Abstract
The ratio of the number of nonsynonymous substitutions per site (Ka) over the number of synonymous substitutions per site (Ks) has often been used to detect positive selection. Investigators now commonly generate Ka/Ks ratio profiles in a sliding window to look for peaks and valleys in order to identify regions under positive selection. Here we show that the interpretation of peaks in the Ka/Ks profile as evidence for positive selection can be misleading. Genie regions with Ka/Ks > 1 in the MRG gene family, previously claimed to be under positive selection, are associated with a high frequency of polar amino acids with a high mutability. This association between an increased Ka and a high proportion of polar amino acids appears general and not limited to the MRG gene family or the sliding-window approach. For example, the sites detected to be under positive selection in the HIV1 protein-coding genes with a high posterior probability turn out to be mostly occupied by polar amino acids. These findings caution against invoking positive selection from Ka/Ks ratios and highlight the need for considering biochemical properties of the protein domains showing high Ka/Ks ratios. In short, a high Ka/Ks ratio may arise from the intrinsic properties of amino acids instead of from extrinsic positive selection.
1. INTRODUCTION
Positive selection is one of the sculptors of biological adaptation. To detect positive selection on protein-coding sequences, it is common to calculate the number of synonymous substitutions per site (Ks) and nonsynonymous substitutions per site (Ka) and test the null hypothesis of Ka − Ks = 0, based on the neutrality principle1–5. If the null hypothesis is rejected and Ka/Ks > 1 (or Ka ñ Ks > 0), then the presence of positive selection may be invoked 6–9. Statistical methods frequently used for detecting positive selection at the sequence level include the distance-based method for pairwise comparisons 10–13, and the maximum parsimony 14 and maximum likelihood ML methods 5,15–17 used for phylogeny-based inferences. A number of inherent biases and problems in some of these methods have been outlined by various authors 4,18–22.
A previous study has shown that (Ka/Ks >1) need not be a signature of positive Darwinian selection 23. Here, we illustrate one particular bias associated with the heterogeneous distribution of polar amino acids along the linear protein sequence. Our results suggest that peaks in Ka/Ks profiles can arise from an increased frequency of polar amino acids and consequently may not be taken as evidence for positive selection. The generality of the association between the increased Ka/Ks ratio and a high proportion of polar amino acids is further demonstrated with the protein-coding genes in the HIV1 genome.
2. SITES OF ìPOSITIVE SELECTIONî CODE FOR A RELATIVELY HIGH FREQUENCY OF POLAR AMINO ACIDS
We illustrate the problem by using sequence data from the MRG gene family, which belongs to the G-protein-coupled receptor superfamily, is expressed specifically in nociceptive neurons, and is implicated in the modulation of nociception 8. Using the Pamilo-Bianchi-Li method11,12 with a sliding-window approach (window width of 90 base pairs and step length of 15 base pairs) to generate the Ka/Ks profile along the sequence, it has been reported that the peaks (Ka/Ks > 1) in the profile coincided with the extracellular domain boundaries, and the valleys (Ka/Ks < 1) coincided with the transmembrane and cytoplasmic domains 8. This observation prompted the conclusion that the extracellular domains of the MRG receptor family have experienced strong positive selection.
The PBL method is based on the number of transitional and transversional substitutions on the 0-fold degenerate sites (where any nucleotide substitution leads to a nonsynonymous substitution, e.g., the second codon position), 2-fold degenerate sites (where one nucleotide substitution, typically a transition, is synonymous, and the other two nucleotide substitutions are nonsynonymous; e.g., the third codon position of lysine codons AAA and AAG), and 4-fold degenerate sites (where any nucleotide substitution is synonymous, e.g., the third codon position of glycine codons GGA, GGC, GGG, and GGU). The equations for computing the window-specific Kaw and Ksw under PBL method are as follows:
| (1) |
where L0w, L2w, and L4w are the numbers of 0-fold, 2fold, and 4-fold degenerate sites, and Aiw and Biw are the numbers of transitional and transversional substitutions per i-fold degenerate site, respectively, in the given window, w.
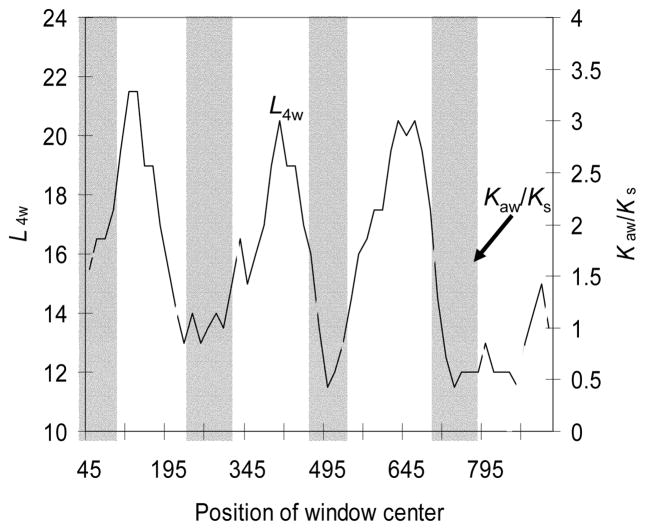
In the practical application of the PBL method, Ksw may often become 0 when the window size is small and/or when the closely-related sequences are compared. In this case, investigators will compute the Kaw/Ks ratio (where Ks is estimated from the whole sequence comparison), rather than the window-specific Kaw/Ksw ratios. The Kaw/Ks profile for the MRG gene family was obtained in this way 8. Thus, any fluctuation seen in Kaw/Ks is simply the fluctuation of Kaw (Fig. 1).
Fig. 1.
Kaw/Ks increases with decreasing L4W. The Kaw/Ks curve is identical to that in Fig. 2a in ref. 8 for MRGX1 vs MRGX4. The number of 4-fold degenerate sites (L4) is superimposed for easy comparison. Shaded areas mark the extracellular domains. The analysis is performed with DAMBE24, 25
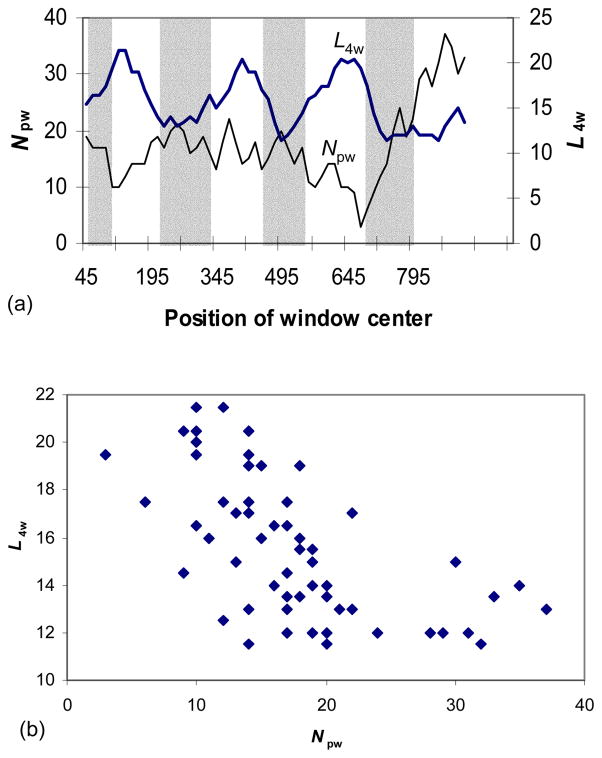
We observed that Kaw/Ks fluctuation for MRG sequences was associated negatively with the number of 4-fold degenerate sites in a window (L4w). This negative correlation is highly significant (Pearson r = − 0.45246, P = 0.003; Fig 1) and means that codons in the extracellular domains contain a rather small number of 4-fold degenerate 3rd codon positions. A survey of the amino acid composition of extracellular domains provides the answer: extracellular domains contain (and require) hydrophilic (polar) amino acids that are mostly coded by 2-fold degenerate codons (we restrict “polar amino acids” to refer to the eight strongly polar amino acids only, i.e., Arg, Asn, Asp, Glu, Gln, Ser, Lys, and His). The negative correlation between the window-specific L4w and the number of polar amino acids (Npw) is also statistically highly significant (Fig. 2; Pearson r = −0.5946, P < 10−5).
Fig. 2.
The increased number of polar amino acids (Npw) in the extracellular domains results in low L4w values (a), leading to significant negative correlation between L4w and Npw (b). Based on comparisons between MRGX1 and MRGX4. Only strongly polar amino acids (Arg, Asn, Asp, Glu, Gln, Ser, Lys, His) were included. The shaded areas mark extracellular domains.
The negative association between L4w and Kaw in the extracellular domains, which implies that nonsynonymous substitutions at the extracellular domains mainly occur at 2-fold degenerate sites, points to a reason for the higher Kaw in the extracellular domains. The nonsynonymous substitutions at the 2-fold degenerate sites involve amino acids that are biochemically more similar to each other than those at the 0-fold degenerate positions. This can be seen by considering the extent of amino acid dissimilarity, which can be measured by Grantham’s 26 or Miyata’s biochemical distance 27. Grantham’s distance is based on the chemical composition of the side chain, the volume and the polarity of the amino acid residues, whereas Miyata’s distance is based on the volume and polarity only. It is well established that amino acid pairs with a small Grantham’s or Miyata’s distance replace each other more often than those with a large Grantham’s or Miyata’s distance 28. With this we address the question of whether amino acid substitutions at the 2-fold degenerate sites have smaller Grantham’s or Miyata’s distance. Among the 196 possible codon substitutions involving a single nucleotide change for the universal genetic code 28, 58 are transversions at the 2nd codon position (i.e., 0-fold degenerate site), with the average Grantham’s 26 distance between the two involved amino acids equal to 102.48. In contrast, 24 nonsynonymous transversions at the third codon position and 56 nonsynonymous transversions at the first codon position (i.e., 2-fold degenerate site) have an average Grantham’s distance equal to only 67.67 and 69.27, respectively. A similar trend is observed with Miyata’s distance 27. These Grantham dissimilarity values are close to those reported for interspecific variation in many different proteins 28–30.
The observations mentioned above suggest that an increase in the number of 2-fold degenerate positions in a sliding window increases the opportunity for nonsynonymous substitutions involving more similar amino acids that would be subjected to less intense purifying selection and would yield elevated fixation rates of nonsynonymous mutations. This possibility is supported by the fact that some of the polar amino acids (present in high frequency in the extracellular domains) have high substitution rates. For example, Serine is known to be the fastest-evolving amino acid in the PAM and JTT substitution matrices 31,32.
In the windows with the highest Kaw/Ks peak in Fig. 1 (corresponding to the second shaded extracellular domain), six of the eight serine residues are involved in nonsynonymous substitutions. The MRGX1 and MRGX4 sequences code for 194 strongly polar amino acids, with 50 (25.8%) involved in nonsynonymous substitutions, which is in contrast to the 450 non-polar or weakly-polar amino acids with only 86 (19.1%) involved in the nonsynonymous substitutions. In short, the high Kaw/Ks peaks associated with the extracellular domains in the MRG gene family may be attributed, at least partially, to the biochemical constraint that extracellular domains need to have a high frequency of polar amino acids, i.e., it may not be necessary to invoke positive selection.
3. SIMULATIONS CONFIRMING THE ASSOCIATION BETWEEN HIGHER Kaw/Ks RATIO AND INCREASED FREQUENCY OF POLAR AMINO ACIDS
While the above-mentioned properties of extracellular domains explain the elevation of Kaw, they do not explain why Kaw/Ks ratio is greater than 1 for some peaks. In order to investigate how this can happen, we examined the effects of the overabundance of codons coding for polar amino acids (hereafter referred to as PAA-coding codons) on the estimation of Ks and Ka values. We simulated the evolution of protein-coding genes with codon frequencies derived from MRGX1 and MRGX4 sequences by using the Evolver program in PAML (abacus.gene.ucl.ac.uk/software/paml.html).
We set transition/transversion ratio (κ) = 2, sequence length = 90, the branch length = 1.5 nucleotide substitutions per codon, and omega = 1 (i.e., no differential selection against synonymous and nonsynonymous substitutions). We performed two types of simulations, designated MorePolarAA and FewerPolarAA, that differ only in the frequencies of codons coding for the polar amino acids. The codon frequencies used in these two types of simulations differ as follows. First, the codon frequencies for the MRGX1 and MRGX4 sequences used in Choi and Lahn 8 were obtained. Second, designating PPAA.i.obs as the observed frequency of ith PAA-coding codon in the two sequences, the PPAA.i value equals (10/11)*PPAA.i.obs in the MorePolarAA simulation and (1/11)* PPAA.i.obs in the FewerPolarAA simulation. Thus, the PAA-coding codons are 10-fold more frequent in the MorePolarAA simulation than in the FewerPolarAA simulation. A 10fold difference such as this is not drastic because, for the window-specific codon frequencies, the extreme values for the frequencies of PAA-coding codons are 3.3% and 63.3%, respectively (a nearly 20-fold difference) in MRG genes 8. The codon frequencies for non PAA-coding frequencies are the same for the two types of simulations.
Each simulation was repeated 150 times, and the Ka, Ks and Ka/Ks ratios were calculated. The use of PBL method on these simulated data produced a mean Ka/Ks of 1.22 for the MorePolarAA simulation, and 0.79 for the FewPolarAA simulation (t = 5.1372, df = 298, P = 0.0000). Thus, those Kaw/Ks peaks may be caused at least partially by the presence of high frequencies of codons coding for polar amino acids in the extracellular domains. Therefore, we conclude that the heterogeneous distribution of polar amino acids along the protein sequences and the problem with estimating Ka/Ks for short sequences may generate spurious peaks and valleys in the Ka/Ks profiles not indicative of positive selection.
The association between the extracellular domains of the MRG gene family and the high Kaw/Ks peaks 8 may be interpreted in two ways. First, it is possible that these domains are under positive selection, but it is also possible that these domains carry high frequencies of polar amino acids because of the hydrophilic necessity of being extracellular. In the second case, the high Kaw/Ks peaks may simply arise because of higher intrinsic mutability of polar amino acids. Unless we can exclude the second possibility, it is prudent to refrain from interpreting the existence of high Kaw/Ks peaks as evidence in favor of positive selection.
4. DISCUSSION
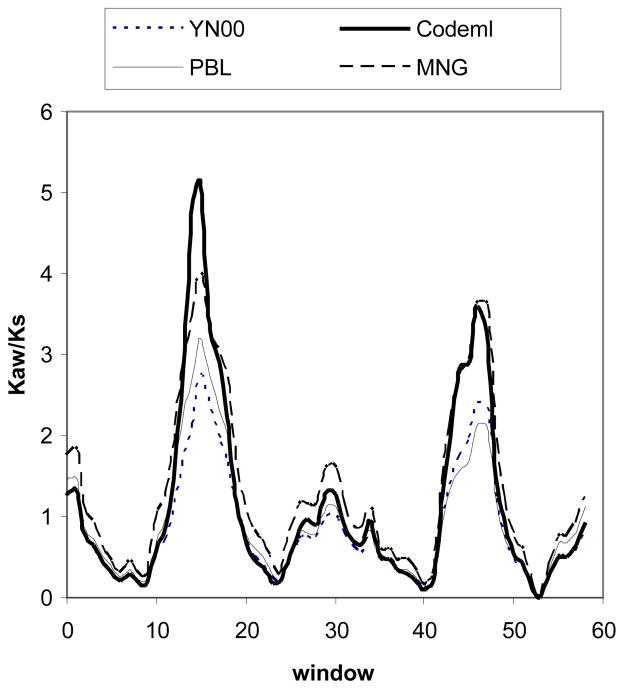
How robust are our conclusions drawn from the analysis of the MRG genes using the PBL method? In particular, do other methods suffer the same problem as the PBL method? Our answer is positive because the high Kaw/Ks peaks for the extracellular domains of the MRG genes are also recovered when other statistical methods are used (Fig. 3). Therefore, the potentially erroneous interpretation that extracellular domains (which contain an overabundance of polar amino acids) are subject to positive selection will be made using many different existing methods, as they all suffer from the similar biases caused by the heterogeneous distribution of polar amino acids along the protein sequences.
Fig. 3.
The window-specific Kaw/Ks values between the MRGX1 vs MRGX4 sequences, estimated by four different methods: YN0013, Codeml 34, PBL 11, 12 and modified Nei-Gojobori 4, pp. 57–59.
The association between the statistically-detected positive selection and polar amino acids is not restricted to the MRG gene family. This is evident from results of our examination of data from a recent study in which positive selection was inferred in protein-coding genes from HIV1 genomes 33. Amino acid sites statistically inferred to be under positive selection tend to be occupied by polar amino acids. In particular, amino acid sites inferred with a greater posterior probability have a greater chance of being occupied by polar amino acids. For example, polar amino acids account for 41.88% of all amino acids coded in the reference HIV1 sequence HXB233, but 49.55% of all amino acids at the positively selected sites detected with the posterior probability P ≥ 0.90, and 59.52% of all amino acids at the positively-selected sites detected with the posterior probability P ≥ 0.95 (data from Table 3 in ref. 33). The pattern is even stronger for the env gene, which harbours the overwhelming majority of the statistically-detected positively-selected sites. Polar amino acids account for 40.54% of all amino acids coded in this gene, but 52.46% of all amino acids at the positively selected sites detected with the posterior probability P ≥ 0.90, and 68.97% of all amino acids at the positively selected sites detected with the posterior probability P ≥ 0.95 (data from Table 3 in ref. 33). These results well exemplify the association between an increased Ka and a high frequency of polar amino acids.
The result from the HIV protein-coding genes is particularly noteworthy because the method used to detect positive selection is not the sliding-window approach, but a more recently developed site-specific approach 35. We may therefore conclude that positively selected sites detected by current statistical methods should be interpreted cautiously. In particular, we suggest that statistically detected “positively selected sites” by qualified with the word “putative”.
Acknowledgments
This work was supported in part by the Discovery Grant, Strategic Grant and RTI Grant from the Natural Science and Engineering Research Council of Canada to X. Xia and an NIH grant to S. Kumar. We thank Masatoshi Nei and S. Aris-Brosou for helpful comments on the previous versions. Two anonymous reviewers provide helpful comments and suggestions that reduced the ambiguity of the paper and improved the generality of our conclusions.
Contributor Information
Xuhua Xia, Email: xxia@uottawa.ca, Department of Biology, University of Ottawa 30 Marie Curie, P.O. Box 450, Station A, Ottawa, Ontario, Canada, KIN 6N.
Sudhir Kumar, Email: s.kumar@asu.edu, Center for Evolutionary Functional Genomics, The Biodesign Institute and The School of Life Sciences, Arizona State University, Tempe AZ, 85287-5301 USA.
References
- 1.Hughes AL, Nei M. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature. 1988;335:167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- 2.Hughes AL, Ota T, Nei M. Positive Darwinian selection promotes charge profile diversity in the antigen-binding cleft of class I major-histocompatibility-complex molecules. Mol Biol Evol. 1990;7:515–524. doi: 10.1093/oxfordjournals.molbev.a040626. [DOI] [PubMed] [Google Scholar]
- 3.Li W-H. Molecular evolution. Sunderland, Massachusetts; Sinauer: 1997. [Google Scholar]
- 4.Nei M, Kumar S. Molecular evolution and phylogenetics. New York: Oxford University Press; 2000. [Google Scholar]
- 5.Yang Z, Bielawski JP. Statistical methods for detecting molecular adaptation. Trends In Ecology And Evolution. 2000;15:496–503. doi: 10.1016/S0169-5347(00)01994-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thornton K, Long M. Excess of Amino Acid Substitutions Relative to Polymorphism between X-linked Duplications in Drosophila melanogaster. Mol Biol Evol. 2004;13:13. doi: 10.1093/molbev/msi015. [DOI] [PubMed] [Google Scholar]
- 7.Skibinski DO, Ward RD. Average allozyme heterozygosity in vertebrates correlates with Ka/Ks measured in the human-mouse lineage. Mol Biol Evol. 2004;21:1753–1759. doi: 10.1093/molbev/msh193. [DOI] [PubMed] [Google Scholar]
- 8.Choi SS, Lahn BT. Adaptive evolution of MRG, a neuron-specific gene family implicated in nociception. Genome Res. 2003;13:2252–2259. doi: 10.1101/gr.1431603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang HY, Tang H, Shen CK, Wu CI. Rapidly evolving genes in human. I. The glycophorins and their possible role in evading malaria parasites. Mol Biol Evol. 2003;20:1795–1804. doi: 10.1093/molbev/msg185. [DOI] [PubMed] [Google Scholar]
- 10.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 11.Li WH. Unbiased estimation of the rates of synonymous and nonsynonymous substitution. J Mol Evol. 1993;36:96–99. doi: 10.1007/BF02407308. [DOI] [PubMed] [Google Scholar]
- 12.Pamilo P, Bianchi NO. Evolution of the ZFX and ZFY genes: Rates and interdependence between the genes. Mol Biol Evol. 1993;10:271–281. doi: 10.1093/oxfordjournals.molbev.a040003. [DOI] [PubMed] [Google Scholar]
- 13.Yang Z, Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol. 2000;17:3243. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki Y, Gojobori T. A method for detecting positive selection at single amino acid sites. Mol Biol Evol. 1999;16:1315–1328. doi: 10.1093/oxfordjournals.molbev.a026042. [DOI] [PubMed] [Google Scholar]
- 15.Goldman N, Yang Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol. 1994;11:725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- 16.Muse SV, Gaut BS. A likelihood approach for comparing synonymous and nonsynonymous nucleotide substitution rates, with application to the chloroplast genome. Mol Biol Evol. 1994;11:715–724. doi: 10.1093/oxfordjournals.molbev.a040152. [DOI] [PubMed] [Google Scholar]
- 17.Aris-Brosou S. Determinants of adaptive evolution at the molecular level: the extended complexity hypothesis. Mol Biol Evol. 2005;22:200–209. doi: 10.1093/molbev/msi006. [DOI] [PubMed] [Google Scholar]
- 18.Bierne N, Eyre-Walker A. The genomic rate of adaptive amino acid substitution in Drosophila. Mol Biol Evol. 2004;21:1350–1360. doi: 10.1093/molbev/msh134. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki Y, Nei M. Reliabilities of parsimony-based and likelihood-based methods for detecting positive selection at single amino acid sites. Mol Biol Evol. 2001;18:2179–2185. doi: 10.1093/oxfordjournals.molbev.a003764. [DOI] [PubMed] [Google Scholar]
- 20.Suzuki Y, Nei M. Simulation study of the reliability and robustness of the statistical methods for detecting positive selection at single amino acid sites. Mol Biol Evol. 2002;19:1865–1869. doi: 10.1093/oxfordjournals.molbev.a004010. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki Y, Nei M. False-positive selection identified by ML-based methods: examples from the Sig1 gene of the diatom Thalassiosira weissflogii and the tax gene of a human T-cell lymphotropic virus. Mol Biol Evol. 2004;21:914–921. doi: 10.1093/molbev/msh098. [DOI] [PubMed] [Google Scholar]
- 22.Wong WS, Yang Z, Goldman N, Nielsen R. Accuracy and power of statistical methods for detecting adaptive evolution in protein coding sequences and for identifying positively selected sites. Genetics. 2004;168:1041–1051. doi: 10.1534/genetics.104.031153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hughes AL, Friedman R. Variation in the Pattern of Synonymous and Nonsynonymous Difference between Two Fungal Genomes. Mol Biol Evol. 2005 doi: 10.1093/molbev/msi120. [DOI] [PubMed] [Google Scholar]
- 24.Xia X, Xie Z. DAMBE: Software package for data analysis in molecular biology and evolution. J Hered. 2001;92:371–373. doi: 10.1093/jhered/92.4.371. [DOI] [PubMed] [Google Scholar]
- 25.Xia X. Data analysis in molecular biology and evolution. Boston: Kluwer Academic Publishers; 2001. [Google Scholar]
- 26.Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185:862–864. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- 27.Miyata T, Miyazawa S, Yasunaga T. Two types of amino acid substitutions in protein evolution. J Mol Evol. 1979;12:219–236. doi: 10.1007/BF01732340. [DOI] [PubMed] [Google Scholar]
- 28.Xia X, Li WH. What amino acid properties affect protein evolution? J Mol Evol. 1998;47:557–564. doi: 10.1007/pl00006412. [DOI] [PubMed] [Google Scholar]
- 29.Briscoe AD, Gaur C, Kumar S. The spectrum of human rhodopsin disease mutations through the lens of interspecific variation. Gene. 2004;332:107–118. doi: 10.1016/j.gene.2004.02.037. [DOI] [PubMed] [Google Scholar]
- 30.Miller MP, Kumar S. Understanding human disease mutations through the use of interspecific genetic variation. Hum Mol Genet. 2001;10:2319–2328. doi: 10.1093/hmg/10.21.2319. [DOI] [PubMed] [Google Scholar]
- 31.Dayhoff MO, Schwartz RM, Orcutt BC. A model of evolutionary change in proteins. In: Dayhoff MO, editor. Atlas of Protein Sequence and Structure. National Biomedical Research Foundation; Washington D.C: 1978. pp. 345–352. [Google Scholar]
- 32.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 33.Yang W, Bielawski JP, Yang Z. Widespread adaptive evolution in the human immunodeficiency virus type 1 genome. J Mol Evol. 2003;57:212–221. doi: 10.1007/s00239-003-2467-9. [DOI] [PubMed] [Google Scholar]
- 34.Yang Z. Phylogenetic analysis by maximum likelihood (PAML). Version 3.12. London: University College; 2002. [Google Scholar]
- 35.Yang Z, Swanson WJ, Vacquier VD. Maximum-likelihood analysis of molecular adaptation in abalone sperm lysin reveals variable selective pressures among lineages and sites. Mol Biol Evol. 2000;17:1446–1455. doi: 10.1093/oxfordjournals.molbev.a026245. [DOI] [PubMed] [Google Scholar]