Abstract
The characterization of the molecular structure and physical properties of self-assembling peptides is an important aspect of optimizing their utility as scaffolds for biomaterials and other applications. Here we report the formation of autofluorescent fibrils by an octapeptide (GVGVAGVG) derived via a single amino acid substitution in one of the hydrophobic repeat elements of human elastin. This is the shortest and most well-defined peptide so far reported to exhibit intrinsic fluorescence in the absence of a discrete fluorophore. Structural characterization by FTIR and solid-state NMR reveals a predominantly β-sheet conformation for the peptide in the fibrils, which are likely assembled in an amyloid-like cross-β structure. Investigation of dynamics and the effects of hydration on the peptide are consistent with a rigid, water excluded structure, which has implications for the likely mechanism of intrinsic fibril fluorescence.
Introduction
Self-assembling peptides are finding increased application as building blocks for the development of biomaterials and other nanostructures.1−4 This is a result of the wide range of chemical and physical properties accessible to peptides based on their amino acid sequence and the type of self-assembly that they undergo. In particular, fibril-forming peptides derived from amyloidogenic sequences, or from the fundamental (AG)n motifs found in silk fibroin, have been put forward as useful scaffolds for creating novel biomaterials.2,4,5 Similarly, sequences derived from the hydrophobic repeats of elastin have been widely investigated as prospective biomimetic polymers with potentially useful properties in terms of temperature-controlled assembly, and elasticity.(6) In each case, the properties of the assembled peptides are typically defined either by altering the primary sequence and solution conditions to change physical properties, such as strength and type of assembly, or through the addition of ligands or cargo, for instance, the deposition of metal nanowires or other polymers templated by amyloid fibril formation.7,8
Because of the highly sequence-dependent behavior of self-assembling peptides, it is important to develop a detailed understanding of the molecular basis for the adoption of different structures and for the material properties of the final product. This is of particular importance in the case of amyloid fibrils because amyloid formed by naturally occurring sequences is linked to the pathogenesis of a number of human diseases, such as Alzheimer’s, Parkinson’s, and prion diseases. The link between structure and disease in these cases is not clearly understood, such that potential risks arising from amyloid-based materials are poorly defined. Thus, understanding the structural properties of potential materials and the link between their molecular structure and physical properties is also of significant biological importance.
The structural details of several biologically relevant amyloid fibrils have been determined using solid-state NMR and other spectroscopic techniques, showing that each contains a variation on the cross-β architecture consistent with X-ray fiber diffraction studies.9−13 Similarly, the local structure of nanotubes formed by the Alzheimer’s β-amyloid protein fragment KLVFFAE has been investigated, providing insight into the self-assembly of this peptide.14,15 In addition to amyloid-based sequences, the molecular assembly of the silk-derived (AG)n peptides has been extensively studied by solid state NMR, although a high-resolution structure for fibrils formed by this peptide has not yet been reported. Recent X-ray structures of amyloid-like peptide crystals have illustrated the diversity of peptide−peptide interactions accessible even to relatively simple systems.(16) Such studies highlight the importance of characterizing a wider array of fibril forming peptides, particularly those that exhibit interesting material properties, such as the elastin-derived (VGGLG)n fibrils reported by del Mercato et al.17,18 These fibrils were formed from a heterogeneous suspension of mixed n-polymer peptides but exhibited unusual blue fluorescence and charge transport properties, with potential application in the development of new materials. However, because of their undefined peptide polymeric state, detailed structural characterization of these fibrils is likely to be challenging.
Here we describe the secondary structure and dynamic properties of amyloid-like fibrils formed by the octapeptide GVGVAGVG. This sequence (GVA) is derived from the (VPGVG)n repeat of human elastin through a single substitution of proline for alanine. On the basis of sequence analysis of amyloid and elastic proteins and extensive molecular dynamics simulations, it has been proposed that this type of substitution would result in a peptide with greater propensity for assembly into amyloid fibrils than elastic polymers.(19) The parent sequence undergoes an elastin-like temperature-induced assembly, presumably forming a simplified elastic matrix.20−22
We report the formation of autofluorescent amyloid fibrils by the GVA peptide and their characterization by FTIR and solid state NMR spectroscopy. This is the first detailed structural study of an intrinsically fluorescent amyloid fibril. The results are consistent with the adoption of a predominantly cross-β structure in the fibrils, giving rise to rigid, dehydrated peptide assemblies. These data contrast with the relatively elastic nature of the autofluorescent fibrils reported by del Mercato et al.(18) and suggest an alternative mechanism for the optical properties of GVA fibrils. Because of their well-defined sequence and assembly, the GVA fibrils reported here are amenable to ongoing studies of the molecular basis for their intrinsic blue fluorescence. The significance of this work can be attributed to the potential utility of fluorescent peptide assemblies as building blocks for new materials and the related requirement to understand the basis for their unusual physical properties.
Methods and Materials
Peptide Synthesis and Fibrillization
All GVA peptides were prepared by solid-phase peptide synthesis using standard FMOC chemistry. The C- and N-termini were amidated and acetylated, respectively. For NMR experiments, four selective labeling schemes were utilized, with incorporation of 13C and 15N amino acids (Isotec and Cambridge Isotope Laboratories) at the residues indicated in Table 1. The final products were purified by reverse phase HPLC, using an acetonitrile/water gradient (with 0.1% TFA) on an 11 × 300 mm C8 peptide column (Vydac). Purified peptides were freeze-dried, and their identity was confirmed by MALDI mass spectrometry.
Table 1. 13C, 15N Labeling Schemes Used in the Present Studya.
| peptide name | amino acid sequence |
|---|---|
| GVA(G1V4) | GVGVAGVG |
| GVA(V2G6) | GVGVAGVG |
| GVA(G3V4A5) | GVGVAGVG |
| GVA(V7G8) | GVGVAGVG |
Residues with uniform isotope labeling are indicated in bold.
To form GVA fibrils, peptide was dissolved at 25 mg/mL in 1,1,1,3,3,3-hexafluoroisopropanol (HFIP, Fluka) and bath sonicated for 5 min, then allowed to stand at room temperature for 10 min. The HFIP was evaporated under a stream of N2(g), and the resulting peptide film was resuspended in H2O to give a final peptide concentration of 1 mg/mL. The solution was briefly vortexed and sonicated to ensure uniform resuspension of the peptide. A gel-like suspension of fibrils formed almost immediately, and samples were incubated at 25 °C for 7−10 days to ensure complete assembly prior to biophysical characterization. Amyloid fibrils of PrP(106−126) were prepared as previously described.(23)
Transmission Electron Microscopy
Samples for negative stain TEM were deposited on fresh continuous carbon films layered onto copper rhodium grids (Electron Microscopy Sciences). Prior to the addition of samples, the grids were charged using a glow discharger for 15 s at 30 mA negative discharge. Fibril solutions of 1 mg/mL were adsorbed to grids for 2 min prior to rinsing with 10 μL water for 10s. Samples were blotted using No. 2 Whatman filter paper and stained with freshly filtered 2% uranyl acetate for 15 s. TEM images were obtained using a Jeol 1011 microscope operating at 80 kV.
Fluorescence Microscopy
A drop of suspended GVA fibrils was placed on a poly prep glass slide (Sigma). A poly-l-lysine coverslip (BD Biosciences) was placed on top immediately or after the peptide suspension was allowed to dry. The coverslip was sealed to the slide using clear nail polish with care being taken to not allow the polish to get under the coverslip. Images were taken immediately at The Hospital for Sick Children Imaging Facility using a Leica DMIRE2 (Leica Microsystems) inverted fluorescence microscope equipped with a Hamamatsu Back-Thinned EM-CCD camera. Images were acquired using a 1000× objective lens with fluorescence set to the DAPI channel (excitation 358, emission 460) at an exposure time of 70 ms. Images were acquired using the Velocity software package.
Fourier Transform Infrared Spectroscopy
Fourier-transformed infrared spectra were acquired on a Nicolet Nexus 650 spectrometer system equipped with a midrange IR source, a KBr beam splitter, and an MCT-A detector on the Continuum microscope attachment. Thin films of fibril samples were air-dried on 32 mm cesium chloride (CsCl) discs before spectra were collected on the microscope attachment in transmission mode. Spectra were averaged from 256 scans over a range of 4000−650 cm−1 wave numbers at 4 cm−1 resolution and were baseline-corrected using the OMNIC software package (Nicolet/Thermo Electron). The secondary derivative of the amide 1 and 2 region (1800−1500 cm−1) was calculated using the OriginPro 8 software for spectral-deconvolution and secondary structure determination.
Solid-State Nuclear Magnetic Resonance
Lyophilized fibrils were packed into standard 22 μL 3.2 mm MAS rotors. Hydrated samples were prepared by centrifugation of GVA fibrils at 20 000g, followed by packing the dense wet pellets into 22 or 36 μL rotors, depending on sample volume. Solid-state NMR measurements were carried out on a narrow bore Varian VNMRS spectrometer, operating at a 1H frequency of 499.82 MHz. All experiments were carried out using a Varian triple-resonance 3.2 mm T3MAS probe, with a magic-angle spinning frequency of 10−12 kHz. Sample heating was alleviated by delivering high-flow rates of ambient temperature dry air to the sample. All spectra were externally referenced to the downfield 13C resonance of adamantane at 38.56 ppm relative to TMS.(24)
13C and 15N cross-polarization (CP) was implemented using a linear ramped radio frequency (rf) field centered around 40−60 kHz on the low channel, with a 50−80 kHz field on the 1H channel and contact times of 1 to 1.5 ms. π/2 pulse widths on all channels were typically 2.0−4.0 μs. 1H decoupling fields of 125 kHz were applied during all t1 and t2 periods, using the TPPM decoupling scheme.(25) With the exception of T1 relaxation measurements, a 2 s delay was used between scans. Two-dimensional (2D) 13C−13C spectra were obtained using a radio-frequency-assisted diffusion (RAD) recoupling sequence.26,27 A RAD mixing time of 10 ms was used in all cases, and spectra were recorded at an MAS frequency of 11 kHz. 2D 13C−15N spectra were obtained using specific CP from 15N to 13C, as described by Petkova et al.(28)
13C T1 relaxation times were obtained from CP spectra recorded using the spin-temperature inversion method described by Torchia,(29) with the time between the two π/2 pulses arrayed from 0 to 7.5 s. T2 relaxation time measurements were performed using a CPMG spin−echo experiment,30,31 with an echo period (including π-pulse) arrayed from 50 to 5000 μs. In both T1 and T2 experiments, 1H−13C CP was achieved as described above with an MAS frequency of 10 kHz. Peak intensity was plotted as a function of the recovery time and in each case was fit to a single exponential decay. T2-filtered CP spectra were obtained using a 1H echo prior to CP.32,33
Two-dimensioanl 1H−13C and 1H−15N WISE (wide-line separation) experiments were used to measure 1H−1H dipolar couplings, as described by Schmidt-Rohr et al.(34) Following a 1H−1H dipolar evolution period (maximum of 0.25 ms), magnetization was transferred to 13C or 15N using a 100 μs Lee−Goldberg CP step.(35) To facilitate stable spinning of samples in the 3.2 mm rotors, WISE experiments were carried out at an MAS spinning speed of 10 kHz. Simulations of the 1H dipolar spectra were carried out using Spinevolution.(36)
Results and Discussion
Fibril Morphology and Autofluorescence
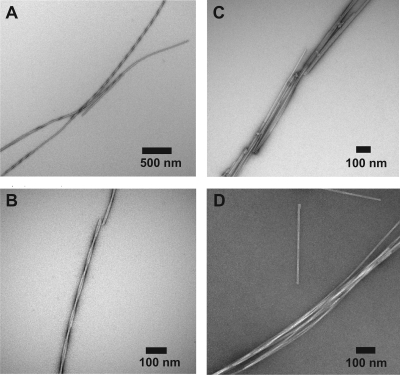
On the basis of reported molecular dynamics simulations of peptides composed of modified hydrophobic repeat sequences of human elastin,(19) we predicted that the peptide sequence GVGVAGVG (GVA) would likely to adopt an extended β-rich structure and assemble into fibrillar aggregates. As anticipated, the GVA peptides rapidly formed a dense gelatinous suspension upon incubation as aqueous solutions, similar to the previously reported fibrillization of (VGGLG)n peptides under similar conditions.(17) Transmission electron microscopy (TEM) images show long fibrillar assemblies of GVA, with two distinct morphologies observed in different samples. The majority of samples contained only the paired twisted filaments shown in Figure 1A,B. These are composed of unbranched, straight protofilaments ∼10 nm wide and up to several micrometers in length, which assemble into pairs with a periodic twist of 125 nm.
Figure 1.
Negative stain transmission electron micrographs of GVA fibrils stained with uranyl acetate. (A,B) Prevalent twisted fibril morphology. (C,D) Representative TEM images from samples containing primarily clumped, untwisted fibrils.
In a small number of samples prepared using the same dissolution conditions, fibrils with a distinct morphology were observed (Figure 1C,D). These appear as flat untwisted filaments, 10 nm wide, which bundle laterally to form bundles up to 50 nm wide. In the present study, we focus exclusively on the more prevalent twisted filaments. Samples displaying the bundled untwisted morphology gave less optimal NMR spectra, suggestive of increased disorder and significant sample heterogeneity, thus inhibiting detailed analysis.
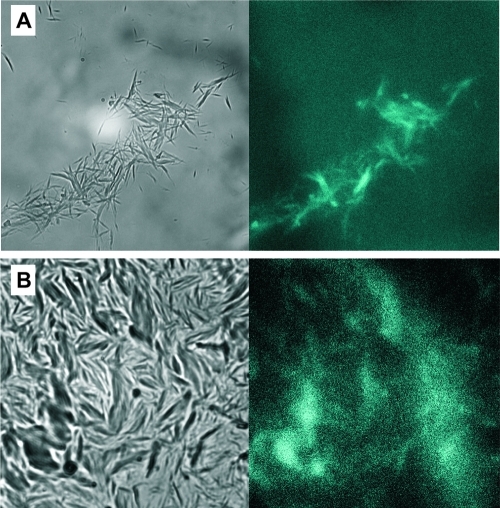
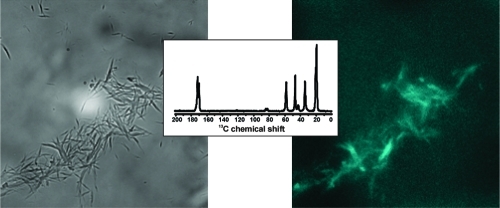
Initial attempts to use binding of the fluorescent dye thioflavin-T to test for the presence of cross-β structure in GVA fibrils gave uncharacteristic emission spectra, which suggested the presence of peptide autofluorescence, as reported for (VGGLG)n fibrils.(18) Fluorescence microscope images are shown in Figure 2, along with bright field images. In wet GVA fibril samples (Figure 2A), small clumps of fibrils are easily visible and clearly show a blue fluorescence (emission filter centered at 460 nm) when excited with 358 nm light. Dry fibril samples gave similar results (Figure 2B), but the images are less clear because of film formation by the GVA fibrils on the slide during drying. Nevertheless, the blue fluorescence colocalizes with the presence of fibrillar aggregates, although the intensity of emission may be lower than that of the wet fibrils. In previous studies of (VGGLG)n autofluorescence, it was suggested that water-mediated backbone hydrogen bonding within the fibrils was important for fluorescence.(18) Whereas this can be used to rationalize the lower intensity of the emission spectrum from dry fibrils, a separate study reported blue fluorescence of protein crystals, in the absence of extensive water hydrogen bonding networks.(37) Instead, the authors proposed a network of delocalized electrons within hydrogen bonds.
Figure 2.
Fluorescence microscopy images of (A) fully hydrated and (B) dry GVA fibrils. The left panel in each case shows a bright field image, whereas the right panel is a fluorescence image of the same field, obtained with excitation at 358 nm and observing emission at 460 nm. Images were acquired using a 1000× objective lens and an exposure time of 70 ms.
Structural Characterization of GVGVAGVG Fibrils
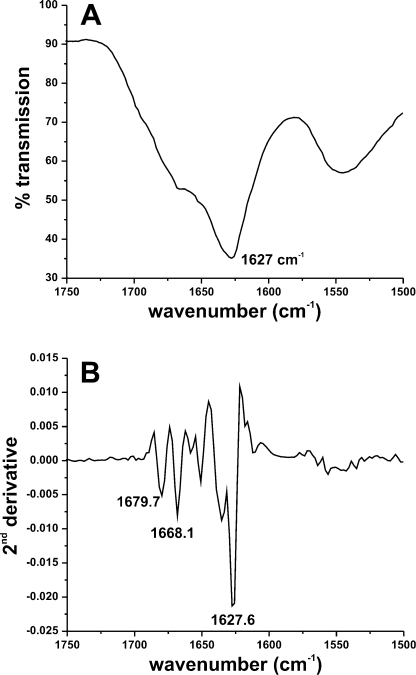
The secondary structure of GVA fibrils was probed using Fourier transform infrared spectroscopy (FTIR). A minimum in the amide I transmission spectrum is observed at 1627 cm−1, indicative of a primarily β-sheet secondary structure (Figure 3). The amide I band does, however, exhibit a small shoulder peak near 1670 cm−1, suggesting the presence of some disordered regions. An additional peak at 1680 cm−1, in combination with the main peak at 1627 cm−1, has been proposed to result from antiparallel β sheets arranged in a cross-β structure.38−40 Whereas there is poor resolution in the amide II region, a broad peak centered at 1545 cm−1 is consistent with a predominantly β-sheet structure. Note that the FTIR spectra were obtained for isolated fibrillar material using a microscope attachment. Therefore, these data likely reflect the presence of some disorder coexisting with β sheet within the fibrils rather than arising from unstructured monomers in solution. Polarized light microscopy of GVA fibrils stained with Congo Red revealed the characteristic apple-green birefringence of amyloid fibrils (not shown), suggesting the presence of a cross-β packing of peptides in GVA fibrils, consistent with a primarily β-sheet structure.(41)
Figure 3.
Fourier transform infrared spectroscopy of unlabeled GVA fibrils. (A) Transmission spectrum encompassing the amide I and amide II regions. (B) Corresponding second derivative spectrum. In each case, the major peak at 1627 cm−1 (corresponding to β sheet) is indicated. Additional weaker features corresponding to random coil or disordered structures are also labeled in part B.
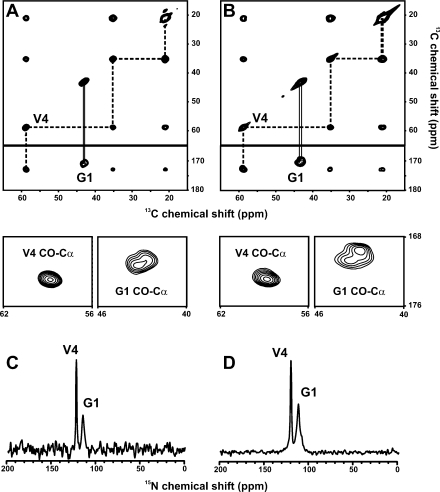
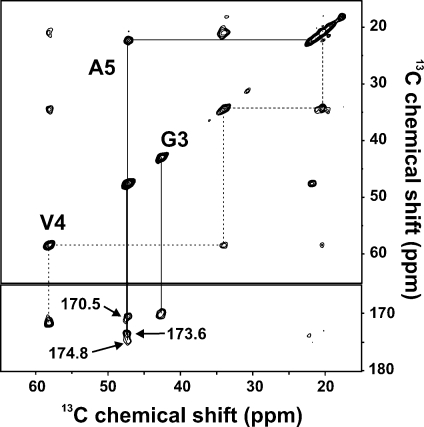
To obtain more detailed insight into the secondary structure of GVA fibrils, we used magic-angle spinning solid-state NMR spectroscopy to obtain isotropic 13C and 15N chemical shifts for this peptide. Resonance assignments were made using 2D chemical shift correlation spectra in all cases based on spin system identification in 13C−13C RAD spectra obtained with 10 ms mixing times. 13C and 15N assignments are shown in Figure 4 for hydrated and lyophilized GVA(G1 V4) fibrils. Note that two distinct populations are observed for Gly1, whereas Val4 has one distinct set of resonances in the same sample. Similar results were obtained for GVA(V2G6) fibrils, in which one set of Val2 resonances is observed, whereas Gly6 has two distinct sets of carbonyl and Cα shifts, although in this case, they are separated by <0.5 ppm. Val4, Val7, and Gly8 also all gave rise to a single set of shifts for each residue. A more dramatic suggestion of conformational polymorphism is observed for Ala5, which has three very distinct carbonyl 13C resonances (Figure 5), with approximately 2:2:1 peak ratios (170.5, 173.6, 174.8 ppm). As is the case for each of the polymorphic glycine residues, all of the Ala5 chemical shifts remain indicative of the same secondary structure, although the resonance at 170.5 ppm exhibits an uncharacteristically large shift relative to the random coil value of 176.1 ppm. The Ala5 carbonyl shifts for the dry fibrils are not reported in Figure 6 because slightly broader lines prevent accurate identification of the resonances near 173 to 174 ppm.
Figure 4.
13C and 15N resonance assignments for GVA(G1 V4) fibrils. (A) Aliphatic and carbonyl regions of a 13C−13C chemical shift correlation spectrum of hydrated GVA(G1 V4) fibrils, obtained with a RAD mixing time of 10 ms. (B) Equivalent spectrum obtained for lyophilized (dry) GVA(G1 V4) fibrils. Expansions of the carbonyl-Cα crosspeaks for Gly1 and Val4 are shown below each 2D spectrum. (C,D) 15N CP spectra of wet and dry GVA(G1 V4) fibrils, respectively. The chemical shift assignments for all resonances are indicated on each spectrum, with both sets of Gly1 carbonyl and Cα resonances identified. All data were obtained at 25 °C, with an MAS rate of 11 kHz MAS.
Figure 5.
2D 13C−13C chemical shift correlation spectrum of GVA(G3 V4A5), obtained using a RAD mixing time of 10 ms, with an MAS rate of 11 kHz. Resonance assignments are shown, with the three carbonyl shifts observed for Ala5 indicated. Note that two distinct, but very similar (<0.25 ppm difference) shifts are also observed for the carbonyl and Cα resonances of Gly3.
Figure 6.
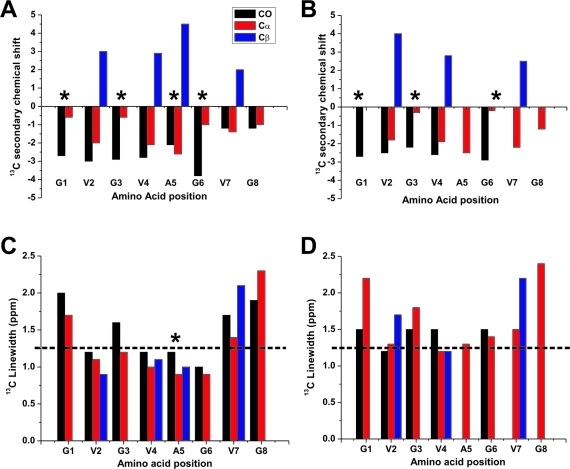
Secondary 13C chemical shifts and NMR linewidths measured from (A,C) fully hydrated and (B,D) lyophilized GVA fibrils. Asterisks indicate the residues that give rise to multiple clearly resolved resonances in hydrated GVA(G1V4) and GVA(G3V4A5) fibrils. The chemical shifts and linewidths for the peaks representing the largest population were used in each case. Note that all populations give rise to similar peak widths and secondary chemical shifts. To facilitate comparison of 13C linewidths, a horizontal line is shown at 1.25 ppm in parts C and D.
As suggested by molecular dynamics simulations of related peptides, the presence of multiple chemical shifts at Ala5 strongly suggests conformational plasticity and adoption of three preferred structures. Given the proposed tendency of similar peptides to adopt hairpin structures, it is likely that some of the GVA peptides contain a β turn at this site.19,42,43 The two glycine resonances observed at several sites may reflect long-range effects of varied Ala5 conformations on the peptide or may be indicative of local flexibility at each site, giving rise to two distinct states. The fact that multiple populations are observed in both the hydrated and lyophilized samples suggests the presence of static structures incorporating local heterogeneity rather than exchange between multiple conformations, unless this occurs on a very slow time scale. No exchange between these sites was observed in 13C RAD spectra obtained with 500 ms mixing such that any exchange would have to occur with a rate<1 s−1.
Even in the presence of some conformational variability, it is informative to examine the secondary 13C chemical shifts, which are calculated as δobs − δrandom coil using the expected shifts for unstructured peptides given by Wishart et al.(44) These are plotted, along with 13C linewidths, in Figure 6 for both hydrated and lyophilized GVA fibrils. Sites exhibiting multiple chemical shifts are indicated by asterisks. The average secondary chemical shifts are plotted for these sites because the predicted secondary structure remains the same in each case (and the differences between populations are small for the glycines). In all cases, negative values for carbonyl and Cα shifts, along with positive Cβ shifts, suggest an extended β structure, although the presence of turns cannot necessarily be excluded by this analysis. The 13C linewidths for most sites in dry and hydrated fibrils are quite narrow, with the only exceptions being the N- and C-terminal residues, where some degree of increased conformational disorder might be expected. There is a very slight broadening of most resonances by 0.2 to 0.5 ppm upon drying the sample, suggesting that some internal motions or increased order may be contributing to line narrowing in the hydrated samples. However, 13C chemical shifts remain largely the same between the two states, and the broadening effect of dehydration is subtle, suggesting relatively small effects of hydration on GVA fibrils.
Dynamic Properties of GVA Fibrils
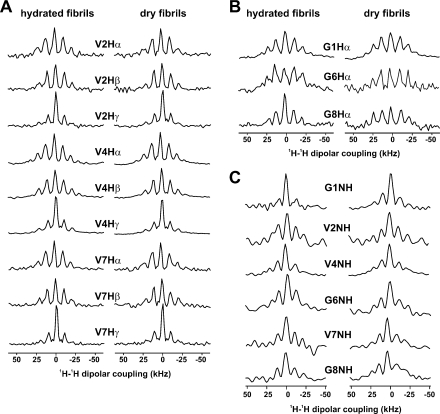
To examine further the potential hydration dependence of GVA fibril internal structure and fluorescence properties, we measured several NMR parameters that report on dynamic processes. In Figure 7, a qualitative analysis of the 1H−1H dipolar couplings in GVA fibrils is presented. The 1H dipolar cross sections extracted from 2D 1H−13C WISE(34) spectra of lyophilized and fully hydrated GVA(G1 V4), GVA(V2G6), and GVA(V7G8) fibrils are shown in Figure 7A,B. Dipolar linewidths estimated from these spectra are presented in Table 2. The presence of mobile segments in the protein will lead to a significant reduction of the 1H−1H dipolar couplings and thus result in a narrowing of the wide-line spectrum − here observed as an increase in the relative intensity of the central component relative to the spinning sidebands. The only site to show significant reduction in dipolar coupling as a function of fibril hydration is the C-terminal glycine (Gly8), for which the 1Hα line width decreased from 49 to 19 kHz, indicating increased mobility for this site relative to the rest of the chain. For all other α and β groups, the dipolar spectra are in close agreement with Spinevolution simulations of static systems. The valine methyl spectra are narrower than those of other sites (12−14 kHz), which is consistent with rapid rotation around the methyl axis. WISE data for GVA(G3V4A5) are not shown because small sample size in this case prevented the acquisition of wide-line data with sufficient signal-to-noise for meaningful analysis.
Figure 7.
1H dipolar spectra extracted from 1H−13C and 1H−15N 2D WISE spectra of GVA fibrils. (A,B) Slices taken from 1H−13C WISE spectra of hydrated (left column) and dry (right column) GVA-1 fibrils. 1H dipolar spectra for valine are shown in part A, whereas those for glycine residues are shown in part B. (C) 1H slices from 2D 1H−15N WISE spectra of the same samples. All spectra were obtained at 25 °C with an MAS frequency of 10 kHz.
Table 2. 1H Dipolar Linewidths (kHz) Measured from Lyophilized and Hydrated GVA Fibrils and Hydrated PrP(106-126) Fibrils.
| hydrated GVA fibrils |
lyophilized GVA fibrils |
|||||||
|---|---|---|---|---|---|---|---|---|
| residue | NH | Hα | Hβ | Hγ | NH | Hα | Hβ | Hγ |
| Gly1 | 12.8 | 41.0 | 16.9 | 45.8 | ||||
| Val2 | 20.7 | 30.1 | 30.3 | 13.5 | 20.1 | 30.1 | 29.9 | 13.6 |
| Val4 | 17.9 | 31.3 | 31.3 | 12.5 | 18.9 | 32.6 | 29.5 | 14.0 |
| Gly6 | 21.8 | 47.6 | 22.4 | 50.4 | ||||
| Val7 | 20.9 | 31.5 | 31.5 | 11.8 | 19.4 | 29.2 | 31.2 | 13.0 |
| Gly8 | 18.2 | 19.3 | 18.9 | 49.2 | ||||
| hydrated PrP(106−126) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Val121 | 30.2 | 9.5 | 7.9 | |||||
| Gly126 | 8.6 | |||||||
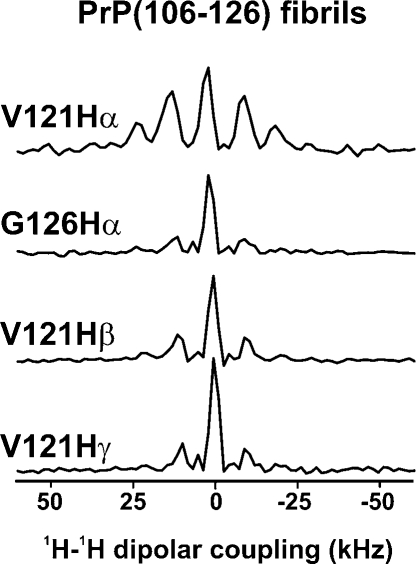
These data are in contrast with the 1H−13C WISE results obtained for hydrated amyloid fibrils formed by the prion-derived peptide PrP(106−126) (Figure 8), whose structure we have previously reported.(23) In this sample, sites known to reside at the surface (V121Cγ and Cβ) show a significant reduction in the 1H−1H dipolar coupling, resulting in linewidths of approximately 8 to 9 kHz, consistent with free rotation of the Val121 side chain. Similarly, the Gly126 α protons exhibit significant reduction in dipolar linewidths (8.5 kHz), which is consistent with their location at the mobile C-terminus of the peptide. The Val121 backbone is locked into the fibril core and exhibits a rigid dipolar spectrum, with an lHα line width of 30 kHz, similar to those observed for Val residues in the GVA fibrils.
Figure 8.
1H dipolar spectra extracted from 1H−13C 2D WISE spectra of fully hydrated amyloid fibrils formed by the prion protein-derived peptide PrP(106−126). The 13C resonance at which the 1H dipolar slice was taken is indicated for each spectrum. All spectra were obtained at 25 °C with an MAS frequency of 10 kHz.
Dipolar slices from 2D 1H−15N WISE spectra are shown in Figure 7C and tabulated in Table 2. These data further support the results from 13C-attached protons in Figures7A,B, with 1H dipolar linewidths of 18−22 kHz observed for most sites, regardless of hydration state, suggesting a lack of increased backbone motions. The amide proton of Gly1 exhibits a slight reduction in dipolar line width upon hydration (from 16.9 to 12.8 kHz), suggesting that the N-termini of at least some GVA peptides are exposed to bulk solvent.
Thus, the results obtained for the GVA fibrils are largely consistent with a highly dehydrated, rigid structure. The 1H dipolar linewidths observed for most sites are similar to those obtained by Ohgo et al., from WISE spectra of lyophilized native elastin (29−34 kHz for all sites).(46) In that study, hydration resulted in a global reduction of the 1H linewidths to 1.8 to 2.3 kHz, indicative of a highly mobile and dynamic system. A similar trend of line width reduction upon sample hydration was reported for the elastin mimetic [(VPGVG)4(VPGKG)]39, again supporting a significantly more rigid and less hydrated structure for GVA fibrils relative to elastic peptide/protein assemblies.(45)
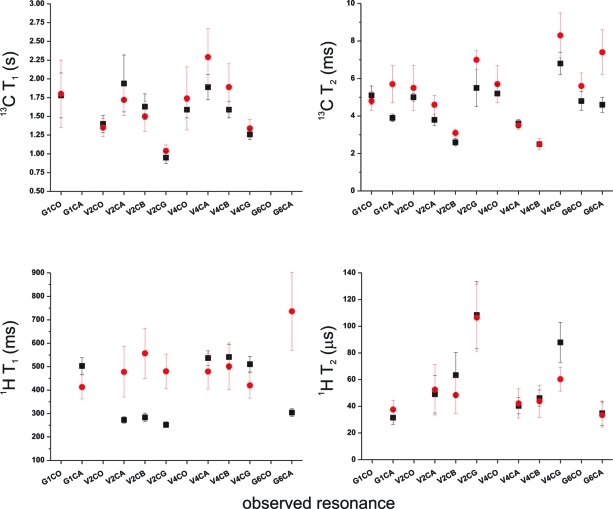
Further evidence of the overall rigidity and insensitivity of GVA fibrils to their hydration state is provided by 1H and 13C T1 and T2 relaxation times, which were measured for several positions within the fibrils (Figure 9). An increased rate of T1 relaxation would be indicative of motions occurring on the nanosecond time scale, whereas T2 relaxation rates are primarily sensitive to dynamic processes occurring on the microsecond to millisecond time scale. In each case, there is relatively little variability across the peptide and little sensitivity to hydration, suggesting little or no change in local mobility. There is a small increase in the 1H T1 relaxation time for Val2 upon hydration, although the dry values at this site are low relative to most other sites.
Figure 9.
1H and 13C NMR relaxation times measured for dry and hydrated GVA fibrils. T1 and T2 relaxation times are plotted for several resonances, as indicated. Data for dry fibrils are indicated by squares, and the wet fibril data are indicated by circles.
The 13C T2 relaxation times also allow us to calculate expected linewidths for completely homogeneous sites as 1/πT2. The expected 13C linewidths range from 0.5 to 1 ppm, which is significantly narrower than the data reported in Figure 6, suggesting the presence of conformational heterogeneity at several sites, primarily at the N-and C-termini, leading to 50−100 Hz of inhomogeneous line broadening. This may alternatively result from heterogeneous packing of the GVA peptides within the larger fibrillar assembly. It is important to note that the linewidths observed for GVA fibrils are consistent with those reported for other peptide amyloid fibrils.23,47−49
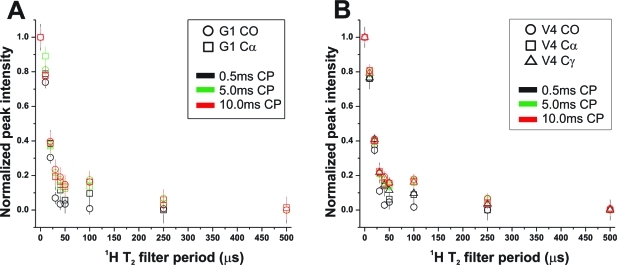
The exposure of various sites within GVA fibrils to water was tested using CP experiments with a 1H T2 filter. Similar experiments have previously been used to look for water-exposed residues in protein microcystals.(32) Signals arising from transfer between water protons and peptide will have much times longer than the 1H T2 of peptide groups. 13C peak intensities for Gly1 and Val4 are plotted in Figure 10 as a function of 1H T2 filter time. Several CP transfer times were used to allow transfer primarily from nearby protons (0.5 ms) or over longer distances (2.5, 5.0, and 10 ms). No change was observed at 2.5 ms, so those data were omitted. A very slight increase in most peptide 13C signals is observed between 25 and 250 μs as CP time increases, but the apparent 1H T2 for all of these experiments remains consistent with values measured for the peptide rather than H2O. This suggests that the majority of the peptide is not exposed to any significant amount of water or that only tightly bound water molecules (which may exhibit 1H T2 values close to that of the protein) contribute to the CP spectrum.
Figure 10.
13C peak intensities as a function of cross-polarization contact time and 1H T2. The signal intensity for several 13C resonances are plotted as a function of a 1H T2 filter applied prior to cross-polarization transfer. Data are shown for contact times of 0.5, 5.0, and 10 ms.
Overall, our data are consistent with relatively rigid fibrils composed of a cross-β arrangement of extended β strands, although the presence of alternative conformations is strongly indicated by the FTIR and 13C chemical shift data. The structure and rigidity of the fibril are insensitive to hydration levels, suggesting a dehydrated assembly composed of several layers of peptide, effectively reducing the amount of surface-exposed peptide. This is supported by the apparent size of the fibrils (even as a fully extended chain, the GVA peptides are only 2 nm long, whereas the protofilaments are 10 nm wide by TEM) and by the T2 filtered CP experiments.
The current study contrasts with the previously reported intrinsic fluorescence of (VGGLG)n fibrils that were shown to be quite elastic and deformable by AFM nanoindentation studies, relative to amyloid fibrils of insulin or diphenylalanine.17,18 Given the ability of those fibrils to transport charge, water-mediated hydrogen bonding within an amyloid-like packing arrangement was proposed as the basis for their physical properties. Whereas we cannot exclude the presence of internally bound water involved in hydrogen bonding in our system, this would be unusual, as most amyloid structures are considered to have a completely dehydrated cross-β structure, which further assembles using hydrophobic steric zipper motifs.(16) In general, bulk water may be expected to surround protofilaments and fibrils but not penetrate the core. Therefore, our data to date support a mechanism similar to that proposed by Shukla et al., in which intrinsic protein fluorescence is the result of delocalized electrons within backbone hydrogen bonds becoming excited by irradiation with UV light in the region of 350 nm wavelength, then undergoing relaxation through a fluorescent pathway. In an extended fibrillar assembly, a large network of this type could potentially exist, leading to intrinsic peptide fluorescence in the absence of true fluorophores.
Conclusions
With increased interest in self-assembling peptides as scaffolds for creating biomaterials with a wide range of physical or chemical properties and as the backbones of other nanostructures, it is important to understand the molecular structure and physical behavior of such systems. To date, relatively few of these potential materials have been subjected to detailed structural analysis. Here we describe the structural and dynamic characterization of fibrillar assemblies formed by the elastin-derived octapeptide, GVGVAGVG (GVA). In addition to forming part of a growing family of self-assembling peptides based on human elastin, fibrils of this peptide exhibit intrinsic fluorescence.
This is only the second reported case of fluorescence emission by an amyloid-like assembly lacking discrete fluorophores and is the first that is readily amenable to detailed structural analysis. Our structural data reveal the presence of predominantly β-sheet conformation, with some local heterogeneity, potentially β turns formed by some of the peptides in the fibril. A solid-state NMR investigation of peptide dynamics and the effect of hydration suggests the adoption of a rigid, water-excluded structure, unlike the relatively soft (GGLGGV)n fibrils previously reported. The latter have been suggested to contain hydrogen-bonded water molecules within the cross-β structure, resulting in fluorescence. Our data provide some evidence of an alternative hypothesis proposed by Shukla et al.,(37) in which a network of delocalized electrons exists within the hydrogen-bonded backbone. Whereas we cannot yet definitively exclude the possibility of static water molecules enclosed within the fibril structure, the GVA fibrils are ideal candidates for our ongoing investigations of the mechanism of peptide fibril autofluorescence.
Acknowledgments
We thank Sonia Ghandi and Anatoli Chkaroubo for assistance in preparing peptides and collection of preliminary NMR data. We also acknowledge Régis Pomès and Fred Keeley for useful discussions and Christopher Yip for assistance in collecting FTIR data. This work was supported by operating grants from the Canadian Institutes of Health Research (CIHR) and the National Sciences and Engineering Research Council of Canada (NSERC). P.W. is supported by an NSERC CGSD scholarship and J.Y. is supported by a CIHR CGSM scholarship. S.S. is a Canada Research Chairs Programme chair holder (Tier II).
References
- Fairman R.; Akerfeldt K. S. Curr. Opin. Struct. Biol. 2005, 15, 453–463. [DOI] [PubMed] [Google Scholar]
- Jung J. P.; Gasiorowski J. Z.; Collier J. H. Pept. Sci. 2010, 94, 49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal K.; Schneider J. P. Curr. Opin. Struct. Biol. 2004, 14, 480–486. [DOI] [PubMed] [Google Scholar]
- Reches M.; Gazit E. Curr. Nanosci. 2006, 2, 105–111. [Google Scholar]
- Childers W. S.; Ni R.; Mehta A. K.; Lynn D. G. Curr. Opin. Chem. Biol. 2009, 13, 652–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacEwan S. R.; Chilkoti A. Pept. Sci. 2010, 94, 60–77. [DOI] [PubMed] [Google Scholar]
- Reches M.; Gazit E. Science 2003, 300, 625–627. [DOI] [PubMed] [Google Scholar]
- Vandermeulen G. W. M.; Kim K. T.; Wang Z.; Manners I. Biomacromolecules 2006, 7, 1005–1010. [DOI] [PubMed] [Google Scholar]
- Jahn T. R.; Makin O. S.; Morris K. L.; Marshall K. E.; Tian P.; Sikorski P.; Serpell L. C. J. Mol. Biol. 2009, 395, 717–727. [DOI] [PubMed] [Google Scholar]
- Margittai M.; Langen R. Q. Rev. Biophys. 2008, 41, 265–297. [DOI] [PubMed] [Google Scholar]
- Tycko R. Q. Rev. Biophys. 2006, 39, 1–55. [DOI] [PubMed] [Google Scholar]
- Wasmer C.; Lange A.; Van Melckebeke H.; Siemer A. B.; Riek R.; Meier B. H. Science 2008, 319, 1523–1526. [DOI] [PubMed] [Google Scholar]
- Heise H.; Celej M. S.; Becker S.; Riedel D.; Pelah A.; Kumar A.; Jovin T. M.; Baldus M. J. Mol. Biol. 2008, 380, 444–450. [DOI] [PubMed] [Google Scholar]
- Lu K.; Jacob J.; Thiyagarajan P.; Conticello V. P.; Lynn D. G. J. Am. Chem. Soc. 2003, 125, 6391–6393. [DOI] [PubMed] [Google Scholar]
- Childers W. S.; Mehta A. K.; Ni R.; Taylor J. V.; Lynn D. G. Angew. Chem., Int. Ed. 2010, 49, 4104–4107. [DOI] [PubMed] [Google Scholar]
- Sawaya M. R.; Sambashivan S.; Nelson R.; Ivanova M. I.; Sievers S. A.; Apostol M. I.; Thompson M. J.; Balbirnie M.; Wiltzius J. J.; McFarlane H. T.; Madsen A. O.; Riekel C.; Eisenberg D. Nature 2007, 447, 453–457. [DOI] [PubMed] [Google Scholar]
- del Mercato L. L.; Maruccio G.; Pompa P. P.; Bochicchio B.; Tamburro A. M.; Cingolani R.; Rinaldi R. Biomacromolecules 2008, 9, 796–803. [DOI] [PubMed] [Google Scholar]
- del Mercato L. L.; Pompa P. P.; Maruccio G.; Della Torre A.; Sabella S.; Tamburro A. M.; Cingolani R.; Rinaldi R. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 18019–18024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauscher S.; Baud S.; Miao M.; Keeley F. W.; Pomes R. Structure 2006, 14, 1667–1676. [DOI] [PubMed] [Google Scholar]
- Nicolini C.; Ravindra B.; Ludolph B.; Winter R. Biophys. J. 2004, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuhn H.; Klok H.-A. Biomacromolecules 2008, 9, 2755–2763. [DOI] [PubMed] [Google Scholar]
- Reiersen H.; Clarke A. R.; Rees A. R. J. Mol. Biol. 1998, 283, 255–264. [DOI] [PubMed] [Google Scholar]
- Walsh P.; Simonetti K.; Sharpe S. Structure 2009, 17, 417–426. [DOI] [PubMed] [Google Scholar]
- Morcombe C. R.; Zilm K. W. J. Magn. Reson. 2003, 162, 479–86. [DOI] [PubMed] [Google Scholar]
- Bennett A. E.; Rienstra C. M.; Auger M.; Lakshmi K. V.; Lansbury P. T.; Griffin R. G. J. Chem. Phys. 1995, 103, 6951–6958. [Google Scholar]
- Takegoshi K.; Nakamura S.; Terao T. Chem. Phys. Lett. 2001, 344, 631–637. [Google Scholar]
- Morcombe C. R.; Gaponenko V.; Byrd R. A.; Zilm K. W. J. Am. Chem. Soc. 2004, 126, 7196–7197. [DOI] [PubMed] [Google Scholar]
- Petkova A. T.; Baldus M.; Belenky M.; Hong M.; Griffin R. G.; Herzfeld J. J. Magn. Reson. 2003, 160, 1–12. [DOI] [PubMed] [Google Scholar]
- Torchia D. A. J. Magn. Reson. 1978, 30, 613–616. [Google Scholar]
- Carr H. Y.; Purcell E. M. Phys. Rev. 1954, 94, 630–638. [Google Scholar]
- Meiboom S.; Gill D. Rev. Sci. Instrum. 1958, 29, 688–691. [Google Scholar]
- Bockmann A.; Juy M.; Bettler E.; Emsley L.; Galinier A.; Penin F.; Lesage A. J. Biomol. NMR 2005, 32, 195–207. [DOI] [PubMed] [Google Scholar]
- Harbison G. S.; Roberts J. E.; Hertzfeld J.; Griffin R. G. J. Am. Chem. Soc. 1988, 110, 7221–7223. [Google Scholar]
- Schmidt-Rohr K.; Clauss J.; Spiess H. W. Macromolecules 1992, 25, 3273–3277. [Google Scholar]
- Lee M.; Goldberg W. I. Phys. Rev. A 1965, 140, 1261–1271. [Google Scholar]
- Veshtort M.; Griffin R. G. J. Magn. Reson. 2006, 178, 248–282. [DOI] [PubMed] [Google Scholar]
- Shukla A.; Mukherjee S.; Sharma S.; Agrawal V.; Kishan K. V. R.; Guptasarma P. Arch. Biochem. Biophys. 2004, 428, 144–153. [DOI] [PubMed] [Google Scholar]
- Clark A. H.; Saunderson D. H. P.; Suggett A. Int. J. Peptide Protein Res. 1981, 17, 353–364. [DOI] [PubMed] [Google Scholar]
- Lefevre T.; Arsenault K.; Pezolet M. Biopolymers 2004, 73, 705–715. [DOI] [PubMed] [Google Scholar]
- Timasheff S. N.; Susi H.; Stevens L. J. Biol. Chem. 1967, 242, 5467–5473. [PubMed] [Google Scholar]
- Glenner G. G.; Eanes E. D.; Bladen H. A.; Linke R. P.; Termine J. D. J. Histochem. Cytochem. 1974, 22, 1141–1158. [DOI] [PubMed] [Google Scholar]
- Glaves R.; Baer M.; Schreiner E.; Stoll R.; Marx D. ChemPhysChem 2008, 9, 2759–2765. [DOI] [PubMed] [Google Scholar]
- Rousseau R.; Schreiner E.; Kohlmeyer A.; Marx D. Biophys. J. 2004, 86, 1393–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D. S.; Bigam C. G.; Holm A.; Hodges R. S.; Sykes B. D. J. Biomol. NMR 1995, 5, 67–81. [DOI] [PubMed] [Google Scholar]
- Yao X. L.; Conticello V. P.; Hong M. Magn. Reson. Chem. 2004, 42, 267–275. [DOI] [PubMed] [Google Scholar]
- Ohgo K.; Niemczura W. P.; Muroi T.; Onizuka A. K.; Kumashiro K. K. Macromolecules 2009, 42, 8899–8906. [Google Scholar]
- Jaroniec C. P.; MacPhee C. E.; Astrof N. S.; Dobson C. M.; Griffin R. G. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 16748–16753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madine J.; Jack E.; Stockley P. G.; Radford S. E.; Serpell L. C.; Middleton D. A. J. Am. Chem. Soc. 2008, 130, 14990–15001. [DOI] [PubMed] [Google Scholar]
- Petkova A. T.; Buntkowsky G.; Dyda F.; Leapman R. D.; Yau W. M.; Tycko R. J. Mol. Biol. 2004, 335, 247–260. [DOI] [PubMed] [Google Scholar]