Abstract
Human apolipoprotein-B mRNA-editing catalytic polypeptide-like 3 (APOBEC3) proteins constitute a family of cytidine deaminases that mediate restriction of retroviruses, endogenous retro-elements and DNA viruses. It is well established that these enzymes are potent mutators of viral DNA, but it is unclear whether their editing activity is a threat to the integrity of the cellular genome. We show that expression of APOBEC3A can lead to induction of DNA breaks and activation of damage responses in a deaminase-dependent manner. Consistent with these observations, APOBEC3A expression induces cell-cycle arrest. These results indicate that cellular DNA is vulnerable to APOBEC3 activity and deregulated expression of APOBEC3A could threaten genomic integrity.
Keywords: APOBEC3A, cytidine deaminase, DNA damage, uracil–DNA glycosylase
Introduction
The apolipoprotein-B mRNA-editing catalytic polypeptide-like 3 (APOBEC3) protein family of cytidine deaminases mediates restriction of retroviruses, endogenous retro-elements and DNA viruses (Chiu & Greene, 2008). These enzymes catalyse deamination of cytosines in DNA, converting them to uracil (Conticello et al, 2007). APOBEC3 (A3) genes are thought to have arisen from duplication of the locus for the activation-induced cytidine deaminase (AID; Jarmuz et al, 2002). AID is a well-characterized DNA mutator (Petersen-Mahrt et al, 2002) that is required for both class-switch recombination and somatic hypermutation in B cells (Delker et al, 2009), two crucial processes of immunoglobulin gene diversification. The precise mechanisms by which AID is targeted to the immunoglobulin locus are unknown, and it has recently become clear that off-target activity of AID can lead to non-specific deamination throughout the cellular genome (Liu et al, 2008). In addition to their antiviral activity (Harris et al, 2003; Bishop et al, 2004; Bogerd et al, 2006; Chen et al, 2006), it has been suggested that APOBEC3 proteins have a role in mediating the degradation of foreign DNA through extensive deamination (Stenglein et al, 2010). Several members of the APOBEC3 family have been shown to localize to and be active in the nucleus (Bogerd et al, 2006; Chen et al, 2006). However, the possibility that these proteins could threaten cellular DNA has not been investigated.
Results And Discussion
A3A expression induces phosphorylation of H2AX
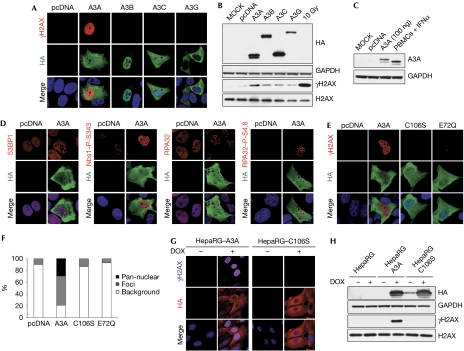
The cellular DNA damage response (DDR) is a highly regulated signal-transduction cascade that detects DNA lesions and replication stress. Detection of DNA breaks by sensor proteins leads to the activation of signalling kinases, which coordinate the amplification of the signal through downstream effector molecules (Ciccia & Elledge, 2010). To evaluate the potential consequences of APOBEC3 protein expression for cellular genomic integrity, we examined the induction of DNA damage signalling. The histone variant H2AX becomes phosphorylated on Ser 139 in response to DNA damage, and the resulting phosphorylated γH2AX is a marker for DNA damage (Rogakou et al, 1998). Human U2OS cells were transfected with plasmids encoding APOBEC3 proteins and analysed by immunofluorescence. A3A and A3C were found throughout the cell, whereas A3B was predominantly localized in the nucleus and A3G was in the cytoplasm (Fig 1A). An antibody against γH2AX showed a strong activation of this DNA damage marker in cells expressing A3A. γH2AX staining was detected in a pan-nuclear pattern or in distinct foci (Fig 1A; supplementary Fig S1A online). By contrast, activation of H2AX was barely detectable in cells transfected with other APOBEC3 family members and AID (Fig 1A; supplementary Fig S1A,B,D,E online). Similar results were observed in HeLa and 293T cell lines (data not shown). Transfection of a nuclear localization signal-containing A3G-expression plasmid resulted in increased accumulation of the protein in the nucleus, but had no effect on its ability to induce H2AX phosphorylation (supplementary Fig S1D,E online). We confirmed these observations by western blot analysis of cell lysates obtained from U2OS cells transfected with APOBEC3 proteins (Fig 1B). Activation of H2AX was exquisitely sensitive to the levels of A3A, and was observed in a dose-dependent manner (supplementary Fig S1C online) at levels that were comparable to induced levels of endogenous A3A in human peripheral blood mononuclear cells (Fig 1C). Together, these results demonstrate that A3A expression can induce a strong DDR.
Figure 1.
APOBEC3A expression can induce DNA damage responses. (A) APOBEC3A induces phosphorylation of histone H2AX. U2OS cells left untreated (MOCK) or transfected with plasmids expressing haemagglutinin (HA)-tagged A3A, A3B, A3C and A3G or the pcDNA3.1 control plasmid were fixed 24 h post-transfection, stained with HA and γH2AX antibodies, and analysed by confocal microscopy. (A,D,E) Nuclei were stained with DAPI. Results are representative of three independent experiments. (B) Western blot detection of phosphorylated H2AX in cells transfected with A3A. U2OS cells were transfected with APOBEC3 expression plasmids or pcDNA3.1 as control. Cells were collected at 48 h and lysates were analysed by western blot using HA, H2AX and γH2AX antibodies. Cells irradiated with 10 Gy were used as a positive control. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control. Results are representative of three independent experiments. (C) Transfected U2OS cells express A3A at physiological levels. A3A in lysates from U2OS transfected with 100 ng of A3A plasmid was compared with levels of endogenous A3A expressed in human peripheral blood mononuclear cells (PBMCs) stimulated with interferon (IFN)α (1,000 U/ml) for 24 h. Cell lysates were analysed by western blotting using A3A antibodies, and GAPDH was used as a loading control. (D) Activation of DNA repair proteins by A3A. U2OS cells were transfected with an A3A expression vector or pcDNA3.1 as control. After 24 h, cells were fixed and stained with antibodies to 53BP1, phosphorylated Nijmegen breakage syndrome 1 (Nbs1, S343), replication protein A32 (RPA32) and phosphorylated RPA32 (S4,8), and analysed by confocal microscopy. (E) Phosphorylated H2AX is not detected in cells transfected with the inactive A3A mutants C106S and E72Q. U2OS cells were transfected with plasmids expressing wild-type A3A, the C106S or E72Q mutants or the pcDNA3.1 control plasmid. After 24 h, cells were fixed, stained with HA and γH2AX antibodies and analysed by confocal microscopy. (F) Analysis of γH2AX staining in cells transfected with A3A or the C106S and E72Q mutants. Transfected cells were scored for γH2AX staining. The graph shows the percentage of cells with pan-nuclear γH2AX staining, γH2AX foci and background staining. (G,H) Analysis of phosphorylated H2AX in stable HepaRG cells expressing inducible A3A or the C106S mutant. Cells were grown in the presence or absence of doxycycline (1 μg/ml) for 24 h. Fixed cells were analysed by confocal microscopy (G) and cells lysates were analysed by western blot (H). Results are representative of three independent experiments. APOBEC3, apolipoprotein-B mRNA-editing catalytic polypeptide-like 3; DAPI, 4,6-diamidino-2-phenylindole.
A3A expression activates DNA damage signalling
To characterize further the events triggered by A3A expression, we analysed activation of other components of the DDR. In undamaged cells, the mediator protein 53BP1 is found diffusely in the nucleus and in a few large domains, but in response to DNA damage it accumulates at numerous foci at damage sites (Schultz et al, 2000). In cells expressing A3A, 53BP1 was detected in distinct foci (Fig 1D; supplementary Fig S1F online). By using antibodies generated to specific phosphorylated residues on proteins that are activated as downstream substrates within the DNA-damage signalling cascade, we also observed phosphorylation of Nijmegen breakage syndrome 1 and checkpoint kinase 2 in the presence of A3A (Fig 1D; supplementary Fig S1F,G online). Replication protein A (RPA) accumulates on single-stranded DNA and is phosphorylated in response to DNA damage. In cells expressing A3A, we observed the presence of several nuclear RPA foci, and staining, with an antibody that recognizes phosphorylated S4,8 residues demonstrated RPA phosphorylation (Fig 1D; supplementary Fig S1F online). Phosphorylation of these proteins indicates that damage kinases are activated by A3A expression. In support of this conclusion, we observed that ataxia telangiectasia mutated was activated, as detected with an antibody generated to the autophosphorylated activation site at S1981 (supplementary Fig S1G online). A3A could induce γH2AX staining in cells deficient in ataxia telangiectasia mutated or DNA-dependent protein kinase, consistent with the known redundancy of phosphoinositide 3-kinase-related kinases in phosphorylating H2AX (supplementary Fig S1H,I online). Together, these results demonstrate strong activation of γH2AX and a cascade of DNA damage signalling events in response to A3A expression.
A3A catalytic activity is required for activation of the DDR
A3A contains a single cytidine deaminase domain with the consensus motif H-X-E-X28-PC-X4-C, in which the histidine and cysteine residues coordinate zinc, and glutamate functions as the catalytic residue. It has been previously demonstrated that mutation of these residues results in catalytically inactive A3A (Bogerd et al, 2006; Chen et al, 2006; Narvaiza et al, 2009). To investigate whether induction of the DDR by A3A requires its catalytic activity, we analysed H2AX phosphorylation in cells transfected with the inactive mutants C106S and E72Q (Fig 1E,F). Mutants in the active site of A3A failed to induce the phosphorylation of H2AX, as detected by immunofluorescence (Fig 1E) and western blotting analyses (supplementary Fig S1J online). In addition, the C106S mutant also failed to activate downstream signalling pathways (supplementary Fig S1G online). As A3A has been suggested to affect transgene expression through extensive deamination of transfected plasmid DNA (Stenglein et al, 2010), we asked whether A3A activates the DDR in the absence of plasmid DNA. To address this question, we generated inducible HepaRG hepatocyte cell lines (Everett et al, 2009) expressing haemagglutinin-tagged A3A and the C106S mutant. When cells were treated with doxycycline, we observed strong γH2AX signals in cells expressing A3A, suggesting that activation of the DDR by A3A does not require the presence of plasmid DNA (Fig 1G,H). As expected, H2AX activation was not detected in cells induced to express the C106S mutant. Similar results were obtained when cells were transfected with mRNA encoding A3A and the C106S mutant (data not shown). Together, these data demonstrate that the catalytic activity of A3A is necessary for activation of the DDR, independently of plasmid DNA.
A3A expression leads to DNA breaks
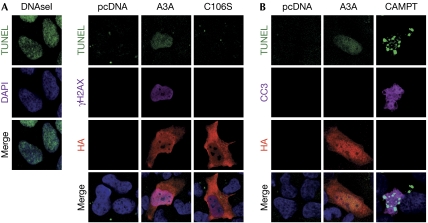
To assess whether the signalling induced by A3A was due to induction of DNA breaks, we used the terminal deoxynucleotidyl transferase dUTP nick end-labelling (TUNEL) assay (Fig 2A; supplementary Fig S2 online), using DNAse1-treated cells as a positive control. In 36% of cells expressing A3A, we observed mostly diffuse positive TUNEL staining, localizing in the nucleus. This signal was absent in control cells and cells transfected with the C106S mutant. In addition, positive TUNEL staining in cells expressing A3A strongly correlated with the presence of pan-nuclear γH2AX staining. To rule out the fact that the observed TUNEL signals were due to labelling of DNA strand breaks generated as a consequence of apoptosis, we used an antibody to the active form of caspase 3 (CC3; Fig 2B). To induce apoptosis, cells were treated with camptothecin, a DNA topoisomerase I inhibitor. Samples treated with camptothecin had a large number of apoptotic cells, which were positive for both CC3 and TUNEL staining. Cells expressing A3A were consistently negative for cleaved CC3 staining. These results indicate that A3A expression induces DNA breaks that do not result from apoptosis.
Figure 2.
Expression of APOBEC3A leads to DNA breaks. (A) Detection of DNA breaks in cells transfected to express A3A. U2OS cells were transfected with plasmids expressing wild-type A3A, the C106S mutant or the pcDNA3.1 control plasmid. Cells were fixed at 24 h, labelled by TUNEL and subsequently incubated with γH2AX and haemagglutinin (HA) antibodies. Cells treated for 10 min with DNAse I (3 U/ml) are shown as a positive control for TUNEL staining. Nuclei were stained with DAPI. Images are representative of three independent experiments. (B) Induction of DNA damage by A3A is not a consequence of apoptosis. U2OS cells were transfected with an A3A expression vector or pcDNA3.1, fixed at 24 h, and labelled by TUNEL and subsequently incubated with HA and the cleaved form of caspase 3 (CC3) antibodies. As a positive control, cells were treated with 20-μM camptothecin (CAMPT) for 20 h to induce apoptosis. Images are representative of three independent experiments. APOBEC3, apolipoprotein-B mRNA-editing catalytic polypeptide-like 3; DAPI, 4,6-diamidino-2-phenylindole; TUNEL, terminal deoxynucleotidyl transferase dUTP nick end-labelling.
UNG is required for induction of DNA damage by A3A
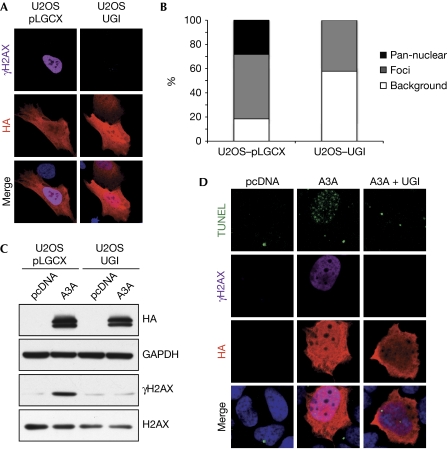
Spontaneous cytosine deamination events generate uracil lesions that are repaired by the base excision repair pathway, a process that is initiated by cellular uracil–DNA glycosylases (UNGs; Kavli et al, 2007). Removal of uracil residues by these enzymes generates abasic sites that are subsequently processed by the apurinic/apyrimidinic endonuclease 1. Base excision repair of uracil primarily relies on the activity of UNG, a main cellular glycosylase that is also required for the recognition of uracil residues in somatic hypermutation and class-switch recombination. To determine whether UNG has a role in induction of DNA breaks and activation of the DDR by A3A expression, we used a retroviral vector expressing the UNG inhibitor (UGI; Kaiser & Emerman, 2006) to generate a U2OS-based stable cell line. By using an oligo cleavage assay, we confirmed that UNG activity was undetectable in these cells (supplementary Fig S3E online). We observed that stable expression of UGI prevented the detection of A3A-mediated H2AX activation by both immunofluorescence and western blotting analyses (Fig 3A,C). Although we could detect some cells with foci of γH2AX induced by A3A, there was no robust A3A-induced pan-nuclear staining in the presence of UGI (Fig 3B). Similar results were obtained with transient co-transfection of A3A and UGI in U2OS cells (supplementary Fig S3A–C online). We also observed that UGI co-transfection prevented induction of DNA breaks by A3A, as detected in the TUNEL assay (Fig 3D; supplementary Fig S3D online). Together, these results demonstrate that UNG is required for activation of the DNA damage response in cells expressing A3A. This suggests that UNG activity might be involved in processing the products of A3A activity into DNA breaks and thereby activating the DDR.
Figure 3.
Uracil–DNA glycosylase activity is required for A3A-induced DNA damage. (A) The UNG inhibitor (UGI) prevents H2AX phosphorylation in cells transfected with A3A. U2OS cells stably expressing UGI or control cells were transfected with A3A. Cells were fixed after 24 h, stained with haemagglutinin (HA) and γH2AX antibodies and analysed by confocal microscopy. Nuclei were stained with DAPI. Images are representative of three independent experiments. (B) Analysis of γH2AX staining in cells stably expressing UGI transfected with A3A. Cells showing HA staining were scored for γH2AX staining. The graph shows the percentage of cells with pan-nuclear H2AX activation, γH2AX foci and background staining. (C) Western blot detection of phosphorylated H2AX in U2OS cells stably expressing UGI transfected with A3A. Cells were transfected with A3A or the pcDNA3.1 control plasmid, collected at 24 h and lysates were analysed by western blotting, using HA, H2AX and γH2AX antibodies. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control. Images are representative of two independent experiments. (D) Induction of DNA breaks by A3A requires UNG activity. U2OS cells were co-transfected with A3A or the pcDNA3.1 control plasmid together with a plasmid expressing UGI (pLPC-UGI) or the empty vector. Cells were fixed after 24 h and labelled by TUNEL and subsequently incubated with γH2AX and HA antibodies. Nuclei were stained with DAPI and cells were analysed by confocal microscopy. Images are representative of three independent experiments. DAPI, 4,6-diamidino-2-phenylindole; TUNEL, terminal deoxynucleotidyl transferase dUTP nick end-labelling; UNG, uracil–DNA glycosylase.
A3A expression induces cell-cycle arrest
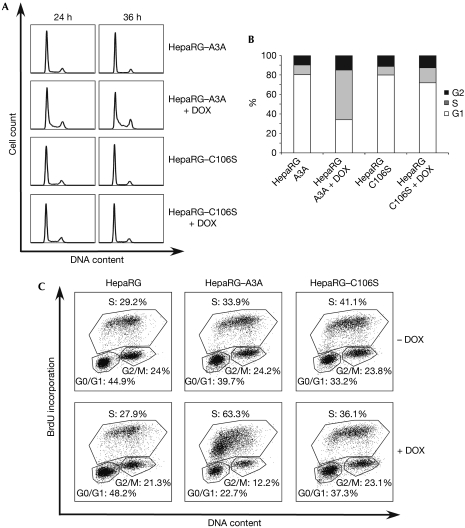
To study the consequences of activation of the DDR in cells expressing A3A, we analysed cell-cycle progression (Fig 4). Inducible A3A cells were grown in the absence or presence of doxycycline, and cell-cycle progression was assessed by flow cytometry at different time points. Propidium iodide staining for total DNA content showed that cells expressing the C106S mutant were unaffected by doxycycline treatment (Fig 4A,B). By contrast, cells induced to express wild-type A3A accumulated in S phase, with the total S-phase fraction reaching 45% by 36 h postinduction. Similar results were observed when inducible cells were synchronized and analysed for cell-cycle progression (data not shown). In addition, we also observed a G1/S arrest in U2OS cells transfected with A3A (supplementary Fig S4 online). To confirm these observations, we evaluated the effect of A3A expression on the cell cycle by monitoring bromodeoxyuridine (BrdU) incorporation. When HepaRG–A3A cells were grown in the presence of doxycycline, a significant number of cells accumulated in early S phase (Fig 4C). At 24 h postinduction, the fraction of induced A3A cells in S phase reached 63.3%, whereas cells expressing C106S and control cells showed 36.1% and 27.9% of cells in S phase, respectively. These results demonstrate that A3A induces a block to cell-cycle progression, which is dependent on its catalytic activity.
Figure 4.
APOBEC3A expression induces cell-cycle arrest. (A) Stable HepaRG cells expressing A3A are blocked in S phase. Inducible cells expressing wild-type and mutant A3A (HepaRG–A3A and HepaRG–C106S) were grown in the presence or absence of doxycycline (DOX, 1 μg/ml) and analysed for cell-cycle progression at different time points by propidium iodide staining. Results are representative of three independent experiments. (B) Graph shows the percentage of cells in G1, S and G2 phase for each sample at 36 h postinduction. (C) HepaRG, HepaRG–A3A and HepaRG–C106S were grown in the presence or absence of doxycycline and analysed for cell-cycle progression 24 h postinduction, using BrdU staining. Results are representative of three independent experiments. APOBEC3, apolipoprotein-B mRNA-editing catalytic polypeptide-like 3; BrdU, bromodeoxyuridine; UNG, uracil–DNA glycosylase.
AID is the only cytidine deaminase whose physiological function includes deamination of specific genomic regions (Delker et al, 2009). Recent studies indicate that efficient DNA repair is required to prevent the off-target activity of AID from generating chromosomal instability (Liu et al, 2008; Hasham et al, 2010). Our results identify A3A as a member of the APOBEC3 family that is distinct in its ability to induce detectable cellular DNA damage and activation of the DNA repair machinery when ectopically expressed. A recent screen for AID mutants with increased catalytic activity suggested that the activity of AID has been limited to minimize the risk of genomic instability (Wang et al, 2009). Many of the selected mutations in this screen not only brought the sequence of AID closer to that found in APOBEC3 proteins, but also increased the rate of chromosomal translocations (Wang et al, 2009). Therefore, our results raise the possibility that the potent antiviral activity of A3A (Chen et al, 2006) might come at the expense of increased risk of modification of the host genome.
The finding that ectopic A3A expression induces cell-cycle arrest in early S phase indicates that cellular DNA undergoing replication might be highly susceptible to A3A activity and/or that A3A leads to stalled replication forks. Few studies have investigated the expression and regulation of APOBEC3 genes in vivo and shown that A3A expression is induced by interferon-α in macrophages and dendritic cells (Koning et al, 2009; Stenglein et al, 2010). Although our results demonstrate that ectopic A3A expression is detrimental for cellular DNA integrity, it remains unclear whether endogenous A3A leads to deamination of genomic DNA. As we were unable to knockdown completely A3A levels in human peripheral blood mononuclear cells, we could not determine whether induction of A3A expression in these cells by interferon-α stimulation leads to H2AX phosphorylation.
Uncontrolled deamination of the cellular genome is potentially harmful and could lead to genomic instability and cancer initiation. Given the emerging links between inflammation and cancer, it is likely that immune responses to infection contribute to malignancy. It will be interesting to determine whether endogenous A3A can be mutagenic and whether DNA repair mechanisms constitute a safeguard against A3A-mediated deamination of cellular DNA. Our results raise the possibility that deregulated activity of APOBEC proteins might contribute to genomic instability and suggest that their potential contribution to malignant transformation should be carefully investigated.
Methods
Cell lines, plasmids and transfections and antibodies. See supplementary information online.
Western blotting and immunofluorescence. For western blotting, lysates were prepared by standard methods. Insoluble fractions were isolated by extracting the pellet with 0.1-M HCl for 30 min at 4°C. For immunofluorescence, U2OS cells were transfected with haemagglutinin-tagged APOBEC3 expression vectors, using lipofectamine 2000 (Invitrogen). Cells were fixed after 24 h with 4% paraformaldehyde and extracted with 0.5% Triton X-100 in PBS for 10 min. Nuclei were visualized by staining with 4,6-diamidino-2-phenylindole. For TUNEL staining, fixed cells were labelled using the in situ Cell Death Detection Kit (Roche), according to the manufacturer's protocol. Images were acquired using a Leica TCS SP2 confocal microscope. Representative images are shown except in cases in which different patterns have been quantified.
Cell-cycle analysis. HepaRG, HepaRG–A3A and HepaRG–C106S cells were grown in the presence or absence of doxycycline (0.1 μg/ml) fixed in 70% ice-cold ethanol, washed in PBS and resuspended in staining solution containing 20 μg/ml propidium iodide (Sigma) and 200 μg/ml RNAse A (Worthington). Experiments were performed in duplicate and 30,000 cells were analysed per sample. For BrdU incorporation, cells were stained using the APC BrdU flow kit (BD Bioscience), according to the manufacturer's instructions.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank R. Everett, M. Emerman, E. Hendrickson, N. Landau, T. Melendy, Y. Shiloh and F. Chisari for reagents. We thank C. Lilley and other members of the Weitzman lab for helpful discussions. We are grateful to R. Bushman, J. Moran and J. Young for comments on the manuscript. Work in the Weitzman lab is partly supported by a Pioneer Developmental Chair from the Salk Institute and by National Institutes of Health grants AI067952 and AI074967 (M.D.W.). This work was also funded by fellowships from the Instituto de Salud ‘Carlos III’/Consejo Superior de Investigaciones Científicas/Salk Institute and the Lynn Streim Postdoctoral Endowment Fellowship (I.N.), and the Natural Sciences & Engineering Research Council of Canada (S.L.).
Footnotes
The authors declare that they have no conflict of interest.
References
- Bishop KN, Holmes RK, Sheehy AM, Davidson NO, Cho SJ, Malim MH (2004) Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr Biol 14: 1392–1396 [DOI] [PubMed] [Google Scholar]
- Bogerd HP, Wiegand HL, Hulme AE, Garcia-Perez JL, O'Shea KS, Moran JV, Cullen BR (2006) Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc Natl Acad Sci USA 103: 8780–8785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Lilley CE, Yu Q, Lee DV, Chou J, Narvaiza I, Landau NR, Weitzman MD (2006) APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr Biol 16: 480–485 [DOI] [PubMed] [Google Scholar]
- Chiu YL, Greene WC (2008) The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu Rev Immunol 26: 317–353 [DOI] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ (2010) The DNA damage response: making it safe to play with knives. Mol Cell 40: 179–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conticello SG, Langlois MA, Neuberger MS (2007) Insights into DNA deaminases. Nat Struct Mol Biol 14: 7–9 [DOI] [PubMed] [Google Scholar]
- Delker RK, Fugmann SD, Papavasiliou FN (2009) A coming-of-age story: activation-induced cytidine deaminase turns 10. Nat Immunol 10: 1147–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett RD, Parsy ML, Orr A (2009) Analysis of the functions of herpes simplex virus type 1 regulatory protein ICP0 that are critical for lytic infection and derepression of quiescent viral genomes. J Virol 83: 4963–4977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH (2003) DNA deamination mediates innate immunity to retroviral infection. Cell 113: 803–809 [DOI] [PubMed] [Google Scholar]
- Hasham MG, Donghia NM, Coffey E, Maynard J, Snow KJ, Ames J, Wilpan RY, He Y, King BL, Mills KD (2010) Widespread genomic breaks generated by activation-induced cytidine deaminase are prevented by homologous recombination. Nat Immunol 11: 820–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmuz A, Chester A, Bayliss J, Gisbourne J, Dunham I, Scott J, Navaratnam N (2002) An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79: 285–296 [DOI] [PubMed] [Google Scholar]
- Kaiser SM, Emerman M (2006) Uracil DNA glycosylase is dispensable for human immunodeficiency virus type 1 replication and does not contribute to the antiviral effects of the cytidine deaminase Apobec3G. J Virol 80: 875–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavli B, Otterlei M, Slupphaug G, Krokan HE (2007) Uracil in DNA—general mutagen, but normal intermediate in acquired immunity. DNA Repair (Amst) 6: 505–516 [DOI] [PubMed] [Google Scholar]
- Koning FA, Newman EN, Kim EY, Kunstman KJ, Wolinsky SM, Malim MH (2009) Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets. J Virol 83: 9474–9485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Duke JL, Richter DJ, Vinuesa CG, Goodnow CC, Kleinstein SH, Schatz DG (2008) Two levels of protection for the B cell genome during somatic hypermutation. Nature 451: 841–845 [DOI] [PubMed] [Google Scholar]
- Narvaiza I, Linfesty DC, Greener BN, Hakata Y, Pintel DJ, Logue E, Landau NR, Weitzman MD (2009) Deaminase-independent inhibition of parvoviruses by the APOBEC3A cytidine deaminase. PLoS Pathog 5: e1000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen-Mahrt SK, Harris RS, Neuberger MS (2002) AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature 418: 99–103 [DOI] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM (1998) DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 273: 5858–5868 [DOI] [PubMed] [Google Scholar]
- Schultz LB, Chehab NH, Malikzay A, Halazonetis TD (2000) p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J Cell Biol 151: 1381–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenglein MD, Burns MB, Li M, Lengyel J, Harris RS (2010) APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nat Struct Mol Biol 17: 222–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Yang Z, Rada C, Neuberger MS (2009) AID upmutants isolated using a high-throughput screen highlight the immunity/cancer balance limiting DNA deaminase activity. Nat Struct Mol Biol 16: 769–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.