI] [PubMed] [Google Scholar]
- Kim JY, van de Wall E, Laplante M, Azzara A, Trujillo ME, Hofmann SM, Schraw T, Durand JL, Li H, Li G, Jelicks LA, Mehler MF, Hui DY, Deshaies Y, Shulman GI, Schwartz GJ, Scherer PE. Obesity-associated improvements in metabolic profile through expansion of adipose tissue. J Clin Invest. 2007;117:2621–2637. doi: 10.1172/JCI31021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota N, Terauchi Y, Kubota T, Kumagai H, Itoh S, Satoh H, Yano W, Ogata H, Tokuyama K, Takamoto I, Mineyama T, Ishikawa M, Moroi M, Sugi K, Yamauchi T, Ueki K, Tobe K, Noda T, Nagai R, Kadowaki T. Pioglitazone ameliorates insulin resistance and diabetes by both adiponectin-dependent and -independent pathways. J Biol Chem. 2006;281:8748–8755. doi: 10.1074/jbc.M505649200. [DOI] [PubMed] [Google Scholar]
- Lehmann JM, Moore LB, Smith-Oliver TA, Wilkison WO, Willson TM, Kliewer SA. An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor gamma (PPAR gamma) J Biol Chem. 1995;270:12953–12956. doi: 10.1074/jbc.270.22.12953. [DOI] [PubMed] [Google Scholar]
- Liu J, Farmer SR. Regulating the balance between peroxisome proliferator-activated receptor gamma and beta-catenin signaling during adipogenesis. A glycogen synthase kinase 3beta phosphorylation-defective mutant of beta-catenin inhibits expression of a subset of adipogenic genes. J Biol Chem. 2004;279:45020–45027. doi: 10.1074/jbc.M407050200. [DOI] [PubMed] [Google Scholar]
- Louet JF, Coste A, Amazit L, Tannour-Louet M, Wu RC, Tsai SY, Tsai MJ, Auwerx J, O’Malley BW. Oncogenic steroid receptor coactivator-3 is a key regulator of the white adipogenic program. Proc Natl Acad Sci U S A. 2006;103:17868–17873. doi: 10.1073/pnas.0608711103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean JA, Tobin G. Animal and human calorimetry. Cambridge [Cambridgeshire] ; New York: Cambridge University Press; 1987. [Google Scholar]
- Moldes M, Zuo Y, Morrison RF, Silva D, Park BH, Liu J, Farmer SR. Peroxisome-proliferator-activated receptor gamma suppresses Wnt/beta-catenin signalling during adipogenesis. Biochem J. 2003;376:607–613. doi: 10.1042/BJ20030426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destree O, Clevers H. XTcf-3 transcription factor mediates beta-catenin-induced axis formation in Xenopus embryos. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- Nguyen H, Rendl M, Fuchs E. Tcf3 governs stem cell features and represses cell fate determination in skin. Cell. 2006;127:171–183. doi: 10.1016/j.cell.2006.07.036. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Pedersen TA, Hagenbeek D, Moulos P, Siersbaek R, Megens E, Denissov S, Borgesen M, Francoijs KJ, Mandrup S, Stunnenberg HG. Genome-wide profiling of PPARgamma:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev. 2008;22:2953–2967. doi: 10.1101/gad.501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- Qi C, Surapureddi S, Zhu YJ, Yu S, Kashireddy P, Rao MS, Reddy JK. Transcriptional coactivator PRIP, the peroxisome proliferator-activated receptor gamma (PPARgamma)-interacting protein, is required for PPARgamma-mediated adipogenesis. J Biol Chem. 2003;278:25281–25284. doi: 10.1074/jbc.C300175200. [DOI] [PubMed] [Google Scholar]
- Roeder RG. Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett. 2005;579:909–915. doi: 10.1016/j.febslet.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Ross SE, Erickson RL, Gerin I, DeRose PM, Bajnok L, Longo KA, Misek DE, Kuick R, Hanash SM, Atkins KB, Andresen SM, Nebb HI, Madsen L, Kristiansen K, MacDougald OA. Microarray analyses during adipogenesis: understanding the effects of Wnt signaling on adipogenesis and the roles of liver X receptor alpha in adipocyte metabolism. Mol Cell Biol. 2002;22:5989–5999. doi: 10.1128/MCB.22.16.5989-5999.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross SE, Hemati N, Longo KA, Bennett CN, Lucas PC, Erickson RL, MacDougald OA. Inhibition of adipogenesis by Wnt signaling. Science. 2000;289:950–953. doi: 10.1126/science.289.5481.950. [DOI] [PubMed] [Google Scholar]
- Schoonjans K, Peinado-Onsurbe J, Lefebvre AM, Heyman RA, Briggs M, Deeb S, Staels B, Auwerx J. PPARalpha and PPARgamma activators direct a distinct tissue-specific transcriptional response via a PPRE in the lipoprotein lipase gene. Embo J. 1996;15:5336–5348. [PMC free article] [PubMed] [Google Scholar]
- Sekiya T, Zaret KS. Repression by Groucho/TLE/Grg proteins: genomic site recruitment generates compacted chromatin in vitro and impairs activator binding in vivo. Mol Cell. 2007;28:291–303. doi: 10.1016/j.molcel.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele R. Influences of glucose loading and of injected insulin on hepatic glucose output. Ann N Y Acad Sci. 1959;82:420–430. doi: 10.1111/j.1749-6632.1959.tb44923.x. [DOI] [PubMed] [Google Scholar]
- Steppan CM, Bailey ST, Bhat S, Brown EJ, Banerjee RR, Wright CM, Patel HR, Ahima RS, Lazar MA. The hormone resistin links obesity to diabetes. Nature. 2001;409:307–312. doi: 10.1038/35053000. [DOI] [PubMed] [Google Scholar]
- Subramaniam N, Treuter E, Okret S. Receptor interacting protein RIP140 inhibits both positive and negative gene regulation by glucocorticoids. J Biol Chem. 1999;274:18121–18127. doi: 10.1074/jbc.274.25.18121. [DOI] [PubMed] [Google Scholar]
- Takahashi N, Kawada T, Yamamoto T, Goto T, Taimatsu A, Aoki N, Kawasaki H, Taira K, Yokoyama KK, Kamei Y, Fushiki T. Overexpression and ribozyme-mediated targeting of transcriptional coactivators CREB-binding protein and p300 revealed their indispensable roles in adipocyte differentiation through the regulation of peroxisome proliferator-activated receptor gamma. J Biol Chem. 2002;277:16906–16912. doi: 10.1074/jbc.M200585200. [DOI] [PubMed] [Google Scholar]
- Tontonoz P, Graves RA, Budavari AI, Erdjument-Bromage H, Lui M, Hu E, Tempst P, Spiegelman BM. Adipocyte-specific transcription factor ARF6 is a heterodimeric complex of two nuclear hormone receptors, PPAR gamma and RXR alpha. Nucleic Acids Res. 1994a;22:5628–5634. doi: 10.1093/nar/22.25.5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tontonoz P, Hu E, Devine J, Beale EG, Spiegelman BM. PPAR gamma 2 regulates adipose expression of the phosphoenolpyruvate carboxykinase gene. Mol Cell Biol. 1995;15:351–357. doi: 10.1128/mcb.15.1.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tontonoz P, Hu E, Graves RA, Budavari AI, Spiegelman BM. mPPAR gamma 2: tissue-specific regulator of an adipocyte enhancer. Genes Dev. 1994b;8:1224–1234. doi: 10.1101/gad.8.10.1224. [DOI] [PubMed] [Google Scholar]
- Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell. 1994c;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- Waki H, Park KW, Mitro N, Pei L, Damoiseaux R, Wilpitz DC, Reue K, Saez E, Tontonoz P. The small molecule harmine is an antidiabetic cell-type-specific regulator of PPARgamma expression. Cell Metab. 2007;5:357–370. doi: 10.1016/j.cmet.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Yamauchi T, Kamon J, Waki H, Terauchi Y, Kubota N, Hara K, Mori Y, Ide T, Murakami K, Tsuboyama-Kasaoka N, Ezaki O, Akanuma Y, Gavrilova O, Vinson C, Reitman ML, Kagechika H, Shudo K, Yoda M, Nakano Y, Tobe K, Nagai R, Kimura S, Tomita M, Froguel P, Kadowaki T. The fat-derived hormone adiponectin reverses insulin resistance associated with both lipoatrophy and obesity. Nat Med. 2001;7:941–946. doi: 10.1038/90984. [DOI] [PubMed] [Google Scholar]
- Zelcer N, Hong C, Boyadjian R, Tontonoz P. LXR regulates cholesterol uptake through Idol-dependent ubiquitination of the LDL receptor. Science. 2009;325:100–104. doi: 10.1126/science.1168974. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.