Abstract
Previous human and animal studies demonstrate that acute nicotine exposure has complicated influences on the function of the nervous system, which may lead to long-lasting effects on the behavior and physiology of the subject. To determine the genes and pathways that might account for long-term changes after acute nicotine exposure, a pathway-focused oligoarray specifically designed for drug addiction research was used to assess acute nicotine effect on gene expression in the neuron-like SH-SY5Y cells. Our results showed that 295 genes involved in various biological functions were differentially regulated by 1 h of nicotine treatment. Among these genes, the expression changes of 221 were blocked by mecamylamine, indicating that the majority of nicotine-modulated genes were altered through the nicotinic acetylcholine receptors (nAChRs)-mediated signaling process. We further identified 14 biochemical pathways enriched among the nicotine-modulated genes, among which were those involved in neural development/synaptic plasticity, neuronal survival/death, immune response, or cellular metabolism. In the genes significantly regulated by nicotine but blocked by mecamylamine, 13 enriched pathways were detected. Nine of these pathways were shared with those enriched in the genes regulated by nicotine, including neuronal function-related pathways such as glucocorticoid receptor signaling, p38 MAPK signaling, PI3K/AKT signaling, and PTEN signaling, implying that nAChRs play important roles in the regulation of these biological processes. Together, our results not only provide insights into the mechanism underlying the acute response of neuronal cells to nicotine but also provide clues to how acute nicotine exposure exerts long-term effects on the nervous system.
Keywords: nicotine, neurons, mecamylamine, gene expression, p38
Introduction
Nicotine is the dependence-inducing constituent underlying tobacco smoking. Through interactions with nicotinic acetylcholine receptors (nAChRs) in the central nervous system (CNS), nicotine exposure not only stimulates the mesocorticolimbic dopamine system in the outer shell of the nucleus accumbens (NAc) and other brain regions (Benwell and Balfour, 1992, 1997; Gaddnas et al., 2001; Tammimaki et al., 2006) but also modulates the release of other neurotransmitters such as norepinephrine, serotonin, and GABA (Kenny et al., 2001; Barik and Wonnacott, 2006, 2009). Prolonged nicotine treatment can regulate the expression of genes/proteins involved in various functions such as ERK1/2 and CREB (Brunzell et al., 2003), as well as their downstream targets such as c-FOS and FOSB (Pagliusi et al., 1996; Nisell et al., 1997; Soderstrom et al., 2007). Further, biochemical pathways underlying various physiological processes; e.g., MAPK signaling, phosphatidylinositol phosphatase signaling, growth factor signaling, and ubiquitin–proteasome pathways, are modulated by nicotine (Tang et al., 1998; Konu et al., 2001; Li et al., 2004). Through its direct or indirect interaction with these genes and biological pathways, nicotine is involved in the regulation of various physiological processes, such as learning and memory, angiogenesis, energy metabolism, synaptic function, response to oxidative stress, and addiction (Harkness and Millar, 2002; Dajas-Bailador and Wonnacott, 2004; Kane et al., 2004; Robinson and Kolb, 2004; Dasgupta and Chellappan, 2006; Dasgupta et al., 2006; Hwang and Li, 2006).
Although nicotine dependence is attributed mainly to repeated and chronic nicotine exposure, acute administration also can evoke noticeable physiological effects. For example, in humans, acute nicotine exposure, while significantly increasing the plasma nicotine concentration (Argacha et al., 2008), modulates a series of physiological processes such as aortic wave reflection, arterial stiffness, and plasma asymmetrical dimethylarginine (ADMA) concentration (Adamopoulos et al., 2009). Experimentally, acute nicotine administration can significantly increase activation in multiple regions of mouse brain (Suarez et al., 2009) and induce spinal motor circuit output in embryonic zebrafish (Thomas et al., 2008). Previous studies have demonstrated the beneficial effect of acute nicotine exposure on cognitive performance (Foulds et al., 1996; Phillips and Fox, 1998; Ernst et al., 2001; Kumari et al., 2003), as well as its antidepressant effect (Tizabi et al., 2009).
Various genes and biochemical processes are involved in the biological response to acute nicotine exposure. These include immediate-early genes such as c-FOS, c-JUN, NURR77, and EGR1 (Ichino et al., 1999, 2002; Salminen et al., 1999) and the dendritically targeted early response genes activity-regulated cytoskeleton-associated protein (ARC) and dendrin in specific brain regions of rats (Schochet et al., 2005, 2008; Schmitt et al., 2008). Acute nicotine exposure not only increases the activity of dopaminergic neurons and promotes dopamine release (Levin and Rose, 1995), but also enhances dopamine turnover and metabolism (Grenhoff and Svensson, 1988). The administration of a single dose of nicotine enhances the synthesis and release of striatal dynorphin, a component of the circuit promoting negative motivational and affective states, possibly under the regulation of dopamine and glutamate (Isola et al., 2009). Acute nicotine exposure may impair nNOS-dependent dilation of cerebral arterioles via a mechanism related to the increase in oxidative stress (Arrick and Mayhan, 2007). Even a few minutes of nicotine exposure can significantly suppress arachidonic acid signaling in various brain regions in rats (Chang et al., 2009). In mice, acute administration of a low dose of nicotine increases phosphor-ERK1/2 immunoreactivity in the NAc, prefrontal cortex, and some other regions innervated by the mesolimbic dopamine pathway (Barik and Wonnacott, 2009).
Clearly, acute nicotine exposure can evoke multiple effects in the neuronal system, but its action mechanism is not completely understood. The neuron-like human neuroblastoma cell line SH-SY5Y endogenously expresses various nAChR subunits (e.g., α3, α5, α7, β2, and β4; Gould et al., 1992; Peng et al., 1994), which are assembled to form different types of nAChRs, such as α3*-nAChR or homomeric α7-nAChR receptors. The cells therefore provide a suitable in vitro model to investigate alterations in nAChRs in the presence of cholinergic ligands and have been widely used in nicotine-related study. In an earlier study, 17 genes with diverse biological functions were found to be significantly regulated in SH-SY5Y cells after exposed to 1 mM nicotine for 1 h (Dunckley and Lukas, 2003). The authors further showed that most of these genes were regulated via nAChRs-mediated mechanisms. However, because of the limitation of array technologies and bioinformatics tools at that time, a detailed analysis, especially at the biological pathway level, was impossible. With the rapid progress in the fields of genomics and bioinformatics, a detailed and comprehensive profiling of the genes and pathways responsive to acute nicotine treatment becomes feasible and necessary. In this study, by using an established and well-tested in vitro model in the field, we investigated the relation between acute nicotine exposure and neuronal function by examining the genes and biochemical pathways differentially regulated by nicotine in SH-SY5Y cells using a customized pathway-focused microarray (Cao et al., 2011; Wei et al., 2011).
Materials and Methods
Cell culture and drug treatment
The human neuroblastoma SH-SY5Y cells were purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA) and cultured in a 1:1 mixture of ATCC-formulated Eagle's Minimal Essential Medium and F12 medium supplemented with 10% fetal bovine serum (GIBCO Invitrogen, Grand Island, NY, USA) at 37°C in a humidified atmosphere of 5% CO2. At about 80% confluence, cells were treated with 1 mM nicotine (calculated as free base), 3 μM mecamylamine (nicotinic receptor antagonist), 1 mM nicotine in combination with 3 μM mecamylamine, or no drug (controls) for 1 h. For each experimental group, six independent cultures were prepared (N = 6).
RNA isolation
Total RNA was isolated separately from each of the drug-treated and control cultures using Trizol (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. To eliminate potential residual DNA contamination, the RNA samples were treated with RNase-free DNase I at 37°C for 30 min followed by inactivation at 65°C for 10 min. The RNA concentration of each sample was measured at an optical density of 260 nm, and the integrity of the RNA was checked by examination of 28S and 18S ribosomal bands using agarose–formaldehyde gel electrophoresis.
Microarray production
A pathway-focused oligoarray designed specifically for the study of drug addiction and related disorders was used. Briefly, 3,565 genes essential to CNS activities for the maintenance of neuronal homeostasis, as well as those associated with the neuron response to addictive substances such as nicotine, alcohol, and cocaine, were selected on the basis of an earlier version of a pathway-focused cDNA microarray (Konu et al., 2004a) and an extensive literature survey. These genes covered most of the major pathways related to cell metabolism, genetic information processing, cellular signaling transduction, neuron-related disease, and cell communication. OligoWiz1 was used to design the oligonucleotide for each gene. The average length of these oligonucleotides was 59.2 ± 3.8 bases (mean ± SD), with a GC content of 0.53 ± 0.05 and a Tm of 76.4 ± 1.7°C. The oligonucleotides and 10 control clones were synthesized by SIGMA Genosys (Sigma-Aldrich, St. Louis, MO, USA) and spotted at a concentration of 40 μM in 3× SSC and 1.5 M betaine buffer onto CMT-GAPS II slides (Corning Life Sciences, Lowell, MA, USA) using an OmniGrid MicroArrayer OGR-03 (GeneMachines, San Carlos, CA, USA).
cDNA probe labeling, hybridization, and scanning
To reduce the potential influence of any gene-specific dye effect, a universal reference design was used in the microarray analysis. For each experimental group, six independent cultures were prepared. Another 12 independent control cultures were used as “universal controls.” For each sample, 10 μg of total RNA was extracted for cDNA synthesis. Following purification with phenol:chloroform extraction and isopropanol precipitation, total RNA was dissolved in 28 μl of water, mixed with 4 μl of 10× buffer, 4 μl of 10 mM dTTP-free dNTP, 1 μl of 10 mM dTTP, 2 μl of 1 mM cyanine 3-dUTP (for either nicotine-treated or control sample) or cyanine 5-dUTP (for universal control sample; Enzo, Farmingdale, NY, USA), and 1 μl of Klenow fragment (50 units/μl), then incubated at 37°C for 3 h prior to purification with a QIAquick PCR kit (Qiagen, Valencia, CA, USA). The cyanine 3-labeled sample (either nicotine-treated or control) cDNA probes were mixed with the cyanine 5-labeled universal control cDNA probe and added to 7.5 μl of 20× SSC, 3 μg of CotI DNA, 3 μg of polyA, and 0.5 μl of 10% SDS adjusted to a final volume of 50 μl. The mixture was applied to the pathway-focused oligonucleotide microarray and hybridized overnight at 60°C. Slides were washed in 1× SSC and 0.2% SDS at 60°C for 5 min followed by washing in 0.1× SSC and 0.2% SDS and in 0.1× SSC at room temperature for 10 min each. Hybridized slides were scanned using the ScanArray Gx microarray scanner, and the intensities of each probe were quantified with the ScanArray Express microarray analysis system (PerkinElmer, Waltham, CA, USA).
Data normalization and statistical analysis
After scanning the arrays, we obtained the raw hybridization intensity for each element printed on the array and then used the background-subtracted median intensity of each spot for further statistical analysis. The two replicates on each chip were initially processed separately. The weakest 5% of spots and the saturated spots in each replicate were discarded. An intensity-dependent method, locally weighed linear regress (Lowess), was used to normalize the data of each replicate (Yang et al., 2002). To minimize experimental error, genes with six or fewer valid measurements were removed from further analysis, and the two technical replicates per chip were averaged to obtain the measurement of each gene for a given sample. The normalized data from each drug treatment group were compared with the normalized data of controls, and significantly regulated genes were identified by unpaired two-tailed Student's t-test. To control the multiple comparison error, the false discovery rate (FDR) was calculated by the method of Benjamini and Hochberg (1995) via MATLAB (The Mathworks Inc., Natick, MA, USA).
The genes significantly regulated by each drug treatment were analyzed by ingenuity pathway analysis (IPA)2 with the goal of revealing the enriched biochemical pathways. The core of IPA is the ingenuity pathways knowledge base (IPKB), which contains the biological function, interaction, and other related information of a curated gene set and more than 330 biochemical pathways. This pathway-based software is designed to identify global canonical pathways, dynamically generated biological networks, and global functions from a given list of genes. Basically, the genes with their symbols, corresponding GenBank Accession Numbers, or both were uploaded into the IPA and compared with the genes included in each canonical pathway using the whole gene set of IPKB as the background. All the pathways with one or more genes overlapping the candidate genes were extracted. In IPA, each of these pathways was assigned a p-value, which denoted the probability of overlap between the pathway and input genes, via Fisher's exact test. Because a relatively large number of pathways were examined, multiple comparison correction for the individually calculated p-values was necessary to make reliable statistical inferences. The FDR was again calculated with the method of Benjamini and Hochberg (1995).
Quantitative real-time RT-PCR
The quantitative real-time RT-PCR (qRT-PCR) was carried out for eight representative genes selected from the aforementioned differentially expressed genes: cyclin d3 (CCND3), cell division cycle 42 (CDC42), death domain-associated protein 6 (DAXX), mitogen-activated protein kinase 14 (MAPK14), mads box transcription enhancer factor 2, polypeptide D (MEF2D), nerve growth factor receptor (NGFR), phospholipase A2, group IVA (PLA2G4A), and transformation-related protein 53 (TRP53). All primers and TaqMan probes for the eight genes were purchased from Applied Biosystems (ABI, Foster City, CA, USA), and qRT-PCR analysis was done as described previously (Gutala et al., 2004; Konu et al., 2004a). Briefly, PCR was carried out in a volume of 25 μl using the TaqMan assay on the ABI 7000 Sequence Detection System. The 18S ribosomal RNA was used as an internal control to normalize the expression patterns of target genes. Data analysis was performed using a comparative Ct method (Winer et al., 1999). Five independent cell cultures were prepared under the same conditions used for the microarray experiments for both the nicotine treatment and control groups (N = 5).
Western blotting analysis
The protein concentration of MAPK14 (p38) was measured in both nicotine-treated (1 mM for 1 h) and control samples. After appropriate treatment, cells were harvested using RIPA buffer followed by incubation on ice for 30 min. The cells were centrifuged at 12,000×g for 15 min to remove debris, and the total protein content of the lysates was determined using the Bio-Rad protein assay (Bio-Rad, Chicago, IL, USA). The samples were boiled at 90°C for 5 min and then cooled on ice. Twenty micrograms of protein was fractionated on 10% SDS-polyacrylamide gel (containing 30% acrylamide and 0.8% bisacrylamide) in Tris–glycine buffer containing SDS at 80 V for approximately 1.5 h. The separated proteins were then transferred to a PVDF membrane (Millipore, Bedford, MA, USA) overnight at 30 V at 4°C. The membrane was blocked for 1 h at room temperature with 5% non-fat dry milk diluted in TBST buffer and incubated overnight with primary antibody at 4°C. After being washed three times in TBST, the membranes were subjected to secondary antibody conjugated to horseradish peroxidase for 1 h at room temperature. Immunoreactivity was detected using an enhanced chemiluminescence kit (Bio-Rad), and the preparations were exposed to X-ray film. The MAPK14 antibody was from Calbiochem (San Diego, CA, USA), and anti-α-tubulin monoclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA) was used to detect the housekeeping protein α-tubulin. The MAPK14 immunoreactivity bands were quantified using densitometry. After the films were developed, they were scanned on a Microtek ScanMaker i800 (Microtek Lab Inc., Cerritos, CA, USA) with ScanWizard 5.5 at a resolution of 600 dpi for quantitative analysis with ImageQuant 5.1 (Molecular Dynamics, Sunnyvale, CA, USA). The relative MAPK14 values were normalized to α-tubulin, and the significance of the difference between the nicotine-treated and control samples was determined by t-test via MATLAB (The Mathworks, Natick, MA, USA). Four independent cell cultures were prepared under the same conditions used for the microarray experiments for both nicotine treatment and control groups (N = 4).
Results
Gene expression changes in response to acute nicotine exposure
In the nicotine-treated group, 295 genes were significantly modulated (p < 0.05; Table A1 in Appendix), with FDR values ≤0.258. Among these genes, 98 had p-values <0.01, which corresponded to an FDR value of 0.155. Of the 295 differentially expressed genes, 221 showed no significant changes (p ≥ 0.05) in the cells treated with nicotine and mecamylamine, suggesting these genes were regulated through the mecamylamine-modulated nicotinic receptors.
Of the 295 genes, 128 were down-regulated by acute nicotine exposure. These included multiple transcription factors; e.g., activating transcription factor 2 (ATF2), general transcription factor IIH polypeptide 2 (GTF2H2), nuclear respiratory factor 1 (NRF1), and transcription factor EB (TRFEB). A few genes related to cell adhesion were also down-regulated by acute nicotine exposure; e.g., ninjurin 1 (NINJ1), kit ligand (KITLG), integrin beta 1 (ITGB1), neurexin 1 (NRXN1), and laminin gamma 1 (LAMC2). Some cell cycle-related genes were suppressed; e.g., cell division cycle 14 homolog B (CDC14B), cell division cycle 2 homolog A (CDC2A), cell division cycle 42 homolog (CDC42), G1 to S phase transition 1 (GSPT1), and meiotic recombination 11 homolog A (MRE11A).
The remaining 167 differentially expressed genes were up-regulated by nicotine treatment. Genes included in this category included those related to: (a) mitochondrial electron transport; e.g., cytochrome c oxidase, subunit VIIC (COX7C), polypeptide 1 of cytochrome p450 family 17 subfamily A (CYP17A1), rieske iron–sulfur protein (RISP), and NADH dehydrogenase (ubiquinone) flavoprotein 1 (NDUFV1); (b) ubiquitination; e.g., ubiquitin-activating enzyme E1 Chr X (UBE1X), ubiquilin 1 (UBQLN1), ubiquitin-specific protease 28 (USP28), and ubiquitin-conjugating enzyme e2a (UBE2A); and (c) transport; e.g., potassium channel TWIK (KCNK1), nucleotide-sensitive chloride channel 1A (CLNS1A), T type alpha 1G subunit of voltage-dependent calcium channel (CACNA1G), solute carrier family 8 (sodium/calcium exchanger) member 2 (SLC8A2), and solute carrier family 2 (facilitated glucose transporter) member 4 (SLC2A4).
Of the 295 genes significantly regulated by nicotine treatment, 74 were differentially expressed (p < 0.05) in the cells treated with nicotine + mecamylamine (Table A1 in Appendix). Most of these genes showed the same trend of regulation as those in the nicotine treatment group, suggesting that mecamylamine may have either no significant effect or a synergistic effect with nicotine on the expression changes in these genes.
Pathways enriched in the genes regulated by nicotine
As shown above, genes and biological processes involved in different functions were regulated by nicotine exposure. To further identify the biological themes underlying these genes, the biochemical pathways enriched in the nicotine-regulated genes were investigated. Two sets of genes were analyzed by IPA; i.e., those regulated significantly by 1 h of nicotine exposure (N = 295), and those with expression significantly changed by 1 h of nicotine with the effect being blocked by mecamylamine (N = 221). The second gene set included only those with p-values ≥0.05 during co-treatment with nicotine and mecamylamine. Although in this set, some genes may have considerable fold changes, the selection was based on p-value only.
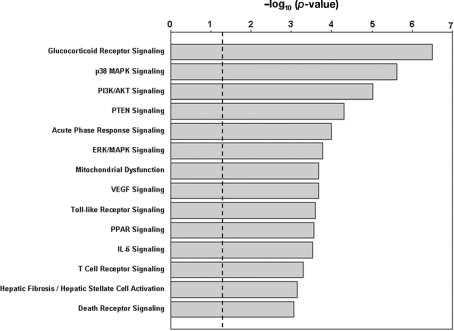
Results from IPA analysis showed that 14 pathways were significantly enriched among the genes regulated by nicotine (p < 0.05; FDR < 0.05; Figure 1; Table 1), and 13 pathways were enriched in the second gene set. Among these pathways, nine were identified for both gene sets; i.e., glucocorticoid receptor signaling, p38 MAPK signaling, PI3K/AKT signaling, PTEN signaling, acute-phase response signaling, ERK/MAPK signaling, Toll-like receptor signaling, VEGF signaling, and T-cell receptor signaling. For the gene set regulated by nicotine treatment, five pathways were uniquely identified; i.e., mitochondrial dysfunction, PPAR signaling, IL-6 signaling, hepatic fibrosis/hepatic stellate cell activation, and death receptor signaling. The pathways uniquely identified in the second gene set were CD28 signaling in T-helper cells, CD40 signaling, role of NFAT in regulation of the immune response, and IL-8 signaling. In the pathways identified for the two gene sets, the genes affected were not identical (Table 1). For the nicotine + mecamylamine-treated samples, the pathways included only genes whose expression was modulated by nicotine exposure but blocked by simultaneous treatment with the two chemicals. For the nicotine-treated samples, the pathways also included genes whose expression was modulated in both nicotine- and nicotine + mecamylamine-treated samples.
Figure 1.
Pathways significantly enriched in the genes differentially regulated by nicotine exposure. The p-values are shown in negative logarithm form at base 10. The dashed line indicates the threshold corresponding to p = 0.05. All 14 pathways shown have an FDR < 0.05.
Table 1.
Pathways enriched in genes significantly regulated by nicotine and genes significantly regulated by nicotine but not by nicotine + mecamylamine.
| Gene regulated by nicotine | Gene regulated by nicotine but blocked by NM* | ||||
|---|---|---|---|---|---|
| Pathway | p-Value | FDR | p-Value | FDR | Genes significantly regulated by nicotine in the pathway** |
| Glucocorticoid receptor signaling | 3.11 × 10−7 | 1.77 × 10−4 | 4.46 × 10−6 | 7.00 × 10−4 | AGT, AR, BCL2, CD3G, DUSP1(MKP1), FGG, FOS, GTF2H2, IKBKG, JAK3, MAP3K7IP1(TAB1), MAPK14, MED14, NFATC3, PIK3CG, SMAD2, STAT5A, TGFB2, TGFBR2, TRAF6, YWHAH |
| p38 MAPK signaling | 2.40 × 10−6 | 6.82 × 10−4 | 6.54 × 10−5 | 2.57 × 10−3 | ATF2, DAXX, DUSP1(MKP1), HMGN1, IRAK3, MAP3K7IP1(TAB1), MAPK14, MEF2D, PLA2G4A, TGFB2, TGFBR2, TRAF6 |
| PI3K/AKT signaling | 9.55 × 10−6 | 1.81 × 10−3 | 3.29 × 10−5 | 1.7 × 10−3 | BCL2, EIF4EBP1, FOXO1, IKBKG, ITGB1, JAK3, PIK3CG, PPM1L, RPS6KB1, SFN, TRP53, YWHAH |
| PTEN signaling | 5.01 × 10−5 | 7.13 × 10−3 | 1.51 × 10−3 | 0.030 | BCL2, BCL2L11, CDC42, FOXO1, IKBKG, ITGB1, NGFR, PIK3CG, RPS6KB1, YWHAH |
| Acute-phase response signaling | 1.02 × 10−4 | 0.012 | 7.06 × 10−4 | 0.018 | AGT, APOA1, FGG, FOS, IKBKG, IL6R, IL6ST, MAP3K7IP1(TAB1), MAPK14, NGFR, PIK3CG, TF, TRAF6 |
| ERK/MAPK signaling | 1.70 × 10−4 | 0.015 | 2.51 × 10−4 | 0.016 | ARAF, ATF2, DUSP1, EIF4EBP1, FOS, ITGB1, PIK3CG, PLA2G4A, PPM1L, PPP1CA, RAPGEF3, YWHAH |
| Mitochondrial dysfunction | 2.09 × 10−4 | 0.015 | – | – | COX6B1, COX7B, COX7C, NDUFA4, NDUFV1, OGDH, SDHB, SDHC, SNCA, UQCRFS1 |
| VEGF signaling | 2.14 × 10−4 | 0.015 | 6.17 × 10−4 | 0.018 | BCL2, EIF2B1, EIF2B5, FOXO1, PIK3CG, SFN, VCL, VEGFA |
| Toll-like receptor signaling | 2.57 × 10−4 | 0.015 | 2.94 × 10−3 | 0.045 | FOS, IKBKG, IRAK3, MAP3K7IP1(TAB1), MAP4K4, MAPK14, TRAF6 |
| PPAR signaling | 2.75 × 10−4 | 0.015 | – | – | FOS, IKBKG, MAP3K7IP1(TAB1), MAP4K4, NGFR, NR2F1, STAT5A, TRAF6 |
| IL-6 signaling | 2.95 × 10−4 | 0.015 | – | – | FOS, IKBKG, IL6R, IL6ST, MAP3K7IP1(TAB1), MAP4K4, MAPK14, NGFR, TRAF6 |
| T-cell receptor signaling | 5.13 × 10−4 | 0.024 | 3.97 × 10−4 | 0.013 | CALM2, CD3G, CSK, FOS, IKBKG, NFATC3, PIK3CG, ZAP70 |
| Hepatic fibrosis/hepatic stellate cell activation | 7.24 × 10−4 | 0.032 | – | – | AGT, BCL2, IGF1R, IL6R, NGFR, SMAD2, TGFB2, TGFBR2, VEGFA |
| Death receptor signaling | 8.71 × 10−4 | 0.035 | – | – | BCL2, CFLAR, DAXX, IKBKG, MAP4K4, TNFRSF10B |
| CD28 signaling in T-helper cells | – | – | 2.90 × 10−5 | 1.72 × 10−3 | ARPC1A, ARPC1B, CALM2, CD3G, CDC42, CSK, FOS, IKBKG, NFATC3, PIK3CG, ZAP70 |
| CD40 signaling | – | – | 1.38 × 10−3 | 0.030 | FOS, IKBKG, JAK3, MAP2K5, MAPK14, PIK3CG, TRAF6 |
| Role of NFAT in regulation of the immune response | – | – | 3.01 × 10−3 | 0.045 | CALM2, CD3G, FOS, GNA15, IKBKG, MEF2D, NFATC3, PIK3CG, ZAP70 |
| IL-8 signaling | – | – | 3.15 × 10−3 | 0.045 | BCL2, CCND3, EIF4EBP1, IKBKG, IRAK3, LIMK1, PIK3CG, TRAF6, VEGFA |
*NM, nicotine + mecamylamine. **For each pathway, all the genes were significantly regulated by nicotine exposure (p < 0.05). The expression changes of the genes were blocked by the co-treatment of nicotine + mecamylamine except those shown in bold font, which means genes shown in bold font were differentially expressed in both the nicotine and nicotine + mecamylamine-treated samples.
We also performed pathway analysis on the 74 genes significantly regulated in both nicotine-treated and the nicotine + mecamylamine-treated samples via IPA. Although a few pathways were assigned p-values <0.05, they are not reported here because of the relatively high FDR value after multiple comparison correction (with the minimum FDR being 0.262).
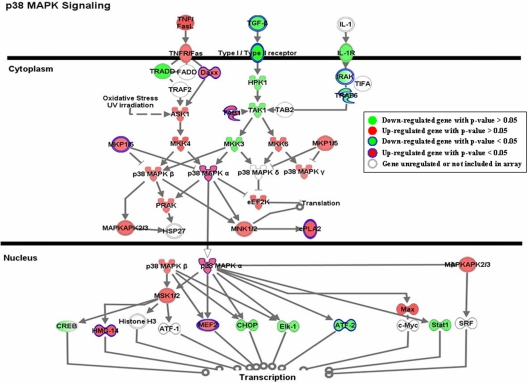
As shown in Table 1, multiple genes in each pathway were significantly regulated by nicotine exposure. Although the expression changes of some genes within each pathway were not statistically significant, they showed consistent patterns with those significantly modulated. One of the pathways, p38 MAPK signaling, is shown in Figure 2. Genes involved in this pathway were extensively regulated in response to nicotine exposure. The expression of a few genes, such as DAXX, MKP1/5, MAPK14, and MEF2, was significantly modulated by acute nicotine exposure (p < 0.05). Although the expression of other genes; e.g., ASK1, MKK4, and HPK1, was not significantly regulated under the experimental conditions, they showed patterns consistent with those of significantly regulated genes.
Figure 2.
Gene expression pattern of the P38 MAPK signaling pathway. Genes shown in green are down-regulated, whereas those shown in red are induced by acute nicotine treatment. The genes significantly regulated by nicotine are those with closed blue edges. To demonstrate the expression pattern of the pathway, all the genes with expression ratios > 1.1 or <0.90 relative to the controls are also shown. The genes in white with light gray edges are either not regulated by nicotine (expression ratios between 0.9 and 1.1) or were not measured in our analysis. The pathway was drawn by the IPA program.
Expression verification of representative genes
In an independent experiment involving the same treatments as in the microarray analysis, qPCR was performed on eight representative genes; i.e., CCND3, CDC42, DAXX, MAPK14, MEF2D, NGFR, PLA2G4A, and TRP53. All these genes were significantly regulated by nicotine exposure (Table A1 in Appendix), and each gene was part of one or more enriched pathways associated with nicotine treatment (Table 1). For example, CCND3 is part of the IL-8 signaling pathway; CDC42 of CD28 signaling and PTEN signaling pathways; DAXX of the p38 MAPK signaling pathway; MAPK14 of multiple pathways such as glucocorticoid receptor signaling, p38 MAPK signaling, acute-phase response signaling, and Toll-like receptor signaling; MEF2D of p38 MAPK signaling that plays a role of NFAT in regulation of the immune response; NGFR of PTEN signaling, acute-phase response signaling, PPAR signaling, and IL-6 signaling; PLA2G4A of p38 MAPK and ERK/MAPK signaling; and TRP53 of the PI3K/AKT signaling pathway.
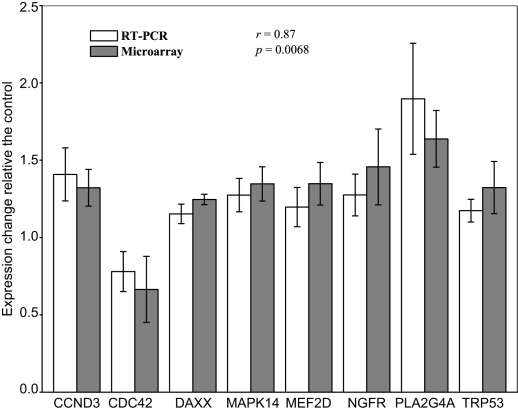
Figure 3 shows the fold change in activity along with the standard deviation of the eight genes in response to nicotine treatment relative to the controls, as detected by microarray and quantitative RT-PCR. These analyses revealed significant up-regulation of seven genes and down-regulation of CDC42, which is consistent with the result from the microarray analysis. Furthermore, a comparison of the fold changes of each gene detected by the two molecular techniques revealed a correlation coefficient of 0.87 (p = 0.0068), indicating overall measurement of gene expression from the two approaches was consistent. Although only eight representative genes were selected for verification by qRT-PCR, a comparison of the two techniques demonstrates that the results from our microarray analyses were highly reproducible and reliable.
Figure 3.
Comparative plot of results from microarray and real-time RT-PCR verification for eight representative genes. Y axis represents the fold change of expression in 1-h nicotine-treated SH-SY5Y cells compared with the average expression in the control sample for that particular gene. For the qRT-PCR results, the expression of each gene was normalized to the average of control samples. Abbreviations: CCND3, cyclin D3; CDC42, cell division cycle 42 homolog; DAXX, Fas death domain-associated protein; MAPK14, mitogen-activated protein kinase 14; MEF2D, myocyte enhancer factor 2d; NGFR, nerve growth factor receptor; PLA2G4A, phospholipase A2, group IVA; TRP53, transformation-related protein 53.
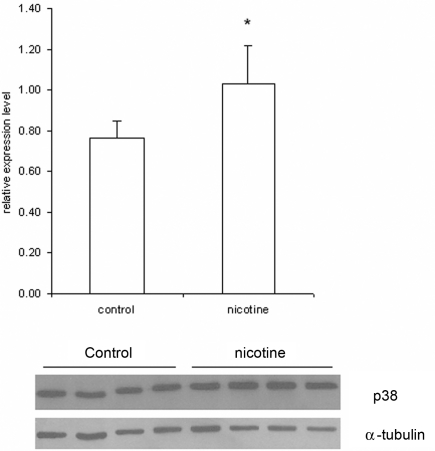
Furthermore, we used Western blotting to determine the total amount of MAPK14 (p38 MAPK) protein in lysates of cells treated with 1 mM nicotine and control samples. The results revealed significant induction (35.5%; p = 0.03) of MAPK14 expression in nicotine-treated samples (Figure 4).
Figure 4.
Expression of MAPK14 (p38 MAPK) protein in SH-SY5Y cells determined by Western blotting. The SH-SY5Y cells were treated for 1 h with or without (Controls) 1 mM nicotine. After treatment, protein was purified from the lysates and analyzed by Western blotting. In this figure, the MAPK14 concentration was determined using a specific monoclonal antibody. The histogram shows the relative expression of MAPK14 normalized to expression of α-tubulin, which demonstrated that the protein concentrations in nicotine-treated cells are significant higher than those in controls (p < 0.03). Data are presented as mean ± SEM (N = 4).
Discussion
In this study, we profiled gene expression in the SH-SY5Y cell line in response to 1 h of 1 mM nicotine exposure using a pathway-focused oligonucleotide microarray. Treatment with 1 mM nicotine maximally induces the up-regulation of numbers of nAChR radioligand-binding sites in SH-SY5Y cells, and this up-regulation has been implicated in the long-lasting effects of nicotine exposure, such as dependence and tolerance (Ke et al., 1998; Dunckley and Lukas, 2003, 2006). Given that the purpose of this study was to determine the pharmacologic effects of nicotine using an in vitro system, we adopted a 1-mM concentration for nicotine, as used in a previous study (Dunckley and Lukas, 2003). However, the current study is neither a simple modeling of the physiological conditions of smokers, nor repeat earlier studies in the same model. Instead, it focuses on exploring the molecular response at the pathway level to acute nicotine exposure that may maximally activate the function of molecules such as nAChRs and their downstream genes in SH-SY5Y cells.
Nicotine evokes its physiological effects mainly by binding with nAChRs. As a broad-spectrum antagonist of nAChRs, mecamylamine at a concentration of 3 μM can block most of the responses of α3*-nAChR to nicotine, as well as a fraction of the α7-nAChR responses (Chavez-Noriega et al., 1997; Papke et al., 2001; Dunckley and Lukas, 2003, 2006). Of the 295 genes significantly regulated by nicotine, 221 were not differentially expressed during co-treatment of nicotine + mecamylamine, providing evidence of direct involvement of nAChR in the process of modulation of SH-SY5Y cells by nicotine. Thus, our results indicate that the changes in 75% (221/295) of the differentially expressed genes in response to nicotine are attributable to direct interaction with nAChRs, especially α3*-nAChR receptors, which is consistent with the conclusions drawn from previous studies (Dunckley and Lukas, 2003, 2006). However, it is difficult to confirm a clear relation between nicotinic receptor subtype and gene expression from our findings because of the existence of different subtypes of nAChRs on the membrane of SH-SY5Y cells. These receptors have different properties, such as sensitivity to nicotine, permeability to calcium, and propensity to desensitize. It is possible that the regulation of some genes is related to multiple subtypes of nAChRs, including those α3- or α7-containing receptors. Partially blocking the function of some types of receptors (e.g., α3*- and α7-containing) may reduce the overall activation of nAChRs, which may lead to more subtle regulation of some genes compared with the response when mecamylamine is not applied. On the other hand, our results do not imply that nAChRs play little or no role in the regulation of genes significantly regulated in both nicotine and nicotine + mecamylamine treatment. The expression changes of these genes may have multiple explanations. A small fraction of α3*-nAChR receptors remain functionally active under the influence of mecamylamine, which may be sufficient to regulate the expression of some genes. At the same time, α7 and other types of nAChR receptors may be responsible for the expression change of some other genes as well. It is also possible that the expression changes of some genes in response to nicotine are caused by mechanisms not directly involving nAChRs, as nicotine can readily cross the plasma membrane and affect intracellular processes through an nAChR-independent mechanism.
Our analysis showed that acute nicotine exposure has effects on the expression of a large number of genes with diverse functions (Table A1 in Appendix). Among them are genes involved in the ubiquitin–proteasome pathway, such as proteasome subunit beta 4 (PSMB4), proteasome 26S subunit non-ATPase 2 (PSMD2), ubiquitin-activating enzyme E1 Chr X (UBE1X), ubiquitin-conjugating enzyme E2A (UBE2A), ubiquilin 1 (UBQLN1), and ubiquitin-specific protease 28 (USP28). Earlier studies showed that genes involved in the ubiquitin–proteasome pathway were regulated in SH-SY5Y cells by 1 mM nicotine after both 1- and 24-h treatment (Dunckley and Lukas, 2003, 2006). In the current study, more genes related to this pathway were found to be modulated by nicotine exposure. The ubiquitin–proteasome pathway is regulated by nicotine in cell lines (Konu et al., 2004b; Ficklin et al., 2005; Wang et al., 2007), animal models (Kane et al., 2004; Li et al., 2004; Rezvani et al., 2007; Vadasz et al., 2007; Wang et al., 2009), and human smokers (Mexal et al., 2005). Thus, the changes in ubiquitin–proteasome-related genes observed in this study may not be specific for the relatively high nicotine concentration; instead, they may be caused by mechanisms similar to those discovered in previous studies.
Multiple genes involved in mitochondrial functions also were found to be regulated by nicotine; these include acyl-coenzyme A thioesterase 3 (ACATE3), ATP synthase subunit g (ATP5L), mitochondrial ribosomal protein S7 (MRPS7), NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 4 (NDUFA4), and RISP. Previous studies have demonstrated that nicotine exposure can modulate the morphology (Onal et al., 2004) and function of mitochondria, such as protein turnover (Katyare and Shallom, 1988), enzyme activity (Xie et al., 2005), and generation of reactive oxygen species (Cormier et al., 2001; Soto-Otero et al., 2002). Recently, we showed that nicotine exposure regulates the electron transport system in multiple brain regions of rats (Wang et al., 2009). The results from the current study indicate that acute nicotine exposure also can regulate the expression of mitochondrial genes, potentially leading to the modulation of mitochondrial functions. In addition, several genes involved in synaptic transmission were identified, such as neuron-specific gene family member 1 (NSG1), synuclein alpha (SNCA), synapsin 2 (SYN2), synaptotagmin 2 (SYT2), synaptotagmin 7 (SYT7), and tachykinin 2 (TAC2). The expression of NSG1 in mouse cortical neurons is regulated by treatment with nicotine, alcohol, or both (Wang et al., 2007). SNCA is suggested as a candidate associated with alcohol, cocaine, and methamphetamine dependence (Kobayashi et al., 2004; Bonsch et al., 2005; Mash et al., 2008; Spence et al., 2009). The phosphorylation of synapsin is regulated by cocaine in a region-specific manner in rat brain (Edwards et al., 2007). The regulation of these synaptic transmission-related genes indicates that even a short nicotine exposure may have important effects on neuronal functions.
Among the 74 genes significantly regulated by both nicotine and nicotine + mecamylamine treatment, some have already been reported to be involved in the cellular response to nicotine exposure. Activating transcription factor 2 (ATF2), a DNA-binding protein that binds to cAMP response elements, was suppressed in both treatment conditions and is involved in mediating the activation of tyrosine hydroxylase transcription under nicotine treatment in PC12 cells (Gueorguiev et al., 2006). Nerve growth factor (NGF), a member of the neurotrophin family, is an essential mediator of neuronal activity and synaptic plasticity (Levi-Montalcini, 1987; Thoenen et al., 1987). Both NGF and its receptor, NGFR, were up-regulated in the two treatment groups. Nicotine has been reported to regulate the expression of NGF and NGFR in cell lines or tissues (Terry and Clarke, 1994; Jonnala et al., 2002; Garrido et al., 2003; Hernandez and Terry, 2005; French et al., 2006). Neurexin 1 (NRXN1) was down-regulated in both treatment groups. This and another member of the same family, NRXN3, were associated with smoking behaviors in several recent genetic studies (Bierut et al., 2007; Nussbaum et al., 2008; Novak et al., 2009). Although our data provide no further information on the mechanisms by which these genes are regulated, it is clear that acute nicotine exposure is responsible for their expression changes.
Consistently, the pathways enriched in nicotine-regulated genes were involved in many biological processes, such as neural development/synaptic plasticity, cell survival/death, immune response, and cellular metabolism. Of the identified pathways, nine were enriched in genes significantly regulated by nicotine, as well as those regulated by nicotine but blocked by mecamylamine, indicating these pathways were regulated mainly through nAChR-dependent mechanisms. Among these pathways, acute-phase response signaling, Toll-like receptor signaling, and T-cell receptor signaling play important roles in the immune response. Although the other pathways; i.e., glucocorticoid receptor signaling, p38 MAPK signaling, PI3K/AKT signaling, PTEN signaling, ERK/MAPK, and VEGF signaling, are involved in a large range of physiological processes, all of them represent key pathways for maintaining proper neuronal functions, such as cellular signaling and proliferation and differentiation of neurons. The modulation of the neuronal function-related pathways may have relatively long-lasting consequences. One example is that adolescent rats display different long-term neuroadaptive responses to acute nicotine than do adult animals, which is suggested to be related to the immature or still-developing plasticity mechanisms in the prefrontal cortex of young animals (Schochet et al., 2004). Among these pathways, particularly interesting are the glucocorticoid receptor and p38 MAPK signaling.
For the glucocorticoid receptor signaling pathway, 21 genes were significantly regulated by acute nicotine treatment (Table 1). The glucocorticoid receptor, GCCRα (also known as nuclear receptor subfamily 3, group c, member 1; NR3C1) was slightly suppressed by nicotine (fold change 0.83; p-value = 0.09). The regulation of these genes clearly indicates that the glucocorticoid receptor signaling pathway is modulated by acute nicotine exposure. Glucocorticoids, cortisol in humans, and corticosterone in the rodent, are secreted by the adrenal cortex and belong to the family of steroid hormones. Glucocorticoids and their receptors are essential for the development and survival of vertebrates (Cole et al., 1995). All the cellular responses to glucocorticoids are attributed to their binding to the intracellular glucocorticoid receptor (GCCR) and the translocation of the complexes to the nucleus, which then modulates gene expression through diverse mechanisms (Dittmar et al., 1997, 1998; Robinson-Rechavi et al., 2001). Although the activity of the GCCR is directly related to gene transactivation, considerable cross-talk occurs between the GCCR and a cohort of molecules to mediate their functions as transcriptional regulators. Studies on animals and humans have outlined the importance of glucocorticoids in mediating drug-seeking behavior and the rewarding properties of psychostimulant drugs (Piazza and Le Moal, 1997; Marinelli and Piazza, 2002). For example, in mice, activation of glucocorticoid receptors increases the propensity to self-administer cocaine by acting on the postsynaptic dopaminoceptive neurons of the dopaminergic system (Ambroggi et al., 2009). It was also found that learning and memory deficits in cocaine-dependent humans are associated with higher cortisol concentrations (Fox et al., 2009). The neuronal circuitry adaptations induced by heroin self-administration were correlated with elevated glucocorticoid production in certain brain regions of rats (Weber et al., 2009). Corticosteroid receptor modulation of the mesoaccumbens dopamine neurotransmission is believed to be a key neurobiological mechanism mediating the effects of stress in addiction (Tye et al., 2009). Nicotine can also interact with glucocorticoid receptor signaling cascades directly or indirectly. Furthermore, the relations between nicotine and glucocorticoids are bidirectional; i.e., not only can nicotine affect the hypothalamic–pituitary–adrenal (HPA) axis and circulating glucocorticoids, but adrenal steroids can modulate nicotine's behavioral and physiological actions (Caggiula et al., 1998). Nicotine administration elevates plasma corticosterone in animals (Caggiula et al., 1998). Similarly, both cigaret smoking and nicotine infusion rapidly increase circulating cortisol concentrations in humans (Caggiula et al., 1998). In human smokers, the degree of nicotine addiction is reflected by changes in the sensitivity of central glucocorticoid receptors (Reuter et al., 2004). Nicotine administration to pregnant animals can lead to fetal overexposure to maternal glucocorticoid and result in fetal adrenocortical dysfunction and other diseases after birth (Chen et al., 2007). The significant regulation of the corticosteroid receptor signaling pathway by acute nicotine exposure provides additional evidence of the importance of this pathway in nicotine addiction, and the significantly regulated genes in this pathway are excellent candidates for further investigation. At the same time, the cross-talk between glucocorticoid receptor signaling and other pathways, such as MAPK signaling and PI3K/AKT signaling (Clark and Lasa, 2003; Gupta et al., 2007), implies that the regulation of this pathway indirectly induces modulations in other biological processes.
The MAPK signaling pathway is a major modulator of cell growth, division, and differentiation (Adams and Sweatt, 2002) and plays a key role in mediating neuronal activation induced by dopamine, glutamate, and drugs of abuse (Jiao et al., 2007). Nicotine activates the MAPK signaling pathway in a variety of tissues and cell types, including SH-SY5Y cells (Heusch and Maneckjee, 1998; Tang et al., 1998; Dajas-Nakayama et al., 2001; Wang et al., 2001; Bailador et al., 2002). Furthermore, many cellular processes are affected by both nicotine and MAPK signaling, such as cell survival and memory processing (Levin and Simon, 1998). Despite these known interactions between MAPK signaling and nicotine exposure, our work has shown a more detailed pattern of the effect of acute nicotine exposure on this pathway. A number of genes were significantly regulated by nicotine exposure (Table 1). There were also genes in this pathway that were regulated to a lesser extent (Figure 2). Whereas nicotine exposure decreases the amount of mRNA for TGF-beta 2 (TGFB2), TGFBR2, interleukin 1 receptor (IL1R), and interleukin 1 receptor-associated kinase 1 (IRAK), as well as a couple downstream genes, it also induces the expression of a number of genes, such as death-associated protein 6 (DAXX), MAPK14 (p38 MAPK), and mads box transcription enhancer factor 2 polypeptide D (MEF2D). The regulation pattern of this pathway clearly indicates the cross-talk and interaction between different cascades. Earlier studies showed that nicotine treatment decreases the amount of TGF-beta mRNA in different tissues. For example, nicotine treatment significantly suppresses the release of TGF-beta 1 in bovine aortic endothelial cells (Cucina et al., 1999) and several other cell lines (Lane et al., 2005). This protein has a bimodal dose-dependent effect on the proliferation of various cell types, being stimulatory at low concentration and inhibitory at high concentration (Pollman et al., 1999; Ghosh et al., 2005). The observed mRNA decrease of TGFB2 and its receptor, TGFBR2, may indicate an increase in cell proliferation, although this remains to be established. Our analysis further showed a moderate enhancement of MAPK14 as judged by both mRNA and protein expression. MEF2 is one of the many downstream targets regulated by MAPK14. In our analysis, the transcription of two members of the MEF2 family was induced in the nicotine-treated samples; i.e., MEF2D (fold change 1.35; p = 0.008) and MEF2C (fold change 1.27; p = 0.177). MEF2 genes are highly expressed in neurons of the CNS (Heidenreich and Linseman, 2004; Shalizi and Bonni, 2005) and play important roles in neuronal differentiation and maintenance. Blocking the function of MEF2 genes may result in the death of neurons, whereas their expression can promote neuronal survival (Mao et al., 1999; Li et al., 2001; Gaudilliere et al., 2002). Although the activity of MEF2 is regulated mainly via its phosphorylation by MAPK14 or other kinases, considering the expression induction of MAPK14 and MEF2 genes, it may be reasonable to assume the activity of MEF2 is enhanced by acute nicotine exposure in SH-SY5Y cells.
In an earlier study, Dunckley and Lukas (2003) analyzed the gene expression patterns in SH-SY5Y cells treated with nicotine under similar experimental conditions (i.e., 1 mM nicotine for 1 h) with different microarray platforms. Among the 17 genes identified, eight had valid expression measurements in our analysis, and their expression patterns were largely consistent in the two studies. While Dunckley and Lukas (2003) performed a more detailed investigation of the relation between the nicotine-regulated genes and nAChRs, we here provide a much more broad and comprehensive analysis and description of the biological processes/pathways underlying responses to acute nicotine exposure. Results from the two studies also indicate that more detailed and comprehensive information about the interaction between acute nicotine and SH-SY5Y cell line could be obtained.
Conclusion
By profiling the gene expression pattern in SH-SY5Y cell lines treated acutely with nicotine, we have identified a set of significantly modulated genes. Further, we showed that the majority of these differently expressed genes were regulated through the nAChR-mediated mechanism. From these significantly regulated genes, we have detected the enriched biochemical pathways, which included those potentially involved in the neuronal response to addictive drugs such as nicotine, including glucocorticoid receptor and p38 MAPK signaling. These findings can help us to understand the molecular mechanisms underlying the development of nicotine addiction. On the other hand, the identification of these genes and pathways further indicates that the physiological consequences of acute nicotine exposure are complicated. Thus, further investigation is necessary in order to obtain a more detailed and comprehensive understanding of the interaction between acute nicotine treatment and the nervous system.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This project was in part supported by NIH grants DA-13783 and DA-12844 to Ming D. Li. The authors thank Dr. David L. Bronson for his excellent editing of this manuscript.
Appendix
Table A1.
Genes significantly expressed in nicotine-treated group.
| Nicotine | Nicotine and mecamylamine | ||||
|---|---|---|---|---|---|
| Gene symbol | Gene name | p-Value | Ratio | p-Value | Ratio |
| ABCD3 | ATP-binding cassette, subfamily D (ALD), member 3 | 0.047 | 1.17 | 0.129 | 1.24 |
| ACAT2 | Acetyl-coenzyme A acyltransferase 2 | 0.046 | 1.22 | 7.03 × 10−3 | 1.34 |
| ACATE3 | Acyl-coenzyme A thioesterase 3 | 9.09 × 10−3 | 0.78 | 0.052 | 0.86 |
| ADCY8 | Adenylyl cyclase 8 | 0.034 | 0.80 | 0.045 | 0.82 |
| ADRA1B | Adrenergic receptor, alpha 1B | 4.88 × 10−3 | 1.31 | 0.361 | 1.17 |
| AGT | Angiotensinogen | 0.039 | 1.50 | 0.593 | 1.10 |
| ALG2 | Asparagine-linked glycosylation 2 homolog (yeast, alpha-1,3-mannosyltransferase) | 2.75 × 10−3 | 1.38 | 0.036 | 1.37 |
| APC | Adenomatosis polyposis coli | 0.046 | 0.83 | 0.780 | 0.96 |
| APG4C | APG4 (ATG4) autophagy-related homolog C (S. cerevisiae) | 0.045 | 0.83 | 0.702 | 1.08 |
| APOA1 | Apolipoprotein A-I | 0.044 | 1.68 | 0.087 | 1.20 |
| AR | Androgen receptor | 0.016 | 0.76 | 0.939 | 1.01 |
| ARAF1 | V-raf murine sarcoma 3611 viral oncogene homolog 1 | 0.030 | 1.24 | 0.016 | 1.15 |
| ARPC1A | Suppressor of profilin/p41 of actin-related complex 2/3 | 0.033 | 1.58 | 0.051 | 1.34 |
| ARPC1B | Actin-related protein 2/3 complex, subunit 1B | 0.042 | 1.26 | 0.282 | 1.11 |
| ASPA | Aspartoacylase | 0.041 | 0.75 | 0.131 | 1.57 |
| ATF2 | Activating transcription factor 2 | 0.016 | 0.80 | 0.026 | 0.93 |
| ATP5L | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit g; F1F0-ATP | 0.048 | 0.74 | 0.218 | 1.41 |
| ATP6V1A1 | ATPase, H+ transporting, V1 subunit A, isoform 1; ATPase, H+ transporting | 0.012 | 1.28 | 0.209 | 1.29 |
| B3GALT3 | UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 3 | 0.034 | 0.70 | 0.017 | 0.50 |
| BCL2 | B-cell leukemia/lymphoma 2 | 0.033 | 1.24 | 0.473 | 1.15 |
| BCL2L11 | Bcl-12-like 11 (apoptosis facilitator) | 0.023 | 0.65 | 0.381 | 0.91 |
| BEGAIN | Brain-enriched guanylate kinase-associated protein 1 mRNA, complete cds | 0.030 | 1.26 | 0.111 | 1.15 |
| BRCA1 | Breast cancer 1 | 0.033 | 0.71 | 0.150 | 0.79 |
| CACNA1G | Calcium channel, voltage-dependent, T type, alpha 1g subunit | 0.019 | 1.38 | 0.842 | 1.03 |
| CALCR | Calcitonin receptor | 0.041 | 1.25 | 0.926 | 1.01 |
| CALCRL | Calcitonin receptor-like receptor | 0.026 | 1.63 | 0.033 | 1.87 |
| CALD1 | Caldesmon 1 | 0.020 | 1.27 | 0.068 | 1.34 |
| CALM2 | Calmodulin 2 | 0.043 | 1.39 | 0.187 | 1.11 |
| CALR | Calreticulin | 0.039 | 1.25 | 0.769 | 1.02 |
| CAMK1 | CaM-like protein kinase | 3.08 × 10−3 | 0.84 | 0.235 | 0.95 |
| CATNA1 | Catenin alpha 1 | 0.043 | 1.25 | 0.074 | 1.19 |
| CATNB | Catenin beta | 0.011 | 1.46 | 0.021 | 1.20 |
| CCNA2 | Cyclin A2 | 0.022 | 1.22 | 0.482 | 1.05 |
| CCND3 | Cyclin D3 | 8.99 × 10−3 | 1.32 | 0.696 | 0.96 |
| CCNE1 | Cyclin E1 | 0.030 | 1.64 | 0.122 | 1.34 |
| CD3G | CD3 antigen, gamma polypeptide | 0.029 | 1.19 | 0.489 | 1.13 |
| CDC14B | CDC14 cell division cycle 14 homolog B (S. cerevisiae) | 0.024 | 0.56 | 0.110 | 0.68 |
| CDC25A | Cell division cycle 25 homolog A (S. cerevisiae) | 6.02 × 10−3 | 0.75 | 7.82 × 10−4 | 0.74 |
| CDC2A | Cell division cycle 2 homolog A (S. pombe) | 0.031 | 0.79 | 0.812 | 0.96 |
| CDC42 | Cell division cycle 42 homolog | 9.85 × 10−3 | 0.67 | 0.798 | 1.03 |
| CDIPT | Phosphatidylinositol synthase | 0.030 | 1.50 | 0.104 | 1.31 |
| CDKL2 | Cyclin-dependent kinase-like 2 (CDC2-related kinase) | 0.046 | 1.33 | 0.198 | 1.44 |
| CDRT1 | CMT1A duplicated region transcript 1 | 0.012 | 1.40 | 0.784 | 0.95 |
| CFLAR | CASP8 and FADD-like apoptosis regulator | 0.010 | 1.35 | 0.313 | 0.95 |
| CIAO1 | WD40 protein Ciao1 | 0.015 | 0.64 | 0.666 | 0.94 |
| CLCN5 | Chloride channel 5 | 0.011 | 0.64 | 9.13 × 10−4 | 0.80 |
| CLNS1A | Chloride channel, nucleotide-sensitive, 1A | 0.023 | 1.31 | 0.583 | 1.08 |
| CLSTN3 | Calsyntenin 3 | 0.033 | 1.33 | 0.093 | 1.22 |
| COPS3 | COP9 (constitutive photomorphogenic) homolog, subunit 3 (Arabidopsis thaliana) | 0.046 | 1.45 | 0.734 | 1.03 |
| COX6B | Cytochrome c oxidase, subunit VIb | 6.98 × 10−3 | 1.56 | 0.344 | 1.10 |
| COX7B | Cytochrome c oxidase, subunit VIIb | 0.032 | 1.22 | 0.028 | 1.21 |
| COX7C | Cytochrome c oxidase, subunit VIIc | 1.71 × 10−3 | 1.31 | 0.150 | 1.12 |
| COX8B | Cytochrome c oxidase, subunit VIIIb | 3.37 × 10−3 | 1.93 | 0.019 | 1.84 |
| CREG | Cellular repressor of E1A-stimulated genes 1 | 0.023 | 1.25 | 0.621 | 1.06 |
| CRSP2 | Cofactor required for Sp1 transcriptional activation subunit 2 (150 kDa) | 4.19 × 10−3 | 0.67 | 0.108 | 0.82 |
| CRYZ | Crystallin, zeta | 0.035 | 1.14 | 0.078 | 1.17 |
| CSK | C-src tyrosine kinase | 0.044 | 1.45 | 0.110 | 1.29 |
| CTCF | CCCTC-binding factor | 0.029 | 1.18 | 0.014 | 1.31 |
| CTNF | Ciliary neurotrophic factor receptor | 4.85 × 10−3 | 0.74 | 0.169 | 1.34 |
| CYP17A1 | Cytochrome P450, family 17, subfamily a, polypeptide 1 | 0.043 | 1.33 | 0.283 | 1.23 |
| CYP2F1 | Cytochrome P450, subfamily IIF, polypeptide 1 | 0.022 | 0.68 | 0.082 | 0.77 |
| CYP2R1 | Cytochrome P450, family 2, subfamily r, polypeptide 1 | 0.030 | 0.84 | 0.517 | 1.09 |
| DAB1 | Disabled homolog 1 (Drosophila) | 0.036 | 0.55 | 0.245 | 0.76 |
| DAXX | Death domain-associated protein 6 | 9.34 × 10−3 | 1.25 | 0.501 | 1.11 |
| DFFA | DNA fragmentation factor, 40 kD, beta polypeptide (caspase-activated DNase) | 9.62 × 10−3 | 0.88 | 0.522 | 1.03 |
| DIO2 | Deiodinase, iodothyronine, type II | 0.040 | 1.27 | 0.583 | 1.14 |
| DNAJA4 | DnaJ (Hsp40) homolog, subfamily A, member 4 | 4.48 × 10−3 | 0.66 | 0.481 | 1.11 |
| DNCIC2 | Dynein, cytoplasmic, intermediate polypeptide 2 | 0.024 | 1.38 | 0.294 | 0.96 |
| DOCK7 | Dedicator of cytokinesis 7 | 3.57 × 10−3 | 1.39 | 0.256 | 1.26 |
| DRAP1 | Dr1 associated protein 1 (negative cofactor 2 alpha) | 6.61 × 10−3 | 0.75 | 0.025 | 0.79 |
| DRD4 | Dopamine receptor D4 | 0.013 | 1.23 | 0.714 | 1.02 |
| DRPLA | Dentatorubral-pallidoluysian atrophy | 5.89 × 10−6 | 0.70 | 0.043 | 1.08 |
| DSTN | Destrin | 0.012 | 1.26 | 0.543 | 1.04 |
| DUSP1 | Dual specificity phosphatase 1 (or protein-tyrosine phosphatase, non-receptor-type, 10) | 0.016 | 1.17 | 0.138 | 1.10 |
| EDG4 | Similar to endothelial differentiation, lysophosphatidic acid G-protein-coupled receptor 4 | 2.53 × 10−3 | 1.34 | 0.225 | 0.85 |
| EIF2B | Eukaryotic translation initiation factor 2B | 8.54 × 10−5 | 1.31 | 0.856 | 0.99 |
| EIF2B5 | Eukaryotic translation initiation factor 2B, subunit 5 epsilon | 7.80 × 10−3 | 1.20 | 0.029 | 1.09 |
| EIF4EBP1 | Eukaryotic translation initiation factor 4E binding protein 1 | 0.014 | 0.65 | 0.501 | 0.83 |
| EPHA1 | Eph receptor a1 | 0.033 | 0.56 | 0.046 | 0.61 |
| FASN | Fatty acid synthase | 0.017 | 1.21 | 0.695 | 0.97 |
| FGF13 | Fibroblast growth factor 13 | 0.034 | 1.37 | 0.028 | 1.26 |
| FGG | Fibrinogen, gamma polypeptide | 0.014 | 1.22 | 0.583 | 1.04 |
| FKRP | Fukutin related protein | 0.040 | 0.70 | 0.737 | 0.95 |
| FOS | Fos oncogene | 0.031 | 1.28 | 0.414 | 1.14 |
| FOXO1 | Forkhead box 1 | 8.70 × 10−4 | 0.52 | 0.627 | 1.12 |
| FXC1 | Similar to Mitochondrial import inner membrane translocase subunit TIM9 B (fracture callus protein 1) | 0.043 | 1.23 | 0.852 | 0.97 |
| GABARAP | Gamma-aminobutyric acid receptor-associated protein | 0.012 | 0.79 | 0.906 | 1.01 |
| GABRA3 | Gamma-aminobutyric acid (GABA_A) receptor, subunit alpha 3 | 6.52 × 10−4 | 0.77 | 0.113 | 0.83 |
| GABRD | GABA(A) receptor delta subunit | 0.026 | 0.63 | 0.861 | 1.03 |
| GAL | Galanin | 0.021 | 1.39 | 0.329 | 1.07 |
| GALNT10 | UDP-N-acetyl-alpha-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 | 1.37 × 10−3 | 1.33 | 0.662 | 1.05 |
| GEMIN5 | Gem (nuclear organelle) associated protein 5 | 0.015 | 0.66 | 0.511 | 0.95 |
| GLCCI1 | Glucocorticoid induced transcript 1 | 0.014 | 0.71 | 0.039 | 0.55 |
| GLI1 | GLI-Kruppel family member GLI1 | 0.010 | 1.33 | 0.869 | 0.99 |
| GLI3 | GLI-Kruppel family member Gli 3 | 0.037 | 0.87 | 0.012 | 0.67 |
| GLUD1 | Glutamate dehydrogenase | 0.047 | 1.28 | 1.73 × 10−3 | 1.36 |
| GNA15 | Guanine nucleotide binding protein, alpha 15 | 0.012 | 1.41 | 0.276 | 1.29 |
| GPAA1 | GPI anchor attachment protein 1 | 0.018 | 0.83 | 0.602 | 0.89 |
| GRIK1 | Glutamate receptor, ionotropic, kainate 1 | 0.034 | 0.67 | 0.074 | 0.79 |
| GRM2 | Glutamate receptor, metabotropic 2 | 5.59 × 10−3 | 0.68 | 0.152 | 0.76 |
| GSPT1 | G1 to phase transition 1 | 0.035 | 0.89 | 0.754 | 1.02 |
| GSTM6 | Glutathione S-transferase, mu6 | 7.76 × 10−3 | 1.41 | 1.17 × 10−3 | 1.42 |
| GTF2H2 | General transcription factor IIH, polypeptide 2 (44 kDa subunit) | 0.011 | 0.54 | 0.023 | 0.64 |
| GTF3C5 | General transcription factor IIIC, polypeptide 5 | 0.036 | 1.36 | 0.018 | 1.36 |
| GUK1 | Guanylate kinase 1 | 0.019 | 0.57 | 0.821 | 1.06 |
| H1F0 | H1 histone family, member 0 | 0.016 | 1.22 | 0.561 | 1.07 |
| HBA | Hemoglobin X, alpha-like embryonic chain in Hba complex | 0.029 | 0.72 | 0.030 | 1.15 |
| HBB | Hemoglobin beta chain complex | 7.12 × 10−4 | 0.73 | 0.153 | 0.61 |
| HBG1 | Hemoglobin, gamma A | 4.84 × 10−3 | 1.45 | 3.32 × 10−4 | 1.71 |
| HHL | RAB GTPase activating protein 1-like | 0.042 | 1.27 | 0.034 | 1.27 |
| HIATL2 | Hippocampus abundant gene transcript-like 2 | 0.033 | 0.69 | 0.507 | 0.88 |
| HIS4 | Histone 4 protein | 0.045 | 1.19 | 0.084 | 1.25 |
| HMGN1 | High mobility group nucleosomal binding domain 1 | 0.014 | 1.42 | 0.851 | 1.01 |
| HRMT1L2 | Heterogeneous nuclear ribonucleoproteins methyltransferase-like 2 (S. cerevisiae) | 0.019 | 0.79 | 0.457 | 0.91 |
| HTR3A | 5-Hydroxytryptamine receptor 3a (Htr3a) | 1.77 × 10−3 | 1.65 | 0.783 | 1.07 |
| IDG3G | Isocitrate dehydrogenase 3 (NAD + ), gamma | 0.038 | 1.41 | 0.020 | 1.31 |
| IGF1R | Insulin like growth factor receptor 1 | 0.035 | 0.70 | 0.340 | 1.26 |
| IGSF4A | Cell adhesion molecule 1 | 6.48 × 10−4 | 1.21 | 0.957 | 1.00 |
| IKBKG | Inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma | 0.031 | 1.27 | 0.255 | 1.15 |
| IL6R | Interleukin 6 receptor | 5.48 × 10−3 | 1.17 | 0.265 | 0.94 |
| IL6RA | Interleukin 6 receptor, alpha | 0.044 | 0.62 | 0.721 | 1.05 |
| IL6ST | Interleukin 6 signal transducer | 9.39 × 10−3 | 0.74 | 0.182 | 0.89 |
| IL7R | Interleukin 7 receptor | 0.047 | 1.37 | 0.026 | 1.36 |
| IMPA1 | Lithium-sensitive myo-inositol monophosphatase A1 | 0.015 | 1.25 | 0.388 | 1.22 |
| ING4 | Inhibitor of growth family, member 4 | 0.024 | 1.25 | 0.602 | 0.96 |
| IRAK3 | Interleukin-1 receptor-associated kinase 3 | 0.041 | 0.67 | 0.490 | 1.10 |
| ITGB1 | Integrin beta 1 | 0.043 | 0.86 | 0.776 | 0.97 |
| ITPKA | Inositol 1,4,5-trisphosphate 3-kinase A | 7.66 × 10−3 | 1.14 | 1.91 × 10−4 | 1.50 |
| ITPR5 | Inositol 1,4,5-triphosphate receptor 5 | 2.67 × 10−3 | 0.81 | 0.015 | 0.83 |
| JAK3 | Janus kinase 3 | 0.024 | 0.80 | 0.206 | 0.84 |
| KCMF1 | Potassium channel modulatory factor 1 | 0.034 | 0.76 | 0.347 | 0.86 |
| KCNB2 | Potassium voltage-gated channel, Shab-related subfamily, member 2 | 0.039 | 0.80 | 3.60 × 10−3 | 0.62 |
| KCNC2 | Potassium voltage-gated channel, Shaw-related subfamily, member 3 | 0.017 | 0.86 | 0.164 | 0.91 |
| KCNK1 | Putative potassium channel TWIK | 0.012 | 1.20 | 0.095 | 1.15 |
| KCNMA1 | Potassium large conductance calcium-activated channel, subfamily M, alpha member 1 | 1.93 × 10−3 | 1.45 | 0.443 | 1.28 |
| KITL | Kit ligand | 1.05 × 10−3 | 0.77 | 0.108 | 0.83 |
| KPNB1 | Karyopherin (importin) beta 1 | 5.08 × 10−3 | 1.20 | 0.086 | 1.19 |
| LAG3 | Lymphocyte-activation gene 3 | 0.012 | 0.78 | 0.038 | 0.81 |
| LAMB2 | Laminin beta | 0.043 | 1.54 | 0.539 | 1.06 |
| LAMC1 | Lamnin gamma | 5.02 × 10−3 | 0.76 | 0.205 | 0.87 |
| LAMR1 | Laminin receptor 1 | 0.044 | 1.21 | 0.045 | 1.20 |
| LIMK1 | Lim kinase LIM motif-containing protein kinase 1 | 0.040 | 0.71 | 0.147 | 0.69 |
| LMO1 | LIM domain only 3 | 0.023 | 0.82 | 0.451 | 0.86 |
| LPL | Lipoprotein lipase | 0.020 | 0.74 | 0.990 | 1.00 |
| LRP1 | Low density lipoprotein receptor-related protein 1 | 8.86 × 10−3 | 0.74 | 0.800 | 0.98 |
| LRP4 | Low density lipoprotein receptor-related protein 4 | 2.93 × 10−3 | 1.41 | 0.748 | 1.03 |
| LTBP3 | Latent transforming growth factor beta binding protein 3 | 0.030 | 1.27 | 0.428 | 1.06 |
| MAF | Avian musculoaponeurotic fibrosarcoma (v-maf) AS42 oncogene homolog | 0.012 | 1.34 | 0.041 | 0.90 |
| MAP2K5 | Mitogen-activated protein kinase kinase 5 | 2.96 × 10−3 | 0.86 | 0.660 | 0.96 |
| MAP3K7IP1 | Mitogen-activated protein kinase kinase kinase 7 interacting protein 1 | 7.63 × 10−3 | 1.45 | 0.950 | 0.99 |
| MAP4K4 | Mitogen-activated protein kinase kinase kinase kinase 4 | 3.33 × 10−4 | 0.69 | 1.48 × 10−3 | 0.64 |
| MAPK14 | Mitogen-activated protein kinase 14 | 8.65 × 10−3 | 1.35 | 0.445 | 1.12 |
| MAS1 | MAS1 oncogene | 0.014 | 0.79 | 0.944 | 0.99 |
| MATR3 | Matrin 3 | 0.011 | 0.73 | 0.512 | 0.90 |
| MCM5 | Minichromosome maintenance deficient 5, cell division cycle 46 (S. cerevisiae) | 0.041 | 1.10 | 0.181 | 1.12 |
| MEF2D | Myocyte enhancer factor 2d | 7.73 × 10−3 | 1.35 | 0.152 | 1.31 |
| MKLN1 | Muskelin 1, intracellular mediator containing kelch motifs | 4.12 × 10−5 | 0.68 | 0.056 | 0.85 |
| MLYCD | Malonyl-CoA decarboxylase | 8.54 × 10−3 | 1.66 | 0.023 | 1.46 |
| MME | Membrane metallo endopeptidase | 0.024 | 1.21 | 0.473 | 1.02 |
| MRE11A | Meiotic recombination 11 homolog A (S. cerevisiae) | 0.036 | 0.70 | 0.300 | 0.85 |
| MRPL39 | Mitochondrial ribosomal protein L39 | 1.22 × 10−3 | 0.78 | 1.20 × 10−3 | 0.69 |
| MRPS14 | Mitochondrial ribosomal protein S14 | 5.45 × 10−3 | 0.71 | 0.945 | 1.01 |
| MRPS17 | Mitochondrial ribosomal protein S17 | 0.019 | 0.82 | 0.695 | 0.91 |
| MRPS7 | Mitochondrial ribosomal protein S7 | 7.10 × 10−3 | 1.62 | 0.852 | 0.98 |
| MTC | MT-protocadherin (KIAA 1775) | 3.55 × 10−4 | 1.29 | 0.984 | 1.00 |
| MTEFR | Transcription termination factor, mitochondrial-like | 0.025 | 0.66 | 0.542 | 0.87 |
| MYO7A | Myosin VIIA | 7.50 × 10−3 | 0.64 | 0.361 | 0.88 |
| NCBP1 | Nuclear cap binding protein subunit 1, 80 kDa | 2.15 × 10−3 | 1.36 | 0.750 | 1.06 |
| NDUFA4 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4 | 3.41 × 10−3 | 1.40 | 0.909 | 1.01 |
| NDUFV1 | NADH dehydrogenase (ubiquinone) flavoprotein 1 | 0.022 | 1.65 | 0.043 | 1.37 |
| NEU3 | Neurotrophic tyrosine kinase, receptor, type 1 | 0.034 | 1.25 | 0.040 | 1.36 |
| NEUD4 | Neuronal d4 domain family member | 0.018 | 1.70 | 0.029 | 1.36 |
| NFATC3 | Nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 | 0.010 | 0.74 | 0.084 | 0.77 |
| NFE2L2 | Nuclear factor, erythroid derived 2, like 2 | 1.38 × 10−4 | 1.12 | 0.315 | 0.85 |
| NGFA | Nerve growth factor, alpha | 0.029 | 1.20 | 0.036 | 1.15 |
| NGFR | Nerve growth factor receptor | 4.51 × 10−3 | 1.46 | 0.018 | 1.30 |
| NINJ1 | Ninjurin 1 | 0.031 | 0.72 | 0.111 | 0.73 |
| NLK1 | Nemo like kinase | 3.33 × 10−3 | 0.55 | 0.022 | 0.70 |
| NOL5 | Nucleolar protein 5 | 3.10 × 10−3 | 0.80 | 0.071 | 0.82 |
| NR2F1 | Nuclear receptor subfamily 2, group F, member 1 | 1.74 × 10−3 | 1.33 | 0.211 | 1.10 |
| NRF1 | Nuclear respiratory factor 1 | 0.040 | 0.83 | 0.166 | 0.66 |
| NRXN1 | Neurexin1 | 7.69 × 10−3 | 0.78 | 1.73 × 10−4 | 0.74 |
| NSG1 | Neuron-specific gene family member 1 | 0.039 | 0.84 | 0.309 | 0.95 |
| NUCB | Calcium-binding protein | 0.023 | 1.35 | 0.126 | 1.12 |
| NXPH4 | Neurexophilin4 | 3.87 × 10−3 | 1.37 | 0.048 | 1.46 |
| OGDH | Oxoglutarate dehydrogenase | 0.031 | 1.27 | 0.947 | 0.99 |
| OGN | Osteoglycin | 1.48 × 10−3 | 0.57 | 0.261 | 0.84 |
| PBX4 | Pre-B-cell leukemia transcription factor 4 | 7.99 × 10−4 | 1.59 | 8.04 × 10−3 | 1.38 |
| PCP4 | Purkinje cell protein 4 | 0.011 | 0.75 | 0.030 | 0.67 |
| PDLIM2 | PDZ and LIM domain 2 | 4.91 × 10−3 | 1.99 | 0.014 | 1.55 |
| PHYHIP | Phytanoyl-CoA hydroxylase interacting protein | 5.61 × 10−3 | 0.88 | 0.023 | 1.04 |
| PIGQ | Glycosylphosphatidylinositol 1 homolog | 0.013 | 1.36 | 0.035 | 1.18 |
| PIK3CG | Phosphoinositide-3-kinase, catalytic, gamma polypeptide | 0.049 | 1.30 | 0.657 | 1.08 |
| PIR51 | RAD51 associated protein 1 | 0.050 | 1.28 | 0.631 | 0.96 |
| PLA2G4A | Phospholipase A2, group IVA (cytosolic, calcium-dependent) | 3.26 × 10−3 | 1.42 | 0.221 | 1.25 |
| PMP22 | Peripheral myelin protein 22 | 5.96 × 10−3 | 1.17 | 0.118 | 1.07 |
| POLE3 | Polymerase (DNA directed), epsilon 3 (p17 subunit) | 9.98 × 10−3 | 1.39 | 0.124 | 1.18 |
| PPIA | Peptidylprolyl isomerase A | 0.040 | 1.38 | 0.130 | 1.17 |
| PPM1L | Protein phosphatase 1 (formerly 2C)-like | 0.032 | 1.35 | 0.212 | 1.31 |
| PPP1CA | Protein phosphatase 1, catalytic subunit, alpha isoform | 0.020 | 1.21 | 9.25 × 10−3 | 1.21 |
| PPT | Palmitoyl-protein thioesterase | 0.050 | 1.56 | 0.166 | 1.10 |
| PRDX6 | Peroxiredoxin 6 | 0.023 | 1.22 | 0.341 | 1.13 |
| PRRX1 | Paired related homeobox 1 | 0.050 | 1.16 | 0.805 | 1.03 |
| PSCDBP | Pleckstrin homology, Sec7, and coiled-coil domains, binding protein | 9.26 × 10−3 | 0.89 | 0.052 | 0.81 |
| PSMB4 | Proteasome (prosome, macropain) subunit, beta type, 4 | 2.14 × 10−3 | 1.23 | 0.235 | 1.09 |
| PSMD10 | Proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 | 0.025 | 1.17 | 0.875 | 1.01 |
| PSMD14 | Proteasome (prosome, macropain) 26S subunit, non-ATPase, 14 | 0.029 | 0.70 | 0.154 | 0.79 |
| PSMD2 | 26S proteasome, non-ATPase, 2 | 2.37 × 10−3 | 1.54 | 0.033 | 1.25 |
| PTH | Parathyroid hormone | 0.040 | 1.19 | 0.608 | 0.96 |
| PTP4A2 | Protein–tyrosine phosphatase type 4a, member 2 | 0.027 | 1.24 | 0.011 | 1.24 |
| PTPRZ1 | Protein–tyrosine phosphatase, receptor type, Z1 | 0.041 | 0.90 | 0.597 | 1.05 |
| RAB1 | Ras-related protein | 0.036 | 1.23 | 0.029 | 1.36 |
| RAB7 | Ras 7member of RAS oncogene family | 0.042 | 0.80 | 0.048 | 0.85 |
| RAB9 | Ras 9 member of RAS oncogene family | 0.036 | 1.32 | 0.377 | 1.15 |
| RAD1 | RAD1 homolog (S. pombe) | 6.01 × 10−4 | 0.69 | 0.046 | 0.78 |
| RAD51 | RAD51 homolog (S. cerevisiae) | 0.013 | 1.45 | 0.131 | 1.48 |
| RALA | V-ral simian leukemia viral oncogene homolog A (Ras-related) | 8.17 × 10−3 | 1.74 | 0.096 | 1.27 |
| RAPGEF3 | Rap guanine nucleotide exchange factor (GEF) 3 | 0.032 | 0.84 | 0.158 | 0.89 |
| RASA2 | RAS p21 protein activator 2 | 0.034 | 0.73 | 0.537 | 1.18 |
| RBL1 | Retinoblastoma-like 1 (p107) | 0.046 | 0.73 | 0.111 | 0.73 |
| RIS2 | Retroviral integration site 2 | 0.024 | 0.74 | 0.244 | 0.88 |
| RISP | Ubiquinol–cytochrome c reductase, Rieske iron–sulfur polypeptide 1 | 0.048 | 0.76 | 0.159 | 0.84 |
| RNF20 | Ring finger protein 20 | 0.017 | 0.69 | 0.058 | 0.76 |
| RPL18 | Ribosomal protein L18 | 0.031 | 0.75 | 0.033 | 0.80 |
| RPL29 | Ribosomal protein L29 | 1.43 × 10−3 | 1.50 | 0.110 | 1.19 |
| RPL36 | Ribosomal protein L36 | 0.031 | 0.85 | 0.979 | 1.00 |
| RPS26 | Ribosomal protein S26 | 0.032 | 1.57 | 0.192 | 1.14 |
| RPS27L | Ribosomal protein S27 | 0.032 | 1.44 | 0.201 | 1.13 |
| RPS6KA2 | Ribosomal protein S6 kinase, 90kD, polypeptide 2 | 5.70 × 10−4 | 0.82 | 0.708 | 0.95 |
| RPS6KB1 | Ribosomal protein S6 kinase, 70kD, polypeptide 1 | 0.037 | 1.28 | 0.017 | 1.25 |
| SCN11A | Sodium channel, voltage-gated, type11, alpha polypeptide | 0.024 | 1.45 | 0.346 | 0.86 |
| SCO1 | Similar to SCO cytochrome oxidase deficient homolog 1 (yeast) | 0.033 | 0.84 | 0.641 | 1.05 |
| SCUBE2 | Signal peptide, CUB domain, EGF-like 2 | 2.96 × 10−3 | 0.67 | 0.010 | 0.70 |
| SCYA20 | Small inducible cytokine subfamily A20 | 4.02 × 10−3 | 0.75 | 0.288 | 0.91 |
| SDHB | Succinate dehydrogenase complex, subunit A, flavoprotein | 6.23 × 10−3 | 1.32 | 0.181 | 0.87 |
| SDHC | Succinate dehydrogenase complex, subunit C | 0.017 | 1.18 | 0.035 | 1.13 |
| SECISBP2 | Selenocysteine insertion sequence-binding protein 2 | 8.27 × 10−3 | 0.76 | 0.367 | 1.03 |
| SERPINC1 | Serine (or cysteine) peptidase inhibitor, clade C (antithrombin), member 1 | 0.040 | 1.35 | 0.152 | 1.37 |
| SFN | Stratifin | 0.022 | 1.14 | 0.635 | 1.04 |
| SKIL | Ski like | 8.52 × 10−3 | 1.31 | 2.44 × 10−4 | 1.17 |
| SLC18A2 | Solute carrier family 18 member 2 | 0.023 | 1.46 | 0.144 | 0.91 |
| SLC29A1 | Solute carrier family 29 (nucleoside transporters), member 1 | 0.021 | 0.73 | 0.164 | 1.41 |
| SLC2A14 | Solute carrier family 2 (facilitated glucose transporter), member 14 | 4.37 × 10−3 | 0.68 | 0.032 | 0.73 |
| SLC2A4 | Solute carrier family 2 (facilitated glucose transporter), member 4 | 0.030 | 1.58 | 0.221 | 1.32 |
| SLC35D2 | Solute carrier family 35, member D2 | 0.024 | 1.39 | 0.016 | 1.61 |
| SLC35F5 | Solute carrier family 35, member F5 | 0.018 | 0.73 | 0.890 | 0.98 |
| SLC5A3 | Solute carrier family 5 (inositol transporters), member 3 | 4.51 × 10−4 | 0.63 | 4.60 × 10−3 | 0.76 |
| SLC6A4 | Solute carrier family 6, member 4 (serotonin transporter) | 3.78 × 10−6 | 0.62 | 0.134 | 1.08 |
| SLC8A2 | Solute carrier family 8 (sodium/calcium exchanger), member 2 | 0.032 | 1.30 | 0.194 | 0.93 |
| SMAD2 | MAD homolog 2 (Drosophila) | 0.035 | 0.83 | 0.323 | 1.09 |
| SNCA | Synuclein, alpha | 5.13 × 10−3 | 0.71 | 0.523 | 0.87 |
| SOX6 | SoxLZ/Sox6 leucine zipper binding protein in testis | 0.027 | 1.17 | 0.937 | 0.99 |
| SPEN | SMART/HDAC1 associated repressor protein | 0.024 | 0.79 | 0.013 | 0.68 |
| STAT5A | Signal transducer and activator of transcription 5a | 0.021 | 1.25 | 0.044 | 1.24 |
| SWAP70 | Switch-associated protein 70 | 0.032 | 0.61 | 0.646 | 0.87 |
| SYN2 | Synapsin 2 | 0.014 | 1.25 | 0.988 | 1.00 |
| SYT2 | Synaptotagmin 2 | 1.36 × 10−3 | 1.24 | 0.192 | 1.17 |
| SYT7 | Synaptotagmin 7 | 5.30 × 10−3 | 1.69 | 0.492 | 1.08 |
| TAC2 | Tachykinin 2 | 3.45 × 10−3 | 0.72 | 0.988 | 1.00 |
| TBC1D17 | TBC1 domain family, member 17 | 0.046 | 1.43 | 0.051 | 1.21 |
| TBP1 | Tat-binding protein-1 | 0.041 | 0.77 | 0.108 | 0.80 |
| TCFEB | Transcription factor EB | 6.61 × 10−5 | 0.76 | 0.518 | 0.97 |
| TDG | Thymine DNA glycosylase | 0.032 | 0.57 | 0.108 | 0.70 |
| TF | Transferrin | 0.028 | 0.73 | 0.045 | 0.70 |
| TFR | Transferrin receptor | 5.66 × 10−3 | 1.32 | 0.893 | 1.02 |
| TGFB2 | Transforming growth factor, beta 2 | 5.58 × 10−5 | 0.74 | 0.019 | 0.71 |
| TGFBR2 | Transforming growth factor, beta receptor 2 | 6.22 × 10−3 | 0.57 | 0.913 | 0.98 |
| TNFRSF10B | Tumor necrosis factor receptor superfamily, member 10b | 0.042 | 1.43 | 0.134 | 1.22 |
| TRAF6 | Tnf receptor-associated factor 6 | 0.035 | 0.75 | 0.654 | 0.94 |
| TRG | Thyroid regulating gene | 0.019 | 0.69 | 0.090 | 0.71 |
| TRNT1 | TRNA nucleotidyl transferase, CCA-adding1 | 0.022 | 0.76 | 0.538 | 0.97 |
| TRP53 | Transformation-related protein 53 | 6.94 × 10−3 | 1.32 | 0.252 | 1.08 |
| TSGA13 | Testis specific gene A13 | 2.29 × 10−3 | 1.07 | 5.98 × 10−3 | 1.41 |
| TUBB5 | Beta-tubulin, 5 | 0.017 | 1.19 | 0.079 | 1.08 |
| UAP1 | Similar to UDP-N-acteylglucosamine pyrophosphorylase 1 homolog | 0.021 | 1.24 | 0.526 | 0.93 |
| UBE1X | Ubiquitin-activating enzyme e1, Chr X | 0.019 | 1.14 | 0.761 | 0.99 |
| UBE2A | Ubiquitin-conjugating enzyme E2A | 0.018 | 1.54 | 0.250 | 1.10 |
| UBE2G1 | Ubiquitin-conjugating enzyme E2G 1 (UBC7 homolog, C. elegans) | 0.023 | 0.88 | 0.425 | 1.22 |
| UBQLN1 | Ubiquilin 1 | 0.030 | 1.18 | 0.763 | 0.96 |
| UBQLN3 | Ubiquilin 3 | 0.016 | 0.58 | 0.036 | 0.60 |
| UQCRFS1 | Ubiquinol–cytochrome c reductase, Rieske iron–sulfur polypeptide 1 | 3.11 × 10−3 | 1.53 | 0.832 | 0.98 |
| USP28 | Ubiquitin-specific protease 28 | 8.60 × 10−3 | 1.29 | 0.062 | 1.29 |
| VAMP3 | Vesicle-associated membrane protein 3 | 0.036 | 1.21 | 9.68 × 10−3 | 1.12 |
| VCL | Vinculin | 2.10 × 10−3 | 1.39 | 3.10 × 10−3 | 1.30 |
| VEGFA | Vascular endothelial growth factor A | 0.038 | 0.84 | 0.513 | 0.92 |
| VGLL2 | Vestigial like 2 homolog (Drosophila) | 0.014 | 1.21 | 0.501 | 1.02 |
| VIL1 | Villin1 | 0.016 | 0.65 | 9.82 × 10−3 | 0.79 |
| WNT1 | Wingless-related MMTV integration site 1 | 0.042 | 1.17 | 0.493 | 1.07 |
| XCL2 | Chemokine (C motif) ligand 2 | 0.023 | 1.38 | 0.073 | 1.33 |
| XTP2 | HBxAg transactivated protein 2 | 0.031 | 0.88 | 0.308 | 0.90 |
| YWHAH | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide | 0.037 | 1.21 | 0.069 | 0.87 |
| ZAP70 | Zeta-chain (TCR) associated protein kinase | 2.07 × 10−4 | 1.36 | 0.139 | 1.09 |
| ZFP98 | Zinc finger protein 98 | 8.22 × 10−4 | 1.21 | 0.616 | 0.97 |
| ZNF189 | Zinc finger protein 189 | 0.035 | 0.79 | 0.264 | 0.80 |
| ZNF286 | Zinc finger protein 286 | 0.012 | 1.18 | 0.381 | 1.09 |
Footnotes
References
- Adamopoulos D., Argacha J. F., Gujic M., Preumont N., Degaute J. P., van de Borne P. (2009). Acute effects of nicotine on arterial stiffness and wave reflection in healthy young non smokers. Clin. Exp. Pharmacol. Physiol. 36, 784–789 [DOI] [PubMed] [Google Scholar]
- Adams J. P., Sweatt J. D. (2002). Molecular psychology: roles for the ERK MAP kinase cascade in memory. Annu. Rev. Pharmacol. Toxicol. 42, 135–163 [DOI] [PubMed] [Google Scholar]
- Ambroggi F., Turiault M., Milet A., Deroche-Gamonet V., Parnaudeau S., Balado E., Barik J., van der Veen R., Maroteaux G., Lemberger T., Schutz G., Lazar M., Marinelli M., Piazza P. V., Tronche F. (2009). Stress and addiction: glucocorticoid receptor in dopaminoceptive neurons facilitates cocaine seeking. Nat. Neurosci. 12, 247–249 [DOI] [PubMed] [Google Scholar]
- Argacha J. F., Adamopoulos D., Gujic M., Fontaine D., Amyai N., Berkenboom G., van de Borne P. (2008). Acute effects of passive smoking on peripheral vascular function. Hypertension 51, 1506–1511 10.1161/HYPERTENSIONAHA.107.104059 [DOI] [PubMed] [Google Scholar]
- Arrick D. M., Mayhan W. G. (2007). Acute infusion of nicotine impairs nNOS-dependent reactivity of cerebral arterioles via an increase in oxidative stress. J. Appl. Physiol. 103, 2062–2067 [DOI] [PubMed] [Google Scholar]
- Barik J., Wonnacott S. (2006). Indirect modulation by alpha7 nicotinic acetylcholine receptors of noradrenaline release in rat hippocampal slices: interaction with glutamate and GABA systems and effect of nicotine withdrawal. Mol. Pharmacol. 69, 618–628 [DOI] [PubMed] [Google Scholar]
- Barik J., Wonnacott S. (2009). Molecular and cellular mechanisms of action of nicotine in the CNS. Handb. Exp. Pharmacol. 173–207 [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B 57, 289–300 [Google Scholar]
- Benwell M. E., Balfour D. J. (1992). The effects of acute and repeated nicotine treatment on nucleus accumbens dopamine and locomotor activity. Br. J. Pharmacol. 105, 849–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benwell M. E., Balfour D. J. (1997). Regional variation in the effects of nicotine on catecholamine overflow in rat brain. Eur. J. Pharmacol. 325, 13–20 [DOI] [PubMed] [Google Scholar]
- Bierut L. J., Madden P. A., Breslau N., Johnson E. O., Hatsukami D., Pomerleau O. F., Swan G. E., Rutter J., Bertelsen S., Fox L., Fugman D., Goate A. M., Hinrichs A. L., Konvicka K., Martin N. G., Montgomery G. W., Saccone N. L., Saccone S. F., Wang J. C., Chase G. A., Rice J. P., Ballinger D. G. (2007). Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum. Mol. Genet. 16, 24–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsch D., Lederer T., Reulbach U., Hothorn T., Kornhuber J., Bleich S. (2005). Joint analysis of the NACP-REP1 marker within the alpha synuclein gene concludes association with alcohol dependence. Hum. Mol. Genet. 14, 967–971 [DOI] [PubMed] [Google Scholar]
- Brunzell D. H., Russell D. S., Picciotto M. R. (2003) In vivo nicotine treatment regulates mesocorticolimbic CREB and ERK signaling in C57Bl/6J mice. J. Neurochem. 84, 1431–1441 10.1046/j.1471-4159.2003.01640.x [DOI] [PubMed] [Google Scholar]
- Caggiula A. R., Donny E. C., Epstein L. H., Sved A. F., Knopf S., Rose C., McAllister C. G., Antelman S. M., Perkins K. A. (1998). The role of corticosteroids in nicotine's physiological and behavioral effects. Psychoneuroendocrinology 23, 143–159 10.1016/S0306-4530(97)00078-4 [DOI] [PubMed] [Google Scholar]
- Cao J., Dwyer J. B., Mangold J. E., Wang J., Wei J., Leslie F. M., Li M. D. (2011). Modulation of cell adhesion systems by prenatal nicotine exposure in limbic brain regions of adolescent female rats. Int. J. Neuropsychopharmacol. 14, 157–174 10.1017/S1461145710000179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L., Rapoport S. I., Nguyen H. N., Greenstein D., Chen M., Basselin M. (2009). Acute nicotine reduces brain arachidonic acid signaling in unanesthetized rats. J. Cereb. Blood Flow Metab. 29, 648–658 10.1038/jcbfm.2008.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez-Noriega L. E., Crona J. H., Washburn M. S., Urrutia A., Elliott K. J., Johnson E. C. (1997). Pharmacological characterization of recombinant human neuronal nicotinic acetylcholine receptors h alpha 2 beta 2, h alpha 2 beta 4, h alpha 3 beta 2, h alpha 3 beta 4, h alpha 4 beta 2, h alpha 4 beta 4 and h alpha 7 expressed in Xenopus oocytes. J. Pharmacol. Exp. Ther. 280, 346–356 [PubMed] [Google Scholar]
- Chen M., Wang T., Liao Z. X., Pan X. L., Feng Y. H., Wang H. (2007). Nicotine-induced prenatal overexposure to maternal glucocorticoid and intrauterine growth retardation in rat. Exp. Toxicol. Pathol. 59, 245–251 [DOI] [PubMed] [Google Scholar]
- Clark A. R., Lasa M. (2003). Crosstalk between glucocorticoids and mitogen-activated protein kinase signalling pathways. Curr. Opin. Pharmacol. 3, 404–411 [DOI] [PubMed] [Google Scholar]
- Cole T. J., Blendy J. A., Monaghan A. P., Krieglstein K., Schmid W., Aguzzi A., Fantuzzi G., Hummler E., Unsicker K., Schutz G. (1995). Targeted disruption of the glucocorticoid receptor gene blocks adrenergic chromaffin cell development and severely retards lung maturation. Genes Dev. 9, 1608–1621 10.1101/gad.9.13.1608 [DOI] [PubMed] [Google Scholar]
- Cormier A., Morin C., Zini R., Tillement J. P., Lagrue G. (2001) In vitro effects of nicotine on mitochondrial respiration and superoxide anion generation. Brain Res. 900, 72–79 10.1016/S0006-8993(01)02254-5 [DOI] [PubMed] [Google Scholar]
- Cucina A., Corvino V., Sapienza P., Borrelli V., Lucarelli M., Scarpa S., Strom R., Santoro-D'Angelo L., Cavallaro A. (1999). Nicotine regulates basic fibroblastic growth factor and transforming growth factor beta1 production in endothelial cells. Biochem. Biophys. Res. Commun. 257, 306–312 [DOI] [PubMed] [Google Scholar]
- Dajas-Bailador F., Wonnacott S. (2004). Nicotinic acetylcholine receptors and the regulation of neuronal signalling. Trends Pharmacol. Sci. 25, 317–324 [DOI] [PubMed] [Google Scholar]
- Dajas-Bailador F. A., Soliakov L., Wonnacott S. (2002). Nicotine activates the extracellular signal-regulated kinase 1/2 via the alpha7 nicotinic acetylcholine receptor and protein kinase A, in SH-SY5Y cells and hippocampal neurones. J. Neurochem. 80, 520–530 10.1046/j.0022-3042.2001.00725.x [DOI] [PubMed] [Google Scholar]
- Dasgupta P., Chellappan S. P. (2006). Nicotine-mediated cell proliferation and angiogenesis: new twists to an old story. Cell Cycle 5, 2324–2328 10.4161/cc.5.20.3366 [DOI] [PubMed] [Google Scholar]
- Dasgupta P., Rastogi S., Pillai S., Ordonez-Ercan D., Morris M., Haura E., Chellappan S. (2006). Nicotine induces cell proliferation by beta-arrestin-mediated activation of Src and Rb-Raf-1 pathways. J. Clin. Invest. 116, 2208–2217 10.1172/JCI28164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar K. D., Banach M., Galigniana M. D., Pratt W. B. (1998). The role of DnaJ-like proteins in glucocorticoid receptor.hsp90 heterocomplex assembly by the reconstituted hsp90.p60.hsp70 foldosome complex. J. Biol. Chem. 273, 7358–7366 [DOI] [PubMed] [Google Scholar]
- Dittmar K. D., Demady D. R., Stancato L. F., Krishna P., Pratt W. B. (1997). Folding of the glucocorticoid receptor by the heat shock protein (hsp) 90-based chaperone machinery. The role of p23 is to stabilize receptor.hsp90 heterocomplexes formed by hsp90.p60.hsp70. J. Biol. Chem. 272, 21213–21220 [DOI] [PubMed] [Google Scholar]
- Dunckley T., Lukas R. J. (2003). Nicotine modulates the expression of a diverse set of genes in the neuronal SH-SY5Y cell line. J. Biol. Chem. 278, 15633–15640 [DOI] [PubMed] [Google Scholar]
- Dunckley T., Lukas R. J. (2006). Nicotinic modulation of gene expression in SH-SY5Y neuroblastoma cells. Brain Res. 1116, 39–49 10.1016/j.brainres.2006.07.111 [DOI] [PubMed] [Google Scholar]
- Edwards S., Graham D. L., Bachtell R. K., Self D. W. (2007). Region-specific tolerance to cocaine-regulated cAMP-dependent protein phosphorylation following chronic self-administration. Eur. J. Neurosci. 25, 2201–2213 [DOI] [PubMed] [Google Scholar]
- Ernst M., Heishman S. J., Spurgeon L., London E. D. (2001). Smoking history and nicotine effects on cognitive performance. Neuropsychopharmacology 25, 313–319 10.1016/S0893-133X(01)00257-3 [DOI] [PubMed] [Google Scholar]
- Ficklin M. B., Zhao S., Feng G. (2005). Ubiquilin-1 regulates nicotine-induced up-regulation of neuronal nicotinic acetylcholine receptors. J. Biol. Chem. 280, 34088–34095 [DOI] [PubMed] [Google Scholar]
- Foulds J., Stapleton J., Swettenham J., Bell N., McSorley K., Russell M. A. (1996). Cognitive performance effects of subcutaneous nicotine in smokers and never-smokers. Psychopharmacology (Berl.) 127, 31–38 [DOI] [PubMed] [Google Scholar]
- Fox H. C., Jackson E. D., Sinha R. (2009). Elevated cortisol and learning and memory deficits in cocaine dependent individuals: relationship to relapse outcomes. Psychoneuroendocrinology 34, 1198–1207 10.1016/j.psyneuen.2009.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French K. L., Granholm A. C., Moore A. B., Nelson M. E., Bimonte-Nelson H. A. (2006). Chronic nicotine improves working and reference memory performance and reduces hippocampal NGF in aged female rats. Behav. Brain Res. 169, 256–262 [DOI] [PubMed] [Google Scholar]
- Gaddnas H., Pietila K., Piepponen T. P., Ahtee L. (2001). Enhanced motor activity and brain dopamine turnover in mice during long-term nicotine administration in the drinking water. Pharmacol. Biochem. Behav. 70, 497–503 [DOI] [PubMed] [Google Scholar]
- Garrido R., King-Pospisil K., Son K. W., Hennig B., Toborek M. (2003). Nicotine upregulates nerve growth factor expression and prevents apoptosis of cultured spinal cord neurons. Neurosci. Res. 47, 349–355 [DOI] [PubMed] [Google Scholar]
- Gaudilliere B., Shi Y., Bonni A. (2002). RNA interference reveals a requirement for myocyte enhancer factor 2A in activity-dependent neuronal survival. J. Biol. Chem. 277, 46442–46446 [DOI] [PubMed] [Google Scholar]
- Ghosh J., Murphy M. O., Turner N., Khwaja N., Halka A., Kielty C. M., Walker M. G. (2005). The role of transforming growth factor beta1 in the vascular system. Cardiovasc. Pathol. 14, 28–36 [DOI] [PubMed] [Google Scholar]
- Gould J., Reeve H. L., Vaughan P. F., Peers C. (1992). Nicotinic acetylcholine receptors in human neuroblastoma (SH-SY5Y) cells. Neurosci. Lett. 145, 201–204 [DOI] [PubMed] [Google Scholar]
- Grenhoff J., Svensson T. H. (1988). Selective stimulation of limbic dopamine activity by nicotine. Acta Physiol. Scand. 133, 595–596 10.1111/j.1748-1716.1988.tb08450.x [DOI] [PubMed] [Google Scholar]
- Gueorguiev V. D., Cheng S. Y., Sabban E. L. (2006). Prolonged activation of cAMP-response element-binding protein and ATF-2 needed for nicotine-triggered elevation of tyrosine hydroxylase gene transcription in PC12 cells. J. Biol. Chem. 281, 10188–10195 [DOI] [PubMed] [Google Scholar]
- Gupta V., Awasthi N., Wagner B. J. (2007). Specific activation of the glucocorticoid receptor and modulation of signal transduction pathways in human lens epithelial cells. Invest. Ophthalmol. Vis. Sci. 48, 1724–1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutala R., Wang J., Kadapakkam S., Hwang Y., Ticku M., Li M. D. (2004). Microarray analysis of ethanol-treated cortical neurons reveals disruption of genes related to the ubiquitin-proteasome pathway and protein synthesis. Alcohol. Clin. Exp. Res. 28, 1779–1788 [DOI] [PubMed] [Google Scholar]
- Harkness P. C., Millar N. S. (2002). Changes in conformation and subcellular distribution of alpha4beta2 nicotinic acetylcholine receptors revealed by chronic nicotine treatment and expression of subunit chimeras. J. Neurosci. 22, 10172–10181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidenreich K. A., Linseman D. A. (2004). Myocyte enhancer factor-2 transcription factors in neuronal differentiation and survival. Mol. Neurobiol. 29, 155–166 10.1385/MN:29:2:155 [DOI] [PubMed] [Google Scholar]
- Hernandez C. M., Terry A. V., Jr. (2005). Repeated nicotine exposure in rats: effects on memory function, cholinergic markers and nerve growth factor. Neuroscience 130, 997–1012 10.1016/j.neuroscience.2004.10.006 [DOI] [PubMed] [Google Scholar]
- Heusch W. L., Maneckjee R. (1998). Signalling pathways involved in nicotine regulation of apoptosis of human lung cancer cells. Carcinogenesis 19, 551–556 10.1093/carcin/19.4.551 [DOI] [PubMed] [Google Scholar]
- Hwang Y. Y., Li M. D. (2006). Proteins differentially expressed in response to nicotine in five rat brain regions: identification using a 2-DE/MS-based proteomics approach. Proteomics 6, 3138–3153 10.1002/pmic.200500745 [DOI] [PubMed] [Google Scholar]
- Ichino N., Ishiguro H., Yamada K., Nishii K., Sawada H., Nagatsu T. (1999). Nicotine withdrawal up-regulates c-Fos transcription in pheochromocytoma cells. Neurosci. Res. 35, 63–69 [DOI] [PubMed] [Google Scholar]
- Ichino N., Yamada K., Nishii K., Sawada H., Nagatsu T., Ishiguro H. (2002). Increase of transcriptional levels of egr-1 and nur77 genes due to both nicotine treatment and withdrawal in pheochromocytoma cells. J. Neural Transm. 109, 1015–1022 10.1007/s007020200084 [DOI] [PubMed] [Google Scholar]
- Isola R., Zhang H., Tejwani G. A., Neff N. H., Hadjiconstantinou M. (2009). Acute nicotine changes dynorphin and prodynorphin mRNA in the striatum. Psychopharmacology (Berl.) 201, 507–516 [DOI] [PubMed] [Google Scholar]
- Jiao H., Zhang L., Gao F., Lou D., Zhang J., Xu M. (2007). Dopamine D(1) and D(3) receptors oppositely regulate NMDA- and cocaine-induced MAPK signaling via NMDA receptor phosphorylation. J. Neurochem. 103, 840–848 10.1111/j.1471-4159.2007.04840.x [DOI] [PubMed] [Google Scholar]
- Jonnala R. R., Terry A. V., Jr., Buccafusco J. J. (2002). Nicotine increases the expression of high affinity nerve growth factor receptors in both in vitro and in vivo. Life Sci. 70, 1543–1554 10.1016/S0024-3205(01)01529-6 [DOI] [PubMed] [Google Scholar]
- Kane J. K., Konu O., Ma J. Z., Li M. D. (2004). Nicotine coregulates multiple pathways involved in protein modification/degradation in rat brain. Brain Res. Mol. Brain Res. 132, 181–191 10.1016/j.molbrainres.2004.09.010 [DOI] [PubMed] [Google Scholar]
- Katyare S. S., Shallom J. M. (1988). Altered cerebral protein turnover in rats following prolonged in vivo treatment with nicotine. J. Neurochem. 50, 1356–1363 10.1111/j.1471-4159.1988.tb03016.x [DOI] [PubMed] [Google Scholar]
- Ke L., Eisenhour C. M., Bencherif M., Lukas R. J. (1998). Effects of chronic nicotine treatment on expression of diverse nicotinic acetylcholine receptor subtypes. I. Dose- and time-dependent effects of nicotine treatment. J. Pharmacol. Exp. Ther. 286, 825–840 [PubMed] [Google Scholar]
- Kenny P. J., File S. E., Rattray M. (2001). Nicotine regulates 5-HT(1A) receptor gene expression in the cerebral cortex and dorsal hippocampus. Eur. J. Neurosci. 13, 1267–1271 [DOI] [PubMed] [Google Scholar]
- Kobayashi H., Ide S., Hasegawa J., Ujike H., Sekine Y., Ozaki N., Inada T., Harano M., Komiyama T., Yamada M., Iyo M., Shen H. W., Ikeda K., Sora I. (2004). Study of association between alpha-synuclein gene polymorphism and methamphetamine psychosis/dependence. Ann. N. Y. Acad. Sci. 1025, 325–334 [DOI] [PubMed] [Google Scholar]
- Konu O., Kane J. K., Barrett T., Vawter M. P., Chang R., Ma J. Z., Donovan D. M., Sharp B., Becker K. G., Li M. D. (2001). Region-specific transcriptional response to chronic nicotine in rat brain. Brain Res. 909, 194–203 10.1016/S0006-8993(01)02685-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konu O., Xu X., Ma J. Z., Kane J., Wang J., Shi S. J., Li M. D. (2004a). Application of a customized pathway-focused microarray for gene expression profiling of cellular homeostasis upon exposure to nicotine in PC12 cells. Brain Res. Mol. Brain Res. 121, 102–113 10.1016/j.molbrainres.2003.11.012 [DOI] [PubMed] [Google Scholar]
- Konu O., Xu X., Ma J. Z., Kane J. K., Wang J., Shi S. J., Li M. D. (2004b). Application of a customized pathway-focused microarray for gene expression profiling of cellular homeostasis upon exposure to nicotine in PC12 cells. Brain Res. Mol. Brain Res. 110, 102–113 10.1016/j.molbrainres.2003.11.012 [DOI] [PubMed] [Google Scholar]
- Kumari V., Gray J. A., Ffytche D. H., Mitterschiffthaler M. T., Das M., Zachariah E., Vythelingum G. N., Williams S. C., Simmons A., Sharma T. (2003). Cognitive effects of nicotine in humans: an fMRI study. Neuroimage 19, 1002–1013 10.1016/S1053-8119(03)00110-1 [DOI] [PubMed] [Google Scholar]
- Lane D., Gray E. A., Mathur R. S., Mathur S. P. (2005). Up-regulation of vascular endothelial growth factor-C by nicotine in cervical cancer cell lines. Am. J. Reprod. Immunol. 53, 153–158 [DOI] [PubMed] [Google Scholar]
- Levi-Montalcini R. (1987). The nerve growth factor 35 years later. Science 237, 1154–1162 10.1126/science.3306916 [DOI] [PubMed] [Google Scholar]
- Levin E. D., Rose J. E. (1995). Acute and chronic nicotinic interactions with dopamine systems and working memory performance. Ann. N. Y. Acad. Sci. 757, 245–252 [DOI] [PubMed] [Google Scholar]
- Levin E. D., Simon B. B. (1998). Nicotinic acetylcholine involvement in cognitive function in animals. Psychopharmacology (Berl.) 138, 217–230 [DOI] [PubMed] [Google Scholar]
- Li M., Linseman D. A., Allen M. P., Meintzer M. K., Wang X., Laessig T., Wierman M. E., Heidenreich K. A. (2001). Myocyte enhancer factor 2A and 2D undergo phosphorylation and caspase-mediated degradation during apoptosis of rat cerebellar granule neurons. J. Neurosci. 21, 6544–6552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M. D., Kane J. K., Wang J., Ma J. Z. (2004). Time-dependent changes in transcriptional profiles within five rat brain regions in response to nicotine treatment. Brain Res. Mol. Brain Res. 132, 168–180 10.1016/j.molbrainres.2004.09.009 [DOI] [PubMed] [Google Scholar]
- Mao Z., Bonni A., Xia F., Nadal-Vicens M., Greenberg M. E. (1999). Neuronal activity-dependent cell survival mediated by transcription factor MEF2. Science 286, 785–790 10.1126/science.286.5440.785 [DOI] [PubMed] [Google Scholar]
- Marinelli M., Piazza P. V. (2002). Interaction between glucocorticoid hormones, stress and psychostimulant drugs. Eur. J. Neurosci. 16, 387–394 [DOI] [PubMed] [Google Scholar]
- Mash D. C., Adi N., Duque L., Pablo J., Kumar M., Ervin F. R. (2008). Alpha synuclein protein levels are increased in serum from recently abstinent cocaine abusers. Drug Alcohol Depend. 94, 246–250 10.1016/j.drugalcdep.2007.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mexal S., Frank M., Berger R., Adams C. E., Ross R. G., Freedman R., Leonard S. (2005). Differential modulation of gene expression in the NMDA postsynaptic density of schizophrenic and control smokers. Brain Res. Mol. Brain Res. 139, 317–332 10.1016/j.molbrainres.2005.06.006 [DOI] [PubMed] [Google Scholar]
- Nakayama H., Numakawa T., Ikeuchi T., Hatanaka H. (2001). Nicotine-induced phosphorylation of extracellular signal-regulated protein kinase and CREB in PC12h cells. J. Neurochem. 79, 489–498 10.1046/j.1471-4159.2001.00602.x [DOI] [PubMed] [Google Scholar]
- Nisell M., Nomikos G. G., Chergui K., Grillner P., Svensson T. H. (1997). Chronic nicotine enhances basal and nicotine-induced Fos immunoreactivity preferentially in the medial prefrontal cortex of the rat. Neuropsychopharmacology 17, 151–161 10.1016/S0893-133X(97)00040-7 [DOI] [PubMed] [Google Scholar]
- Novak G., Boukhadra J., Shaikh S. A., Kennedy J. L., Le Foll B. (2009). Association of a polymorphism in the NRXN3 gene with the degree of smoking in schizophrenia: a preliminary study. World J. Biol. Psychiatry 10, 929–935 [DOI] [PubMed] [Google Scholar]
- Nussbaum J., Xu Q., Payne T. J., Ma J. Z., Huang W., Gelernter J., Li M. D. (2008). Significant association of the neurexin-1 gene (NRXN1) with nicotine dependence in European- and African-American smokers. Hum. Mol. Genet. 17, 1569–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onal A., Uysal A., Ulker S., Delen Y., Yurtseven M. E., Evinc A. (2004). Alterations of brain tissue in fetal rats exposed to nicotine in utero: possible involvement of nitric oxide and catecholamines. Neurotoxicol. Teratol. 26, 103–112 [DOI] [PubMed] [Google Scholar]
- Pagliusi S. R., Tessari M., DeVevey S., Chiamulera C., Pich E. M. (1996). The reinforcing properties of nicotine are associated with a specific patterning of c-fos expression in the rat brain. Eur. J. Neurosci. 8, 2247–2256 [DOI] [PubMed] [Google Scholar]
- Papke R. L., Sanberg P. R., Shytle R. D. (2001). Analysis of mecamylamine stereoisomers on human nicotinic receptor subtypes. J. Pharmacol. Exp. Ther. 297, 646–656 [PubMed] [Google Scholar]
- Peng X., Katz M., Gerzanich V., Anand R., Lindstrom J. (1994). Human alpha 7 acetylcholine receptor: cloning of the alpha 7 subunit from the SH-SY5Y cell line and determination of pharmacological properties of native receptors and functional alpha 7 homomers expressed in Xenopus oocytes. Mol. Pharmacol. 45, 546–554 [PubMed] [Google Scholar]
- Phillips S., Fox P. (1998). An investigation into the effects of nicotine gum on short-term memory. Psychopharmacology (Berl.) 140, 429–433 [DOI] [PubMed] [Google Scholar]
- Piazza P. V., Le Moal M. (1997). Glucocorticoids as a biological substrate of reward: physiological and pathophysiological implications. Brain Res. Brain Res. Rev. 25, 359–372 [DOI] [PubMed] [Google Scholar]
- Pollman M. J., Naumovski L., Gibbons G. H. (1999). Vascular cell apoptosis: cell type-specific modulation by transforming growth factor-beta1 in endothelial cells versus smooth muscle cells. Circulation 99, 2019–2026 [DOI] [PubMed] [Google Scholar]
- Reuter M., Hennig J., Netter P. (2004). Do smoking intensity-related differences in vigilance indicate altered glucocorticoid receptor sensitivity? Addict. Biol. 9, 35–41 [DOI] [PubMed] [Google Scholar]
- Rezvani K., Teng Y., Shim D., De Biasi M. (2007). Nicotine regulates multiple synaptic proteins by inhibiting proteasomal activity. J. Neurosci. 27, 10508–10519 10.1523/JNEUROSCI.3353-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson T. E., Kolb B. (2004). Structural plasticity associated with exposure to drugs of abuse. Neuropharmacology 47(Suppl. 1), 33–46 10.1016/j.neuropharm.2004.06.025 [DOI] [PubMed] [Google Scholar]
- Robinson-Rechavi M., Carpentier A. S., Duffraisse M., Laudet V. (2001). How many nuclear hormone receptors are there in the human genome? Trends Genet. 17, 554–556 [DOI] [PubMed] [Google Scholar]
- Salminen O., Seppa T., Gaddnas H., Ahtee L. (1999). The effects of acute nicotine on the metabolism of dopamine and the expression of Fos protein in striatal and limbic brain areas of rats during chronic nicotine infusion and its withdrawal. J. Neurosci. 19, 8145–8151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt H. F., Huang L. Z., Son J. H., Pinzon-Guzman C., Slaton G. S., Winzer-Serhan U. H. (2008). Acute nicotine activates c-fos and activity-regulated cytoskeletal associated protein mRNA expression in limbic brain areas involved in the central stress-response in rat pups during a period of hypo-responsiveness to stress. Neuroscience 157, 349–359 10.1016/j.neuroscience.2008.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schochet T. L., Bremer Q. Z., Brownfield M. S., Kelley A. E., Landry C. F. (2008). The dendritically targeted protein dendrin is induced by acute nicotine in cortical regions of adolescent rat brain. Eur. J. Neurosci. 28, 1967–1979 [DOI] [PubMed] [Google Scholar]
- Schochet T. L., Kelley A. E., Landry C. F. (2004). Differential behavioral effects of nicotine exposure in adolescent and adult rats. Psychopharmacology (Berl.) 175, 265–273 [DOI] [PubMed] [Google Scholar]
- Schochet T. L., Kelley A. E., Landry C. F. (2005). Differential expression of arc mRNA and other plasticity-related genes induced by nicotine in adolescent rat forebrain. Neuroscience 135, 285–297 10.1016/j.neuroscience.2005.05.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalizi A. K., Bonni A. (2005). brawn for brains: the role of MEF2 proteins in the developing nervous system. Curr. Top. Dev. Biol. 69, 239–266 [DOI] [PubMed] [Google Scholar]
- Soderstrom K., Qin W., Williams H., Taylor D. A., McMillen B. A. (2007). Nicotine increases FosB expression within a subset of reward- and memory-related brain regions during both peri- and post-adolescence. Psychopharmacology (Berl.) 191, 891–897 [DOI] [PubMed] [Google Scholar]
- Soto-Otero R., Mendez-Alvarez E., Hermida-Ameijeiras A., Lopez-Real A. M., Labandeira-Garcia J. L. (2002). Effects of (−)-nicotine and (−)-cotinine on 6-hydroxydopamine-induced oxidative stress and neurotoxicity: relevance for Parkinson's disease. Biochem. Pharmacol. 64, 125–135 [DOI] [PubMed] [Google Scholar]
- Spence J. P., Liang T., Liu L., Johnson P. L., Foroud T., Carr L. G., Shekhar A. (2009). From QTL to candidate gene: a genetic approach to alcoholism research. Curr. Drug Abuse Rev. 2, 127–134 10.2174/1874473710902020127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez S. V., Amadon A., Giacomini E., Wiklund A., Changeux J. P., Le Bihan D., Granon S. (2009). Brain activation by short-term nicotine exposure in anesthetized wild-type and beta2-nicotinic receptors knockout mice: a BOLD fMRI study. Psychopharmacology (Berl.) 202, 599–610 [DOI] [PubMed] [Google Scholar]
- Tammimaki A., Pietila K., Raattamaa H., Ahtee L. (2006). Effect of quinpirole on striatal dopamine release and locomotor activity in nicotine-treated mice. Eur. J. Pharmacol. 531, 118–125 [DOI] [PubMed] [Google Scholar]
- Tang K., Wu H., Mahata S. K., O'Connor D. T. (1998). A crucial role for the mitogen-activated protein kinase pathway in nicotinic cholinergic signaling to secretory protein transcription in pheochromocytoma cells. Mol. Pharmacol. 54, 59–69 [DOI] [PubMed] [Google Scholar]
- Terry A. V., Jr., Clarke M. S. (1994). Nicotine stimulation of nerve growth factor receptor expression. Life Sci. 55, PL91–PL98 [DOI] [PubMed] [Google Scholar]
- Thoenen H., Bandtlow C., Heumann R. (1987). The physiological function of nerve growth factor in the central nervous system: comparison with peiphery. Rev. Physiol. Biochem. Pharmacol. 109, 145–178 [DOI] [PubMed] [Google Scholar]
- Thomas L., Welsh L., Galvez F., Svoboda K. (2008). Acute nicotine exposure and modulation of a spinal motor circuit in embryonic zebrafish. Toxicol. Appl. Pharmacol. 239, 1–12 [DOI] [PubMed] [Google Scholar]
- Tizabi Y., Getachew B., Rezvani A. H., Hauser S. R., Overstreet D. H. (2009). Antidepressant-like effects of nicotine and reduced nicotinic receptor binding in the Fawn-Hooded rat, an animal model of co-morbid depression and alcoholism. Prog. Neuropsychopharmacol. Biol. Psychiatry 33, 398–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tye S. J., Miller A. D., Blaha C. D. (2009). Differential corticosteroid receptor regulation of mesoaccumbens dopamine efflux during the peak and nadir of the circadian rhythm: a molecular equilibrium in the midbrain? Synapse 63, 982–990 10.1002/syn.20682 [DOI] [PubMed] [Google Scholar]
- Vadasz C., Saito M., O'Brien D., Zavadil J., Morahan G., Chakraborty G., Wang R. (2007). Ventral tegmental transcriptome response to intermittent nicotine treatment and withdrawal in BALB/cJ, C57BL/6ByJ, and quasi-congenic RQI mice. Neurochem. Res. 32, 457–480 [DOI] [PubMed] [Google Scholar]
- Wang J., Chen Y. B., Zhu X. N., Chen R. Z. (2001). Activation of p42/44 mitogen-activated protein kinase pathway in long-term potentiation induced by nicotine in hippocampal CA1 region in rats. Acta Pharmacol. Sin. 22, 685–690 [PubMed] [Google Scholar]
- Wang J., Gutala R., Sun D., Ma J. Z., Sheela R. C., Ticku M. K., Li M. D. (2007). Regulation of platelet-derived growth factor signaling pathway by ethanol, nicotine, or both in mouse cortical neurons. Alcohol. Clin. Exp. Res. 31, 357–375 [DOI] [PubMed] [Google Scholar]
- Wang J., Kim J. M., Donovan D. M., Becker K. G., Li M. D. (2009). Significant modulation of mitochondrial electron transport system by nicotine in various rat brain regions. Mitochondrion 9, 186–195 10.1016/j.mito.2009.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber R. J., Gomez-Flores R., Smith J. E., Martin T. J. (2009). Neuronal adaptations, neuroendocrine and immune correlates of heroin self-administration. Brain Behav. Immun. 23, 993–1002 [DOI] [PubMed] [Google Scholar]
- Wei J., Wang J., Dwyer J. B., Mangold J., Cao J., Leslie F. M., Li M. D. (2011). Gestational nicotine treatment modulates cell death/survival-related pathways in the brains of adolescent female rats. Int. J. Neuropsychopharmacol. 14, 91–106 10.1017/S1461145710000416 [DOI] [PubMed] [Google Scholar]
- Winer J., Jung C. K., Shackel I., Williams P. M. (1999). Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal. Biochem. 270, 41–49 [DOI] [PubMed] [Google Scholar]
- Xie Y. X., Bezard E., Zhao B. L. (2005). Investigating the receptor-independent neuroprotective mechanisms of nicotine in mitochondria. J. Biol. Chem. 280, 32405–32412 [DOI] [PubMed] [Google Scholar]
- Yang Y. H., Dudoit S., Luu P., Lin D. M., Peng V., Ngai J., Speed T. P. (2002). Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]