Abstract
We report a rapid-cycle, real-time PCR method for identifying six Candida spp. directly from BACTEC blood culture bottles. Target sequences in the noncoding internal transcribed spacer regions of the rRNA operon were simultaneously amplified and interrogated with fluorescent probes to identify Candida albicans, C. glabrata, C. parapsilosis, C. tropicalis, C. krusei, and C. lusitaniae; these account for 88% of the yeast species isolated from positive blood cultures in our laboratory. Any of the first four species can be identified in a single reaction using two fluorescent hybridization probe sets. The antifungal-resistant species C. krusei and C. lusitaniae are detected in a second reaction, also with two probe sets. The assay was validated with DNA extracted from BACTEC blood culture bottles positive for yeasts (n = 62) and was 100% concordant with culture identification based on biochemical and morphological features of C. albicans (n = 22), C. parapsilosis (n = 10), C. tropicalis (n = 1) C. glabrata (n = 22), C. krusei (n = 2), and C. lusitaniae (n = 1). No cross-reactivity was observed in blood culture samples growing yeasts other than the above-mentioned species (n = 4), in those growing bacteria (n = 12), or in the absence of microbial growth. Our assay allows rapid (≤2 h) and specific detection of the most common Candida spp. directly from positive blood cultures and may facilitate delivery of optimal antifungal therapy.
Bloodstream fungal infections are a significant cause of mortality in immunocompromised patients (26). Candida spp. account for 70 to 80% of invasive bloodstream fungal infections, and collectively they represent the fourth most common nosocomial bloodstream infection (1, 9, 23). Among the Candida species causing invasive infections, C. albicans, C. parapsilosis, C. tropicalis, and C. glabrata account for about 80 to 90% of fungal isolates encountered in the clinical laboratory (14, 16). Although C. albicans remains the species most commonly isolated, other newly emerging and antifungal-resistant Candida species, such as C. glabrata, C. krusei, and C. lusitaniae, are increasingly common (16, 23). Hence, rapid identification of these Candida species may facilitate optimal antifungal therapy and patient management (5, 19). However, current methods involving biochemical and phenotypic tests performed on isolated yeast colonies may require up to 96 h for species identification from the initial time of blood culture bottle positivity (25).
Several DNA amplification assays rapidly detect Candida isolates from solid media or blood culture bottles. They include amplification of a target gene(s) followed by a postamplification analysis step to identify the amplicons based on electrophoretic migration or by hybridization with specific nucleotide probes (6, 7, 10, 13, 22). These procedures involve a potential risk of contamination or are labor-intensive due to postamplification analysis. Only a few real-time or direct assays to detect amplicons in the PCR mixture have been developed to identify Candida species. These include a recently described TaqMan assay involving the 5′ exonuclease activity and fluorescent DNA probes and the RNA-directed nucleic acid sequence-based amplification assay (2, 21).
The internal transcribed spacer 1 (ITS1) and ITS2 regions, flanked by coding sequences of the 18S, 5.8S, and 28S rRNA structural genes, were previously demonstrated to reliably identify medically important yeasts on the basis of species-specific DNA sequence polymorphisms (3, 4). In a recent study, we rapidly differentiated two closely related germ tube- positive Candida spp., namely, C. albicans and C. dubliniensis, by designing probes to species-specific regions of ITS2 (20). Here we report a rapid and sensitive genotypic method to identify six Candida spp.: the four most commonly encountered (C. albicans, C. glabrata, C. parapsilosis, and C. tropicalis) and two inherently antifungal-resistant species (C. krusei and C. lusitaniae). Identification of these Candida spp. directly from positive blood culture media is achieved by rapid-cycle nucleic acid amplification of the ITS2 DNA, fluorescent-probe hybridization to specific sequences in the ITS2 amplicons, and melting curve analysis (MCA). We evaluated our assay on template DNA extracted from BACTEC blood culture media (n = 62) that were positive for fungal elements by Gram staining.
MATERIALS AND METHODS
Clinical samples.
Aliquots from positive BACTEC blood culture bottles were Gram stained and subcultured to appropriate fungal medium, including CHROMagar, inhibitory mold agar, Candida Bromcresol Green, and chocolate agar, and the culturable yeasts were identified by phenotypic (germ tube formation) and biochemical (VITEK YBC card) tests and chlamydospore formation on cornmeal agar (25). Sixty-two blood culture bottles received in the clinical laboratory for routine culturing were found to contain yeasts by Gram staining (see Table 3), and 2 ml was frozen at −20C for future extraction of fungal DNA. Negative controls included 12 blood culture bottles growing different bacterial species, including Staphylococcus aureus, coagulase-negative Staphylococcus spp., Enterococcus faecalis, Corynebacterium spp., viridans group streptococci, Streptococcus pyogenes, Serratia marcescens, Escherichia coli, “Helicobacter rappini,” Legionella pneumophila, Fusobacterium necrophorum, and Mycobacterium avium.
TABLE 3.
Identification of Candida spp. in 62 BACTEC blood culture bottles using the LightCycler assay
| Blood cultures (n) | No. of yeasts identified by probe set(s)a
|
|||||
|---|---|---|---|---|---|---|
| CALB | CGLAB | CPAR | CFLUC | CFLUC + CGLAB | CKRU + CLUS | |
| C. albicans (22) | 22/22 | NP | NP | 22/22 | 22/22 | NP |
| C. glabrata (22) | NP | 22/22 | NP | NP | 22/22 | NP |
| C. parapsilosis (10) | NP | NP | 10/10 | 10/10 | 10/10 | NP |
| C. tropicalis (1) | 1/1 | NP | NP | 1/1 | 1/1 | NP |
| C. krusei (2) | NP | NP | NP | NP | NP | 2/2 |
| C. lusitaniae (1) | NP | NP | NP | NP | NP | 1/1 |
| Other speciesb (4) | NP | NP | NP | NP | NP | NP |
Number of bottles positive/total number of bottles tested. NP, no peak.
Other species identified from positive blood cultures in our study served as specificity controls for the hybridization reactions and MCA: C. kefyr (1 isolate), C. guilliermondii (1), Rhodotorula mucilaginosa (1), and Cryptococcus neoformans (1).
DNA extraction from blood culture bottles.
Three different DNA extraction protocols were evaluated to identify the best method for obtaining inhibitor-free nucleic acids from BACTEC blood culture bottles that could be used in conjunction with the LightCycler assay.
QIAamp (silica column) purification.
The total DNA from 0.1 ml of yeast-positive BACTEC blood culture medium was purified according to the manufacturer's directions by using the QIAamp blood kit (Qiagen Corporation, Chatsworth, Calif.) and eluted in 0.1 ml of elution buffer.
Benzyl alcohol-guanidine hydrochloride method (organic extraction).
A previously described method was used (8). Briefly, 0.1 ml of yeast-positive BACTEC blood culture medium was added to 0.1 ml of lysis buffer (5.0 M guanidine hydrochloride-100 mM Tris [pH 8.0] in sterile water) and mixed with a vortex mixer. A total of 0.4 ml of water was added along with 0.8 ml of 99% benzyl alcohol (Sigma Chemical Co.) and mixed again by vortexing. Following centrifugation at 7,000 × g for 5 min, the aqueous supernatant was removed, and DNA was precipitated from it by addition of 0.040 ml of 3.0 M sodium acetate and 0.44 ml of isopropanol. The mixture was centrifuged at 16,000 × g for 15 min at 4°C, the precipitated DNA was washed once with 70% ethanol, and the pellet was air dried. The DNA was resuspended in 0.1 ml of 10 mM Tris-0.1 mM EDTA buffer at pH 8.5.
Mechanical disruption of yeasts.
Mechanical disruption of yeasts was performed as described previously (22). About 0.2 ml of sample was lysed by the addition of 0.8 ml of TXTE buffer (10 mM Tris, 1 mM EDTA [pH 8.0], 1% Triton X-100). Following lysis, Candida blastoconidia were pelleted by centrifugation at 16,000 × g for 5 min, washed three times with 1 ml of TXTE buffer, and resuspended in 0.3 ml of TXTE buffer. About 0.2 ml of 0.5-mm-diameter zirconium beads was added to the mixture, boiled for 15 min, and shaken for 20 min in a mechanical bead beater (Mini-Beadbeater; Biospec Products, Bartlesville, Okla.). After centrifugation for 20 s, the supernatant was stored at 20°C until it was used for PCR amplification.
Capillary based amplification and detection.
Amplification was performed in a 7-μl total reaction volume consisting of 1× LightCycler master hybridization mix (Roche Diagnostics), plus an additional 2.0 mM MgCl2 and 1 μM ITS3-1 forward primer (5′-CATCGATGAAGAACGCAGC-3′) and ITS4 reverse primer (5′-TCCTCCGCTTATTGATATGC-3′). Product identification was achieved by adding a donor probe (0.1 μM; CALB, CFLUC, CGLAB, CPAR, CKRU or CLUS) and the respective acceptor probe (0.2 μM) (Table 1) to the reaction mix along with 1 μl of the template DNA. Thermal cycling conditions were as follows: 95°C initial denaturation for 30 s was followed by 45 cycles of 95°C for 1 s, 55°C for 10 s, and 72°C for 25 s.
TABLE 1.
Probes used in this studya
| Probe | Sequence (5′→3′) | Reference |
|---|---|---|
| CFLUC | TCCCTCAAACCCCTGGGTTT-FLU (donor) | |
| LC-Red640-TGTTGAGCAATACGACTTGGGTTTGp- (acceptor) | 20 | |
| CALB | TCCCTCAAACCGCTGGGTTT-FLU (donor) | |
| LC-Red640-TGTTGAGCAATACGACTTGGGTTTGp- (acceptor) | 20 | |
| CGLAB | ACTTGAAATTGTAGGCCATATCAGTATG-FLU (donor) | |
| LC-Red640-GGACACGAGCGCAAGCTp- (acceptor) | This study | |
| CPAR | TGTTGAGCGATACGCTG-FLU (donor) | |
| LC-Red640-TTTGCTTGAAAGAAAGGCGGAGp- (acceptor) | This study | |
| CKRU | TGAAAGGGAGCGAAGC-FLU (donor) | |
| LC-Red640-GCCGAGCGAACTAGACp- (acceptor) | This study | |
| CLUS | GGTTAGGCGTTGCTCCGAAA-FLU (donor) | |
| LC-Red640-TCAACCGCGCTGTCAAACACp- (acceptor) | This study |
The oligonucleotide probes were coupled with fluorescein (FLU) and LC-Red640 dye for detection. The 3′ end of the acceptor probe is phosphorylated (p) to avoid extension of the probe.
Alternatively, to reduce glass capillary usage, DNA prepared from blood culture bottles was tested in two glass capillary tubes. The CFLUC and CGLAB probe sets identified C. albicans, C. glabrata, C. parapsilosis, and C. tropicalis in the first reaction, and CKRU and CLUS probe sets identified the inherently antifungal-resistant species C. krusei and C. lusitaniae in the second reaction. Template DNA from the corresponding Candida spp. was included as a positive control in each run. The MCA profile was 95°C for 10 s, 40°C for 30 s, and ramping to 70°C. The LightCycler software was used to estimate the peak melting temperature (Tm) by the automatic method. The polynomial with background correction method was selected, and one peak Tm was obtained by using the Gaussian fit analysis.
RESULTS
The LightCycler assay involves rapid PCR thermal cycling in glass capillaries and achieves 45 cycles in 45 min. The amplified products are identified by MCA of various fluorescent hybridization probes binding to amplicons derived from the ITS2 region target DNA sequences. Our method takes advantage of the nucleotide polymorphisms found in the ITS2 region among Candida species, which can be amplified from medically important yeasts using universal primers (4). While individual species can be identified by using individual probe sets, our assay identifies any one of four commonly encountered Candida species in one reaction and two antifungal-resistant Candida species in a second reaction.
The analytical sensitivity of the CFLUC and CGLAB probes for detecting purified and serially diluted C. albicans or C. glabrata DNA was ≤1 fg (data not shown). Accurate identification with each probe set was validated with randomly selected fungal isolates from our well-characterized collection. This collection includes more than 600 isolates of medically important yeasts, comprising 62 different species, encountered in our laboratory. They have been characterized by standard biochemical, morphological, and genetic methods, including ITS1 and ITS2 length polymorphisms and DNA sequence analysis of the ITS1 and ITS2 loci and the D1/D2 variable domain of the 26S rRNA gene (3, 4). ITS2 sequences, the amplification target for the assay described below, were analyzed from multiple isolates of each species and reliably shown to provide species-specific information (3). Further, dendrograms constructed by comparing the sequences of different species provide taxonomically accurate classifications (3, 4). These data directly demonstrate that comparison of ITS2 region DNA sequences with those in the database, validated with hundreds of clinical isolates, can identify or differentiate unknown yeasts. Therefore, the specificity of all probe sets (Table 1) was assessed with DNA isolated from 16 yeast species from the above collection: C. albicans, C. tropicalis. C. glabrata, C. krusei, C. lusitaniae, C. guilliermondii, C. parapsilosis, C. kefyr, C. intermedia, C. lambica, Cryptococcus neoformans, Cryptococcus albidus, Pichia farinosa, Pichia ohmeri, Saccharomyces cerevisiae, and Rhodotorula rubra (Table 2). The CGLAB probe set produced a peak Tm of 60.31 ± 0.64°C (average ± standard deviation) when tested against C. glabrata isolates (n = 19) (Table 2). The CPAR probe set produced a peak Tm of 57.30 ± 0.11°C when tested with C. parapsilosis isolates (n = 15). The CKRU probe set produced a peak Tm of 51.49 ± 0.51°C when tested with C. krusei isolates (n = 19). The CLUS probe set produced a peak Tm of 63.00 ± 0.55°C when tested with C. lusitaniae isolates (n = 18). The CALB probe set was validated in a previous study and identified C. albicans isolates (n = 69) by a peak Tm of 61.04 ± 0.64°C (20). Although this probe set also hybridized with C. tropicalis DNA (n = 19), the 10°C difference in peak Tm (50.17 ± 0.27°C) allowed reliable identification of both species. A previously described hybrid probe, CFLUC (20), hybridized with four Candida spp. and produced characteristic peak Tms that identified each of them: 49.94 ± 0.54°C for C. albicans (n = 69), 54.03 ± 1.01°C for C. dubliniensis (n = 28), 47.19 ± 0.40°C for C. parapsilosis (n = 15), and 52.15 ± 0.32°C for C. tropicalis (n = 19). Only these specific cross hybridizations were detected among the extracted DNAs tested (Table 2).
TABLE 2.
Specificity of probes for detecting medically important yeasts
| Candida sp. tested (n) | Peak Tm with probe seta
|
|||||
|---|---|---|---|---|---|---|
| CALB | CGLAB | CPAR | CKRU | CLUS | CFLUC | |
| C. albicans (69) | 61.04 ± 0.64 | NP | NP | NP | NP | 49.94 ± 0.54 |
| C. glabrata (19) | NP | 60.31 ± 0.64 | NP | NP | NP | NP |
| C. tropicalis (19) | 50.17 ± 0.27 | NP | NP | NP | NP | 52.15 ± 0.32 |
| C. parapsilosis (15) | NP | NP | 57.30 ± 0.11 | NP | NP | 47.19 ± 0.40 |
| C. krusei (19) | NP | NP | NP | 51.49 ± 0.51 | NP | NP |
| C. lusitaniae (18) | NP | NP | NP | NP | 63.00 ± 0.55 | NP |
| Other yeast spp.b | NP | NP | NP | NP | NP | NP |
Individual yeast species were identified by MCA, which produced characteristic peak Tms in the presence of the corresponding hybridization probe set. Values are averages ± standard deviations. NP, no peak.
All six Candida species targeted in our study were tested with all probe sets along with the other medically important yeast species as negative controls: C. guilliermondii, C. kefyr, C. intermedia, C. lambica, Cryptococcus neoformans, Cryptococcus albidus, Pichia farinosa, Pichia ohmeri, Saccharomyces cerevisiae, and Rhodotorula rubra.
To directly apply this technology to patient specimens, we needed to circumvent the possibility that PCR inhibitors can be present in blood culture media (8). Aliquots (100 to 200 μl) from 10 blood cultures positive for yeasts were subjected to DNA extraction by three established protocols. Organic extraction using guanidium hydrochloride (Materials and Methods) produced the most reliable templates for amplification in our LightCycler assay. With Qiagen extraction, six of ten samples produced amplifiable products while the other four were successfully amplified after a 1:5 dilution. A mechanical disruption method worked for nine of ten extractions (see Materials and Methods; also, data not shown).
Using organic extraction, DNA was extracted from 62 blood culture bottles positive for yeasts by Gram staining. The six Candida spp. targeted by our assay were identified with 100% sensitivity and 100% specificity when MCA was used to produce characteristic peak Tms (Table 3). Four blood culture bottles with Cryptococcus neoformans, C. guilliermondii, Rhodotorula mucilaginosa, and C. kefyr were not identified by our assay, nor did they cause false-positive reactions. For these samples, where no melting curve was observed and yeasts were detected in the blood cultures, the PCR mixture from the capillaries was drained and run on a 2% agarose gel (Fig. 1) to confirm positive amplification of the ITS2 region by using universal primers (4). The fluorescent probes did not produce any cross hybridizations in blood culture samples growing bacteria (n = 12) or in the absence of detectable microbial growth (data not shown).
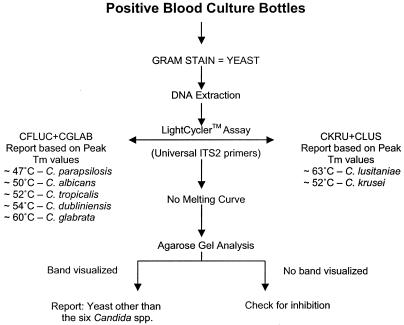
FIG. 1.
Algorithm for rapid detection of Candida species commonly encountered in blood cultures. DNA extracted from blood culture bottles positive for yeasts by Gram staining are used as templates for real-time PCR amplification in a LightCycler. Following amplification of the ITS2 region with universal fungal primers (3, 4), MCA is performed to identify the products based on characteristic peak Tms that identify each Candida spp. Amplicons that produce uninterpretable or no peak Tms are analyzed by gel electrophoresis to confirm nucleic acid amplification. Visualization of amplicon indicates yeast species other than the six Candida spp., and the absence of an amplicon indicates PCR inhibition.
All reactions were performed in a 7-μl total volume, approximately one-third of the manufacturer's recommended 20-μl reaction volume. Further streamlining of the assay and additional cost savings were achieved by using a single capillary to identify more than one species; combining the CFLUC and CGLAB probe sets in one reaction and the CKRUS and CLUS probe sets in another reaction identified six species. The first reaction identified the four most commonly encountered Candida spp. in the clinical laboratory (C. albicans, C. glabrata, C. parapsilosis, and C. tropicalis), and, if necessary, a second reaction identified the antifungal-resistant C. krusei and C. lusitaniae isolates (Table 3).
DISCUSSION
We describe a real-time PCR method to rapidly identify six invasive Candida species: C. albicans, C. glabrata, C. parapsilosis, C. tropicalis, C. krusei, and C. lusitaniae. These Candida spp. account for 80 to 90% of fungemic isolates encountered in the clinical laboratory, including those that are frequently antifungal resistant (14, 16, 23). Unlike growth and phenotypic identification, our assay can be completed in less than 2 h after the initial detection of yeasts by Gram staining. Individual hybridization probes produced characteristic peak Tms specific for each Candida species. Alternately, combining the CFLUC and CGLAB probes identified C. albicans, C. parapsilosis, C. tropicalis, and C. glabrata in a single reaction, and combining the CKRU and CLUS probes identified C. krusei and C. lusitaniae in one additional reaction (Table 3).
Hybridization of fluorescent probes to their target amplicons produced characteristic melting curves with peak Tms that identified each species; following the final amplification cycle, each sample was rapidly heated to 95°C to denature all DNA, cooled below the annealing temperature of the probes to allow their hybridization to target sequences, and slowly heated to melt the hybridization probes away from their target sequences while decreases in fluorescence were monitored to produce characteristic melting curves. Peak Tms represent the first negative derivative of the change in fluorescence over time plotted against temperature, and a characteristic Tm specifically identifies each product. The Tm is dependent on probe length, G+C content of the region of hybridization, and degree of homology between the donor probe and the target sequence.
The ability of the MCA to identify point mutations provides an advantage in the identification of PCR amplicons (12). Theoretically, a probe that has 100% identity with the target allele is more stable than a mismatched probe-allele duplex, and the difference in thermostability due to probe-target mismatch(es) should identify different amplicons based on specific peak Tms revealed during MCA. To take advantage of this property we designed a probe set (CFLUC) to span a region of high sequence homology within the ITS2 regions of the Candida spp. of interest. The ITS2 target sequences of C. albicans, C. tropicalis, and C. parapsilosis had one, one, and two mismatches with the donor probe coupled to fluorescein and zero, three, and two mismatches with the acceptor probe coupled to the LC-Red 640 dye molecule, respectively. Notably, either probe melting away from its target region influences the peak Tm. It is therefore apparent that the peak Tms (Table 2) are not easily predictable based on the number of probe mismatches with target sequences. However, the idea of probe-target mismatches as a means to detect multiple targets with a single probe set is operational, and hybridization and subsequent melting of the CFLUC probes with target amplicons produced characteristic peak Tms that identified each species.
Among the three different nucleic acid extraction formats evaluated, the guanidium hydrochloride extraction method most reliably gave inhibitor-free nucleic acids that were amplifiable. Following DNA extraction our assay can be completed in less than 1 h and demonstrated 100% sensitivity and 100% specificity when compared with standard culture, biochemical, and morphological methods. No cross hybridizations with either bacterial or human DNA were observed.
A large survey recently reported that up to 40% of C. glabrata isolates were resistant to fluconazole and almost all C. krusei isolates were intrinsically resistant (15). Since fluconazole remains the drug of choice to treat invasive candidal infections (18), rapid identification of the fluconazole-resistant species C. glabrata and C. krusei and amphotericin-resistant C. lusitaniae would expedite appropriately targeted antifungal therapy. The use of a genetic approach produces unambiguous and reproducible results in contrast to biochemical methods where phenotypic variations can sometimes obscure an otherwise accurate identification. We did not encounter any mixed candidemias among the blood cultures we evaluated. Experience from our laboratory and others suggests that mixed candidal infections occur at a very low frequency (21, 22). It should be noted that exponential amplification during PCR will bias detection in favor of the predominant yeast population in a mixed infection. Mixed infection would be suspected based on Gram staining or uninterpretable MCA with this assay, and individual species probes would be used to test this hypothesis in repeat testing. Alternately, the adjunctive use of standard culture methods would ensure that small populations of different yeasts are detected in mixed infections.
Several reports of rapid identification of Candida species directly from clinical samples have shown promise in the clinical management of high-risk patients (11, 24). Rapid identification of the fungal pathogen directly from clinical samples and institution of early therapy may help to reduce the hospital stay and high overall costs associated with management of candidemia (17). In conclusion, we have developed a rapid, sensitive, and specific PCR assay to directly detect the most common and antifungal-resistant Candida spp. encountered in a clinical laboratory.
REFERENCES
- 1.Beck-Sague, C., W. R. Jarvis, et al.. 1993. Secular trends in the epidemiology of nosocomial fungal infections in the United States, 1980-1990. J. Infect. Dis. 167:1247-1251. [DOI] [PubMed] [Google Scholar]
- 2.Borst, A., M. A. Leverstein-Van Hall, J. Verhoef, and A. C. Fluit. 2001. Detection of Candida spp. in blood cultures using nucleic acid sequence-based amplification (NASBA). Diagn. Microbiol. Infect. Dis. 39:155-160. [DOI] [PubMed] [Google Scholar]
- 3.Chen, Y. C., J. D. Eisner, M. M. Kattar, S. L. Rassoulian-Barrett, K. Lafe, U. Bui, A. P. Limaye, and B. T. Cookson. 2001. Polymorphic internal transcribed spacer region 1 DNA sequences identify medically important yeasts. J. Clin. Microbiol. 39:4042-4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, Y. C., J. D. Eisner, M. M. Kattar, S. L. Rassoulian-Barrett, K. Lafe, S. L. Yarfitz, A. P. Limaye, and B. T. Cookson. 2000. Identification of medically important yeasts using PCR-based detection of DNA sequence polymorphisms in the internal transcribed spacer 2 region of the rRNA genes. J. Clin. Microbiol. 38:2302-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edwards, J. E., Jr., G. P. Bodey, R. A. Bowden, T. Buchner, B. E. de Pauw, S. G. Filler, M. A. Ghannoum, M. Glauser, R. Herbrecht, C. A. Kauffman, S. Kohno, P. Martino, F. Meunier, T. Mori, M. A. Pfaller, J. H. Rex, T. R. Rogers, R. H. Rubin, J. Solomkin, C. Viscoli, T. J. Walsh, and M. White. 1997. International Conference for the Development of a Consensus on the Management and Prevention of Severe Candidal Infections. Clin. Infect. Dis. 25:43-59. [DOI] [PubMed] [Google Scholar]
- 6.Einsele, H., H. Hebart, G. Roller, J. Loffler, I. Rothenhofer, C. A. Muller, R. A. Bowden, J. van Burik, D. Engelhard, L. Kanz, and U. Schumacher. 1997. Detection and identification of fungal pathogens in blood by using molecular probes. J. Clin. Microbiol. 35:1353-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elie, C. M., T. J. Lott, E. Reiss, and C. J. Morrison. 1998. Rapid identification of Candida species with species-specific DNA probes. J. Clin. Microbiol. 36:3260-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fredricks, D. N., and D. A. Relman. 1998. Improved amplification of microbial DNA from blood cultures by removal of the PCR inhibitor sodium polyanetholesulfonate. J. Clin. Microbiol. 36:2810-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fridkin, S. K., and W. R. Jarvis. 1996. Epidemiology of nosocomial fungal infections. Clin. Microbiol. Rev. 9:499-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujita, S., B. A. Lasker, T. J. Lott, E. Reiss, and C. J. Morrison. 1995. Microtitration plate enzyme immunoassay to detect PCR-amplified DNA from Candida species in blood. J. Clin. Microbiol. 33:962-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hebart, H., J. Loffler, H. Reitze, A. Engel, U. Schumacher, T. Klingebiel, P. Bader, A. Bohme, H. Martin, D. Bunjes, W. V. Kern, L. Kanz, and H. Einsele. 2000. Prospective screening by a panfungal polymerase chain reaction assay in patients at risk for fungal infections: implications for the management of febrile neutropenia. Br. J. Haematol. 111:635-640. [DOI] [PubMed] [Google Scholar]
- 12.Lay, M. J., and C. T. Wittwer. 1997. Real-time fluorescence genotyping of factor V Leiden during rapid-cycle PCR. Clin. Chem. 43:2262-2267. [PubMed] [Google Scholar]
- 13.Luo, G., and T. G. Mitchell. 2002. Rapid identification of pathogenic fungi directly from cultures by using multiplex PCR. J. Clin. Microbiol. 40:2860-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pfaller, M. A. 1996. Nosocomial candidiasis: emerging species, reservoirs, and modes of transmission. Clin. Infect. Dis. 22(Suppl. 2):S89-S94. [DOI] [PubMed] [Google Scholar]
- 15.Pfaller, M. A., D. J. Diekema, R. N. Jones, S. A. Messer, and R. J. Hollis. 2002. Trends in antifungal susceptibility of Candida spp. isolated from pediatric and adult patients with bloodstream infections: SENTRY Antimicrobial Surveillance Program, 1997 to 2000. J. Clin. Microbiol. 40:852-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pfaller, M. A., D. J. Diekema, R. N. Jones, H. S. Sader, A. C. Fluit, R. J. Hollis, and S. A. Messer. 2001. International surveillance of bloodstream infections due to Candida species: frequency of occurrence and in vitro susceptibilities to fluconazole, ravuconazole, and voriconazole of isolates collected from 1997 through 1999 in the SENTRY Antimicrobial Surveillance Program. J. Clin. Microbiol. 39:3254-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rentz, A. M., M. T. Halpern, and R. Bowden. 1998. The impact of candidemia on length of hospital stay, outcome, and overall cost of illness. Clin. Infect. Dis. 27:781-788. [DOI] [PubMed] [Google Scholar]
- 18.Rex, J. H., M. A. Pfaller, A. L. Barry, P. W. Nelson, and C. D. Webb. 1995. Antifungal susceptibility testing of isolates from a randomized, multicenter trial of fluconazole versus amphotericin B as treatment of nonneutropenic patients with candidemia. Antimicrob. Agents Chemother. 39:40-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rex, J. H., T. J. Walsh, J. D. Sobel, S. G. Filler, P. G. Pappas, W. E. Dismukes, and J. E. Edwards. 2000. Practice guidelines for the treatment of candidiasis. Clin. Infect. Dis. 30:662-678. [DOI] [PubMed] [Google Scholar]
- 20.Selvarangan, R., A. P. Limaye, and B. T. Cookson. 2002. Rapid identification and differentiation of Candida albicans and Candida dubliniensis by capillary-based amplification and fluorescent probe hybridization. J. Clin. Microbiol. 40:4308-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shin, J. H., F. S. Nolte, B. P. Holloway, and C. J. Morrison. 1999. Rapid identification of up to three Candida species in a single reaction tube by a 5′ exonuclease assay using fluorescent DNA probes. J. Clin. Microbiol. 37:165-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shin, J. H., F. S. Nolte, and C. J. Morrison. 1997. Rapid identification of Candida species in blood cultures by a clinically useful PCR method. J. Clin. Microbiol. 35:1454-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trick, W. E., S. K. Fridkin, J. R. Edwards, R. A. Hajjeh, and R. P. Gaynes. 2002. Secular trend of hospital-acquired candidemia among intensive care unit patients in the United States during 1989-1999. Clin. Infect. Dis. 35:627-630. [DOI] [PubMed] [Google Scholar]
- 24.van Burik, J. A., D. Myerson, R. W. Schreckhise, and R. A. Bowden. 1998. Panfungal PCR assay for detection of fungal infection in human blood specimens. J. Clin. Microbiol. 36:1169-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warren, N. G., and K. C. Hazen. 1999. Candida, Cryptococcus, and other yeasts of medical importance, p. 1184-1199. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology. American Society for Microbiology, Washington, D.C.
- 26.Wey, S. B., M. Mori, M. A. Pfaller, R. F. Woolson, and R. P. Wenzel. 1988. Hospital-acquired candidemia. The attributable mortality and excess length of stay. Arch. Intern. Med. 148:2642-2645. [DOI] [PubMed] [Google Scholar]