EMBO Rep (2011) advance online publication. doi:; DOI: 10.1038/nature09732
EMBO Rep (2011) advance online publication. doi:; DOI: 10.1038/nature09779
EMBO Rep (2011) advance online publication. doi:; DOI: 10.1038/emboj.2010.112
During mitosis, eukaryotic cells have to properly align their chromosomes. Only after the kinetochore of each chromosome is attached to a polar microtubule can a cell satisfy the ‘spindle assembly checkpoint’, which prevents the mis-segregation of chromosomes. Failure to correctly segregate chromosomes before cell division might contribute to chromosome instability and tumorigenesis. To counteract chromosome aberrations, the cell initiates the apoptotic programme. It has become clear through the use of microtubule-poisoning, chemotherapeutic agents—such as paclitaxel and vincristine—that prolonged activation of the spindle checkpoint can induce mitotic arrest and, subsequently, programmed cell death. The molecular mechanisms responsible for initiating apoptosis during mitotic arrest have remained poorly defined. Two recent papers in Nature (Inuzuka et al, 2011; Wertz et al, 2011) and a report published by the Clarke group last year in The EMBO Journal (Harley et al, 2010) highlight the destruction of MCL1 during prolonged mitotic arrest and shed light on the mechanisms of apoptosis induction.
Myeloid cell leukaemia 1 (MCL1) is an anti-apoptotic member of the B-cell lymphoma 2 (BCL2) family of proteins. MCL1, like BCL2 and BCLxL, prevents the downstream activation of BAX and BAK, which are responsible for mitochondrial outer-membrane permeabilization, initiation of the caspase cascade and induction of apoptosis (Youle & Strasser, 2008). Ubiquitination and proteolysis of MCL1 have been reported, but a mechanism for MCL1 degradation following spindle checkpoint activation remains unknown. Now, the studies referenced above suggest that degradation of MCL1 during prolonged mitotic arrest is essential for the induction of apoptosis. Given its prominent role in driving the cell cycle, as well as in safeguarding the fidelity of this process, it is not surprising that the ubiquitin-proteasome system (UPS) has a key role in dictating the activation of the intrinsic apoptotic pathway in cells arrested in mitosis. However, it is surprising that two E3 ubiquitin ligase complexes simultaneously facilitate this degradation event.
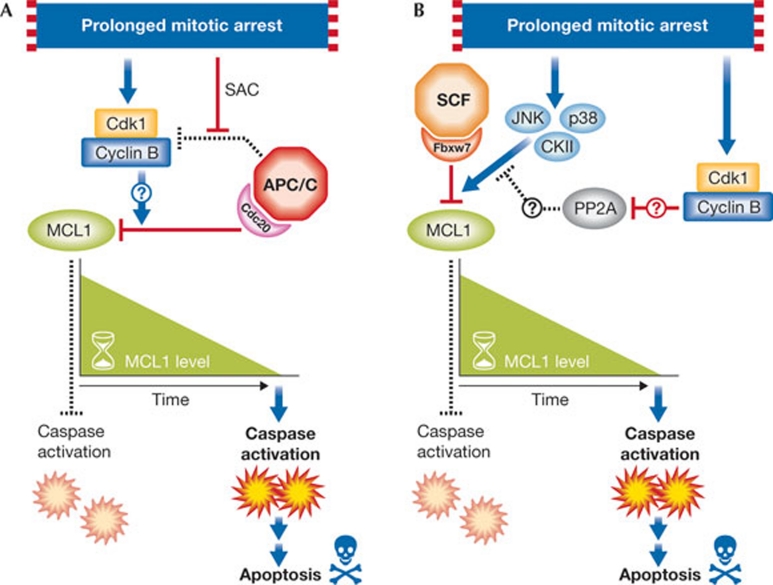
Harley and colleagues describe the regulation of MCL1 by APC/CCdc20(anaphase-promoting complex/cyclosome and its activator Cdc20). This multi-subunit RING E3 ubiquitin ligase is active in mitosis, and ubiquitinates substrates such as securin and cyclin B, thereby allowing progression into anaphase. In their report, Harley and co-workers (2010) demonstrate a Cdk1/cyclin-B-mediated, site-specific phosphorylation (Thr 92 in humans) of MCL1 upon mitotic arrest, followed by its proteolytic destruction by APC/CCdc20. Thus, like the sand of an hourglass flipped at each entry into mitosis, the level of MCL1 steadily decreases. If time ‘runs out’ due to a prolonged mitotic arrest (that is, if MCL1 is completely destroyed), then apoptosis is initiated (Fig 1A). Both phosphorylation at Thr 92 and the presence of a conserved destruction or ‘D’-box motif (a characteristic of APC/C substrates) are required for MCL1 proteolysis, although the precise role of phosphorylation in promoting degradation remains unclear.
Figure 1.
Two models for proteolytic destruction of MCL1 during mitotic arrest. (A) Schematic illustration of the effects of APC/CCdc20 and Cdk1/cyclin B on the degradation of MCL1 during prolonged arrest in mitosis. (B) Schematic illustration of the effects of SCFFbw7, JNK/p38/CKII and Cdk1/cyclin B on MCL1 degradation during mitotic arrest. PP2A is a protein phosphatase that is reported to associate with MCL1. Dashed lines represent inactive processes. Question marks denote unknown mechanisms. APC/CCdc20, anaphase-promoting complex/cyclosome and its activator Cdc20; SAC, spindle assembly checkpoint.
Interestingly, the stability of MCL1 in asynchronous cells seems to be unaffected when the ability of APC/CCdc20 to target MCL1 is compromised by knockdown of Cdc20, or when phosphorylation at Thr 92 is ablated. Although the spindle assembly checkpoint is believed to inhibit APC/CCdc20 activity, the degradation of some targets, such as the CDK-inhibitor p21 and cyclin A, is not affected. Consequently, it is possible that MCL1 can be destroyed through Cdc20 during mitotic arrest.
More recently, in two reports in Nature (Inuzuka et al, 2011; Wertz et al, 2011), it is shown that MCL1 interacts with another E3 ubiquitin ligase, SCFFbxw7. Similarly to the APC/C, the SCF (Skp1/Cul1/F-box protein) is a multi-subunit, RING E3 ubiquitin ligase. The F-box protein provides the specificity for target recognition, often by using specific interaction domains to bind to substrates. In the case of Fbxw7 (also known as Fbw7 and hCdc4), a series of WD40 domains form a pocket that dictates the binding of several substrates. For all known substrates, one or two phosphorylated degradation motifs (phospho-degrons) are recognized by Fbxw7 (Welcker & Clurman, 2008), and MCL1 seems to follow this trend. Briefly, two Fbxw7 degrons—Ser 121/Glu 125 and Ser 159/Thr 163—with different binding affinities were identified in MCL1. Inuzuka and colleagues report that these sites are phosphorylated in a GSK3-dependent manner, supporting a previous report that demonstrated a role for GSK3 in controlling MCL1 degradation (Maurer et al, 2006). They also demonstrate that Fbxw7 affects MCL1 stability during the DNA damage response. Wertz and co-workers provide evidence that, during mitotic arrest, the degrons in MCL1 are instead phosphorylated by JNK, p38 and CKII. Interestingly, when Wertz and colleagues investigated the degradation of MCL1 during mitotic arrest, they discovered a dependence on Fbxw7 similar to that reported for Cdc20 (Fig 1B). Furthermore, a functional Fbxw7–MCL1 interaction was required for the induction of apoptosis in ovarian cancer and T-ALL cell lines treated with microtubule-targeting chemotherapies. This observation presents a dilemma. Which ubiquitin ligase complex—APC/CCdc20 or SCFFbxw7—targets MCL1 for destruction during mitotic arrest? Do they compete or cooperate?
There are several approaches that could be taken to investigate these questions. Perhaps the most promising direction is through understanding the role of various MCL1 phosphorylation events, particularly phosphorylation of Thr 92. The reports collectively demonstrate that Thr 92 and the Fbxw7 degrons are phosphorylated in mitotic cells. It is interesting that Thr 92 phosphorylation is specifically induced at mitosis, and Wertz and colleagues suggest that this event might drive the dissociation of a phosphatase to allow Fbxw7 degron phosphorylation (Fig 1B). However, the results so far are preliminary, and a more complete understanding of the mechanism by which Cdk1/cyclin B phosphorylation of MCL1 promotes proteolysis, and whether this is through Cdc20 and/or Fbxw7, is essential. Although MCL1 degradation after mitotic arrest is unlikely to be associated with the activity of GSK3, is there an induction of GSK3-dependent phosphorylation of MCL1 under other conditions? This important question has been studied previously, but it requires further investigation. Perhaps additional ‘priming’ kinases are involved, as is suspected to be the case for cyclin E and c-Myc, two other substrates of Fbxw7.
The concept of a protein being targeted by two ubiquitin ligases is not new. For example, similarly to MCL1, p21 and MLL are targeted by both APC/CCdc20 and an SCF complex (SCFSkp2). Several APC/CCdh1 substrates (for example, Cdc25A and claspin) are also degraded via SCFβTrCP (Frescas & Pagano, 2008). However, in these instances, APC/C and SCF target the substrates at different phases of the cell cycle. The case of MCL1 is less clear. Wertz and colleagues show that mitotic arrest specifically induces binding of MCL1 to Fbxw7. Conversely, Inuzuka and colleagues provide data suggesting that Fbxw7 loss affects the non-mitotic stability of MCL1. Additionally, in an earlier paper, the Fbxw7 degron was reported to be phosphorylated by GSK3 during cytokine withdrawal (Maurer et al, 2006). Thus, we are left with a picture in which Fbxw7 targets MCL1 during mitotic arrest, but it might also target MCL1 at other points during the cell cycle or in response to external stimuli. With regard to Cdc20-mediated degradation of MCL1, mutation of the D-box seems to stabilize MCL1 only during mitotic arrest, although Cdc20 remains bound to MCL1 in non-mitotic cells. Thus, there might be differences in the conditions for recognition by either Fbxw7 or Cdc20 that merit further investigation. It is also worth mentioning that deubiquitinating enzymes (DUBs) might counteract the activity of Fbxw7, Cdc20, or both. In fact, the DUB USP9X was found to associate with MCL1 (Schwickart et al, 2010).
Assuming that both E3 ligases target the same pool of MCL1 at the same time during mitotic arrest, why are there two modes of regulation? It could be that the ligases cooperate to lower MCL1 levels. It is possible that Fbxw7 and Cdc20 together deplete MCL1 to a point at which apoptosis can be initiated; if either ligase is compromised, apoptotic induction is inefficient. Alternatively, there might be a particularly relevant growth condition or cell-type specificity that favours the activity of one complex over the other. For example, it is possible that in tissues that give rise to human cancers harbouring Fbxw7 mutations (for example, T-ALLs or ovarian carcinomas), SCFFbxw7 acts as the predominant ligase. Finally, there could be redundancy or competition between the different E3 ligases. Perhaps untransformed cells maintain both systems, to protect against apoptosis evasion in the face of spindle dysfunction. Alternatively, one or both of these systems might be compromised in the cell-culture models. Notably, the situation is further complicated by reports indicating that other ligases seem to affect MCL1 stability: Mule/Huwe1 (Zhong et al, 2005) and SCFβTrCP (Ding et al, 2007). Silencing of Mule stabilizes MCL1, although Wertz and colleagues did not observe dramatic changes in MCL1 stability after Mule depletion during mitotic arrest. Instead, three groups did not observe stabilization of MCL1 after βTrCP silencing (Wertz et al, 2011; Inuzuka et al, 2011; Dehan et al, 2009). Moreover, the interaction between MCL1 and βTrCP seems to be mediated by BimEL (a βTrCP substrate), as indicated by increased binding under conditions when BimEL is degraded (rather than under conditions when MCL1 is degraded) and by the fact that some BimEL mutants lose their ability to bind to βTrCP, regardless of their binding to MCL1 (Dehan et al, 2009).
Although the details of MCL1 regulation at mitotic arrest have only begun to unfold, it is clear that this pathway holds promise for furthering our understanding of the regulation of apoptosis. Microtubule-poisoning agents have historically been reliable chemotherapeutics, so, identifying cellular components that regulate MCL1 degradation during mitotic arrest is not only a way to stratify patients for a positive response to such drugs, but might also lead to the identification of novel targets for pharmacological intervention.
References
- Dehan E et al. (2009) Mol Cell 33: 109–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q et al. (2007) Mol Cell Biol 27: 4006–4017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frescas D, Pagano M (2008) Nat Rev Cancer 8: 438–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley ME et al. (2010) EMBO J 29: 2407–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inuzuka H et al. (2011) Nature 471: 104–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer U et al. (2006) Mol Cell 21: 749–760 [DOI] [PubMed] [Google Scholar]
- Schwickart M et al. (2010) Nature 463: 103–107 [DOI] [PubMed] [Google Scholar]
- Welcker M, Clurman BE (2008) Nat Rev Cancer 8: 83–93 [DOI] [PubMed] [Google Scholar]
- Wertz IE et al. (2011) Nature 471: 110–114 [DOI] [PubMed] [Google Scholar]
- Youle RJ, Strasser A (2008) Nat Rev Mol Cell Biol 9: 47–59 [DOI] [PubMed] [Google Scholar]
- Zhong Q et al. (2005) Cell 121: 1085–1095 [DOI] [PubMed] [Google Scholar]