Abstract
RNA interference (RNAi) technology provides a powerful molecular tool to reduce an expression of selected genes in eukaryotic cells. Short interfering RNAs (siRNAs) are the effector molecules that trigger RNAi. Here, we describe siRNAs that discriminate between the wild type and mutant (1174 C→G) alleles of human Presenilin1 gene (PSEN1). This mutation, resulting in L392V PSEN1 variant, contributes to early onset familial Alzheimer's disease. Using the dual fluorescence assay, flow cytometry and fluorescent microscopy we identified positions 8th–11th, within the central part of the antisense strand, as the most sensitive to mismatches. 2-Thiouridine chemical modification introduced at the 3′-end of the antisense strand improved the allele discrimination, but wobble base pairing adjacent to the mutation site abolished the siRNA activity. Our data indicate that siRNAs can be designed to discriminate between the wild type and mutant alleles of genes that differ by just a single nucleotide.
1. Introduction
Alzheimer's disease (AD) is the most common form of a progressive dementia in humans. The most frequently occurring variant is a “late-onset” (LOAD), sporadic form of the disease developing usually after age 65. The LOAD appears as a result of a complex interaction among environmental factors and individual predisposing genetic traits. Almost sixty years ago, Sjögren et al. demonstrated that some patients with AD had an autosomal dominant Mendelian pattern of disease inheritance [1]. Early onset familial cases of Alzheimer's disease (FAD) are rather rare and account for only a few percent of the total population of patients but in this form disease symptoms appear at an unusually early age, between 30–50 year of life. Mutations in the three genes coding Amyloid Precursor Protein (APP), Presenilin 1 (PSEN1), and Presenilin 2 (PSEN2) were identified as responsible for development of the dementia in almost 50% of patients with FAD. Identification of the amyloid-beta (Aβ) peptides isolated from brains of patients with AD and trisomy 21 (Down syndrome) suggested localization of the parent APP gene on chromosome 21 [2–5]. Linkage to disease mapped in FAD families led to the discovery of the first autosomal dominant missense mutations in APP segregating with disease risk [6]. This was followed by identifications of autosomal dominant FAD mutations in the PSEN1 [7] and PSEN2 [8, 9] genes, on chromosomes 14 and 1, respectively. The summary of FAD mutations is maintained at the Alzheimer's Disease and Frontotemporal Dementia Mutation Database [10]. Accordingly, there are currently described totally 227 sequence defects in the human FAD genes: 32 mutations in APP gene, 181 mutations in PSEN1 gene, and 14 mutations in PSEN2 gene, identified, respectively, in 89, 399, and 23 families.
Human Presenilin 1 (PSEN1, called alternatively as PS1, AD3, FAD, S182) is a 467-residues, ~50 kDa protein with 9-TMD (transmembrane domain) topology [11, 12]. PSEN1 together with nicastrin (Nct), anterior pharynx-defective-1 (APH-1) and presenilin enhancer-2 (PEN-2) constitutes a multisubunit membrane-bound protease complex of γ-secretase and is essential for its activity, stability and interaction between all γ-secretase components [13–15]. In the γ-secretase complex, Presenilin 1 is cleaved, in intramolecular autocatalytic event, into N- and C-terminal fragments (~30 kDa NTF and 20 kDa CTF), which form an active heterodimer. Several mutations in PSEN1 gene, detected in FAD patients, can inhibit processing of polypeptide chain. The most well-studied PSEN1 function in γ-secretase complex is catalysis of the intramembranous cleavage of a number of proteins, for example, Notch, APP, N-, and E-cadherins. Due to the fact that only a fraction of cellular protein actually forms the γ-secretase complex [16], it was postulated that Presenilin 1 may have functions beyond those of the γ-secretase complex. For example it is involved in neuronal Ca2+ signaling and homeostasis [17], and it also interacts with and stabilizes β-catenin [18, 19]. The main role of the γ-secretase is participation in the APP processing. The CTF 83 and CTF 99 fragments of APP, formed after its proteolysis by α- or β-secretases, respectively, are cleaved within their transmembrane regions to release the C-terminal domain (AICD) and generate Aβ peptides. Nearly 90% of the secreted amyloid peptides are Aβ40 or shorter forms, while the remaining 10% are Aβ42 or longer peptides. AD-linked mutations in PSENs genes influence the γ-secretase cleavage-site specificity, favouring cleavage at the position 42 relative to the position 40, thus increasing the Aβ42/Aβ40 ratio. Research from the 1990s indicates that mutations in the PSEN1 gene may be responsible for 30–60% of early onset Alzheimer's cases. The hydrophilic loop (amino acids 263–407) of PSEN1, in which many pathogenic mutations are localized, appears to be crucial for the protein function, since it includes the binding domains to different PSEN1 partners [20].
Dominantly inherited disorders constitute particularly attractive targets for allele-specific gene silencing by short interfering RNAs due to the fact that patients with these disorders carry both the wild type gene and mutant allele causing the disease. Specific silencing of a mutant allele that expresses the toxic form of the protein, without reducing the level of the wild type allele, might constitute a promising approach for the cure or prevention of such disorders. Most of the disease-associated genes differ by a single point mutation, making them targets of choice for allele-specific silencing. The list of trials on silencing mutant alleles associated with neurodegenerative disorders includes Huntington disease (HD) [21–27], Parkinson disease (PD) [28], amyotrophic lateral sclerosis (ALS) [29–33], spinocerebellar ataxia (SCA) Type 1 (SCA1) [34], and Type 3 (SCA3) causing Machado-Joseph disease [35, 36], frontotemporal dementia with Parkinsonism linked to chromosome 17 (FTDP-17) [35], slow channel congenital myasthenic syndrome (SCCMS) [37], and prion protein-induced disease [38]. Although several studies have used RNAi to investigate the effect of silencing of PSEN1 and its homolog PSEN2 on γ-secretase processing and cell metabolism [39, 40], to our best knowledge no studies have been reported on the allele-specific silencing of any PSEN1 mutant allele. Mutation L392V in PSEN1 carries the single nucleotide substitution (C→G) at position 1174 of coding sequence in exon 11, followed by amino acid substitution (Leu→Val) at the position 392 of polipeptide chain in TMD 7. Mutant form of PSEN1 is involved in the production of longer forms of the Aβ peptide resulting in its accumulation in the brain, and, in consequence, generation of early-onset Alzheimer's disease. Mean age onset of the disease in patients with L392V PSEN1 mutation was determined as 42.5 years and mean age of death was determined as 52.6 years [10]. According to data from Alzheimer's Disease and Frontotemporal Dementia Mutation Database, this mutation has been already found in 50 patients with FAD in European (Italy, France) and Asian (Japan) population [10, 41–46]. We selected the L392V mutant and wild type alleles of Presenilin 1 as a model system for the evaluation of allele-specificity of siRNA duplexes. A series of siRNA molecules fully complementary to the mutant gene were screened first by a dual fluorescence assay, then by FACS flow cytometry and their activity was confirmed by fluorescence microscopy and observation of reduced level of Aβ42 determined by ELISA. Our data suggest that siRNAs can be designed to discriminate between the wild type and the mutant alleles that differ by just a single nucleotide.
2. Materials and Methods
2.1. Synthesis and Purification of RNA Oligonucleotides
The oligoribonucleotides were synthesized according to the routine phosphoramidite approach [47], using LCA CPG glass support and commercially available nucleoside phosphoramidites (ChemGenes). Synthesis was performed on the Gene World DNA synthesizer under the conditions recommended by the manufacturer. Oligonucleotides were cleaved from the solid support as 5′-DMT-derivatives, then deprotected and purified according to the described procedure [48]. Support-bound oligonucleotides were treated with 33% ethanolic methylamine (Sigma-Aldrich) and DMSO 1 : 1 (v : v) mixture at 65°C for 15 min and then with triethylamine-trihydrofluoride (Sigma-Aldrich) at 65°C for 15 min. The reaction mixture was frozen at −20°C for 30 min, quenched with cold 1.5 M NH4HCO3 and poured into a conditioned SepPak cartridge (Waters). Shorter oligomers were eluted with 14% CH3CN in 50 mM CH3COONa and 50 mM NaCl. The remaining oligomer was treated with 2% aqueous TFA (trifluoroacetic acid) for 15 min at room temperature and washed with water, 1 M NaCl, and water. The product was eluted from the cartridge with 30% CH3CN. The structure and purity of oligomers were confirmed by MALDI-TOF mass spectroscopy (Table 1) and by 20% polyacrylamide/7 M urea gel electrophoresis (data not shown).
Table 1.
Sequences and MALDI-TOF MS data of oligoribonucleotides used for the preparation of siRNAs. The position of mismatch for pairing with the wild type gene is indicated in bold and underlined.
| siRNA | Strand | Sequence | MW | MALDI-TOF MS |
|---|---|---|---|---|
| (calculated) | m/z | |||
| P1 | S: | 5′-GUGGUUGGUAAAGCCUCAGTT-3′ | 6724.1 | 6768.3 |
| As: | 3′-TTCACCAACCAUUUCGGAGUC-5′ | 6587.2 | 6592.1 | |
| P2 | S: | 5′-UGUGGUUGGUAAAGCCUCATT-3′ | 6685.1 | 6681.9 |
| As: | 3′-TTACACCAACCAUUUCGGAGU-5′ | 6611.2 | 6608.2 | |
| P3 | S: | 5′-UUGUGGUUGGUAAAGCCUCTT-3′ | 6662.1 | 6660.3 |
| As: | 3′-TTAACACCAACCAUUUCGGAG-5′ | 6634.2 | 6632.1 | |
| P4 | S: | 5′-GUUGUGGUUGGUAAAGCCUTT-3′ | 6702.1 | 6697.1 |
| As: | 3′-TTCAACACCAACCAUUUCGGA-5′ | 6594.2 | 6592.6 | |
| P5 | S: | 5′-UGUUGUGGUUGGUAAAGCCTT-3′ | 6702.1 | 6699.6 |
| As: | 3′-TTACAACACCAACCAUUUCGG-5′ | 6594.2 | 6590.5 | |
| P6 | S: | 5′-GUGUUGUGGUUGGUAAAGCTT-3′ | 6742.0 | 6736.5 |
| As: | 3′-TTCACAACACCAACCAUUUCG-5′ | 6554.3 | 6552.9 | |
| P7 | S: | 5′-AGUGUU CUGGUUGGUAAAGTT-3′ | 6766.0 | 6764.0 |
| As: | 3′-TTUCACAACACCAACCAUUUC-5′ | 6515.3 | 6514.1 | |
| P8 | S: | 5′-CAGUGUUGUGGUUGGUAAATT-3′ | 6726.1 | 6723.5 |
| As: | 3′-TTGUCACAACACCAACCAUUU-5′ | 6555.3 | 6552.7 | |
| P9 | S: | 5′-ACAGUGUUGUGGUUGGUAATT-3′ | 6726.1 | 6725.1 |
| As: | 3′-TTUGUCACAACACCAACCAUU-5′ | 6555.3 | 6553.2 | |
| P10 | S: | 5′-UACAGUGUUGUGGUUGGUATT-3′ | 6703.0 | 6705.5 |
| As: | 3′-TTAUGUCACAACACCAACCAU-5′ | 6578.3 | 6578.6 | |
| P11 | S: | 5′-CUACAGUGUUGUGGUUGGUTT-3′ | 6679.0 | 6720.2 |
| As: | 3′-TTGAUGUCACAACACCAACCA-5′ | 6617.2 | 6615.5 | |
| P12 | S: | 5′-UCUACAGUGUUGUGGUUGGTT-3′ | 6679.0 | 6679.4 |
| As: | 3′-TTAGAUGUCACAACACCAACC-5′ | 6617.2 | 6617.1 | |
| P13 | S: | 5′-UUCUACAGUGUUGUGGUUGTT-3′ | 6640.1 | 6640.2 |
| As: | 3′-TTAAGAUGUCACAACACCAAC-5′ | 6641.3 | 6664.1 | |
| P14 | S: | 5′-UUUCUACAGUGUUGUGGUUTT-3′ | 6601.1 | 6601.1 |
| As: | 3′-TTAAAGAUGUCACAACACCAA-5′ | 6400.0 | 6701.1 | |
| P15 | S: | 5′-UUUUCUACAGUGUUGUGGUTT-3′ | 6601.1 | 6620.4 |
| As: | 3′-TTAAAAGAUGUCACAACACCA-5′ | 6665.3 | 6686.7 | |
| P16 | S: | 5′-AUUUUCUACAGUGUUGUGGTT-3′ | 6624.1 | 6669.3 |
| As: | 3′-TTUAAAAGAUGUCACAACACC-5′ | 6642.3 | 6640.1 | |
| P17 | S: | 5′-CAUUUUCUACAGUGUUGUGTT-3′ | 6584.1 | 6583.3 |
| As: | 3′-TTGUAAAAGAUGUCACAACAC-5′ | 6682.2 | 6679.5 | |
| P18 | S: | 5′-UCAUUUUCUACAGUGUUGUTT-3′ | 6545.2 | 6583.8 |
| As: | 3′-TTAGUAAAAGAUGUCACAACA-5′ | 6706.2 | 6705.0 | |
| P19 | S: | 5′-UUCAUUUUCUACAGUGUUGTT-3′ | 6545.2 | 6543.6 |
| As: | 3′-TTAAGUAAAAGAUGUCACAAC-5′ | 6706.2 | 6704.4 |
2.2. Assembly of siRNA
siRNA duplexes were assembled in phosphate saline buffer (PBS, without Ca2+ and Mg2+) by mixing equimolar amounts of complementary oligonucleotides, heating at 95°C for 2 min, and slow cooling down to room temperature (~2 hours). Formation of the resulting duplexes was confirmed by 4% agarose gel electrophoresis (data not shown).
2.3. Construction of Plasmids Coding EGFP-PSEN1 Fusion Gene
Human Presenilin 1 (PSEN1, GenBank NM_000021) coding sequence containing 1404 base pair (bp) was isolated from a total RNA, extracted from human cells (SH-SY5Y, human neuroblastoma) by reverse transcription (RT) and polymerase chain reaction (PCR) using One Step RT-PCR Kit (Qiagen). Reactions were performed in the following conditions—RT: 45°C 30 min, 95°C 15 min, and PCR: amplification 40 cycles (95°C 30 s; 55°C 30 s; 68°C 2 min) and termination at 72°C for 10 min. Sequences of primers were as follow: (Fow1) 5′-AAAAAAGAATTCAGATCTATGACAGAGTTACCTGCAC-3′ and (Rev1) 5′-AAAAAAGGATCCCTAGATATAAAATTGATGGAATGC-3′. Primers were designed to introduce EcoR I, Bgl II restriction sites to the 5′ end and BamH I restriction site to the 3′ end of PSEN1 gene, appropriate for cloning into pUC18 (Invitrogen) and pEGFP-C1 (BD Biosciences) plasmids. The cloning process was carried out using E. coli competent cells, strain TOP10 (Invitrogen). Site-directed mutagenesis for introducing a point mutation in the PSEN1 gene (C→G at the position 1174) was performed using QuikChange Site-Directed Mutagenesis Kit (Stratagene) in the conditions recommended by the manufacturer. Sequences of the mutagenic primers were as follow: 5′-CATTTTCTACAGTGTTGTGGTTGGTAAAGCCTCAGC-3′ and 5′-GCTGAGGCTTTACCAACCACAACACTGTAGAAAATG-3′. Correctness of inserts sequences (wild type, Wt-PSEN1, and mutated Mut-PSEN1) was confirmed by the sequencing reaction (IBB, Warsaw). Inserts coding Wt-PSEN1 or Mut-PSEN1 were introduced at the 3′ end of EGFP (Enhanced Green Fluorescent Protein) gene in the pEGFP-C1 expression plasmid. Plasmids pEGFP-PSEN1(1400) and pEGFP-Mut-PSEN1(1400) were used to study PSEN1 gene expression changes of in eukaryotic cells. Due to the low level of the EGFP-PSEN1 (EGFP with full length PSEN1) fusion protein expression in HeLa cells, a new fusion genes coding EGFP with shorter fragment of Wt-PSEN1 or Mut-PSEN1 were constructed. To create appropriate plasmids (pEGFP-Wt-PSEN1(400) and pEGFP-Mut-PSEN1(400)) the fragments of Wt-PSEN1 or Mut-PSEN1 coding region (1061–1404 nt position) were amplified by polymerase chain reaction (PCR) from the pEGFP-Wt-PSEN1(1400) or pEGFP-Mut-PSEN1(1400) plasmids, using the following primers: (Fow2) 5′-AAAAAAGTCGACTAGTAACACCTGAGTCACGAGCTGC-3′ and (Rev1) 5′-AAAAAAGGATCCCTAGATATAAAATTGATGGAATGC-3′. PCR products were digested by Sal I and BamH I restriction enzymes and 344 bp inserts were cloned into pEGFP-C1 plasmid. The accuracy of sequences of both inserts was confirmed by the sequencing reaction (IBB, Warsaw).
2.4. Cell Culture and Transfection Conditions
HeLa (human cervical carcinoma) cells were cultured in RPMI (GIBCO, BRL, Paisley) supplemented with 10% FBS (GIBCO, BRL, Paisley) and antibiotics (penicillin 100 units/mL, streptomycin 100 mg/mL, Polfa) at 37°C and 5% CO2. Twenty-four hours before the experiment, cells were plated in 96-well plate, (plates with black walls and transparent bottom, Perkin-Elmer) at the density of 15 × 103 cells per well. Directly before the transfection, the cell medium containing antibiotics was replaced with the new one, free of antibiotics. Transfection was performed using Lipofectamine 2000 transfection reagent (Invitrogen) at a ratio 1.5 : 1 (1.5 μL of Lipofectamine 2000 per 1 μg of nucleic acid) according to the manufacturer's protocol. For dual fluorescence assay (DFA), HeLa cells were cotransfected with DNA plasmids: reporter plasmid pDsRed-N1 (BD Biosciences) (15 ng/well) and pEGFP-Wt-PSEN1(400) (100 ng/well) or pEGFP-Mut-PSEN1(400) (100 ng/well) and siRNAs (1 nM) dissolved in OPTI-MEM medium (GIBCO, BRL, Paisley). After 5 hours of incubation, transfection mixture was replaced with fresh medium with antibiotics. After next 48 hours of incubation at 37°C in 5% atmosphere of CO2, the cells were washed three times with PBS buffer (without Ca2+ and Mg2+) and lysed with NP-40 buffer (150 mM NaCl, 1% IGEPAL, 50 mM Tris-HCl pH 7.0, 1 mM PMSF) overnight at 37°C. Prepared cell lysates were used for fluorescence determination.
Two cell lines of human fibroblasts (i) expressing wild type PSEN1 (CELMA) and (ii) L382V PSEN1 mutant (NOV4), were cultured in Dulbecco's MEM (Sigma-Aldrich Co., Saint Louis, MO), supplemented with 10% FBS and antibiotics (100 units/mL penicillin and 100 mg/mL streptomycin) in 24-well plate at 37°C and 5% CO2. Before transfection, the culture medium was replaced with fresh medium, free of antibiotics. Transfection was performed using Lipofectamine 2000 transfection reagent (Invitrogen) and appropriate siRNAs at the ratio 2 : 1. To test the effect of siRNA on Aβ level, the cells were transfected with siRNAs (150 or 300 nM). After transfection, cells were incubated for 5-6 hours and then medium with transfection mixture was replaced with the fresh, culturing medium with antibiotics and 5% FBS. After 72-hour incubation at 37°C and 5% CO2, cells were washed three times with PBS buffer (without Ca2+ and Mg2+), the fresh medium with 0.2% FBS was added and cells were incubated for the next 24 hours. Then the culturing medium was collected with 1 mM AEBSF (4-(2-Aminoethyl) benzenesulfonyl fluoride hydrochloride, serine protease inhibitor, Sigma) for determination of Aβ42 level by ELISA (Enzyme-linked immunosorbent assay).
2.5. Dual Fluorescence Assay
Fluorescence values of EGFP and RFP (red fluorescent protein) were determined using a Synergy HT reader (BIO-TEK). Quantification of data was done with KC4 software. Excitation and emission wavelengths for each protein were as follows—EGFP: λEx = 485/20 nm and λEm = 528/20 nm and RFP: λEx = 530/25 nm and λEm = 590/30 nm. The activity of siRNA duplexes was calculated as the ratio of EGFP to RFP fluorescence values according to the following equation. Activity of siRNA (%) = 100% − (sample EGFP-X/RFP : control EGFP-X/RFP) ×100% were (EGFP-X means the fluorescence value of EGFP-X fusion protein, X = Wt-PSEN1 or Mut-PSEN1). Every time an average value of fluorescence was mean of eight repeats calculated after eliminating extreme values. Each siRNA activity value given on the plots is the average of mean values from three independent experiments. The level of fluorescence (EGFP/RFP) in control cells (transfected with pDsRed-N1 and pEGFP-Wt PSEN1(400) or pEGFP-Mut-PSEN1(400) plasmids and control nonsilencing siRNA duplex) was taken as a reference (100%).
2.6. Determination of Aβ42 Level
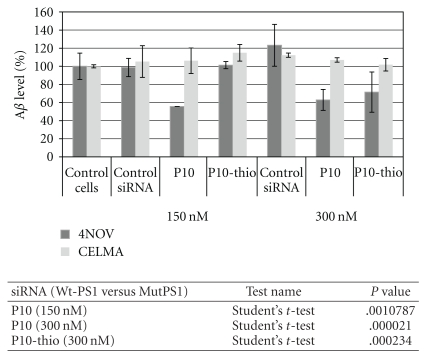
Chromogenic sandwich ELISA kit (Signet Laboratory, Denham, MA) was used according to the manufacturer's procedure. To analyze the level of Aβ42 in conditioned medium (after transfection of siRNAs) collected as described in p.2.4., the 100 μL samples of medium were placed in a 96-well ELISA plate, precoated with a capture antibody (specific to the N-terminus of human Aβ42 peptide) and incubated at 4°C for 16 hours. After washing, a reporting antibody, that binds to the C-terminus of Aβ42, was added. After incubation for the next 2 hours and extensive washing, a horseradish peroxidase conjugated secondary antibody was added. Color developed with substrate o-phenylenediamine was measured at λ492 using a Synergy HT reader, and received data were quantified using KC4 software. The siRNA activity was calculated as the ratio of Aβ42 in experimental and control sample (cells treated with transfection reagent only). Results showed at the graph (Figure 6) are the average of two independent experiments.
Figure 6.
The normalized values of the extracellular Aβ42 levels in medium of cells transfected with the siRNA duplexes. Human fibroblasts expressing endogenously the wild type PSEN1 gene (CELMA) and L392V mutant (NOV4) were cultured in the presence of P10, P10-thio, and control nonsilencing siRNA duplexes (150 and 300 nM). After 48-hour incubation, the culturing medium was collected with 1 mM AEBSF, and the level of Aβ42 was measured by sandwich ELISA test. Data are presented as percentage values to control cells treated with lipofectamine only. The results are mean values from two independent experiments. Statistical analysis of differences between two groups of data (Wt-PSEN1 versus Mut-PSEN1) were calculated by the use of Student's t-test (data with normal distribution). All differences were considered as statistically significant.
2.7. FACS Flow Cytometry
For flow cytometry assay, HeLa cells were seeded on the 6-well plate (3 × 105 cells per well) and cotransfected with appropriate plasmids: pDsRed-N1 (0.5 μg/well) and pEGFP-Wt-PSEN1(400) or pEGFP-Mut-PSEN1(400) (1.5 μg/well) and siRNAs: control nonsilencing siRNA and duplexes targeting the mutated form of PSEN1 transcript such as siRNA no 2, 7, 10, 11, 12 and 16 (denoted as P2, P7, P10, P11, P12, and P16, resp.) (5 nM) using Lipofectamine 2000, according to the manufacturer's protocol. After 48 hour incubation at 37°C and 5% CO2, cells were washed three times with ice cold PBS buffer (without Ca2+ and Mg2+) and collected for flow cytometry assay. Flow cytometry was performed on BD FACS Calibur Flow Cytometry System (Becton Dickinson) using Ar-ion laser (488 nm). Fluorescence dot plots and histograms were generated using Cell Quest software.
2.8. Statistical Analysis
The values of EGFP-PSEN1 fluorescence were normalized to the values of RFP fluorescence for each studied sample. All results are given as a percent of the fluorescence of sample toward fluorescence of control cells (transfected with Lipofectamine 2000 only or with control, nonsilencing siRNA) with standard deviation of mean. The Shapiro-Wilks' W test was used to analyze the normality of distribution of the resulted data. Statistical analysis of differences between two groups of data (Wt-PSEN1 versus Mut-PSEN1) were calculated by the use the Student's t-test (for data with normal distribution) or by the nonparametric Mann-Whitney's U-test. The differences with P < .05 were considered as statistically significant.
3. Results and Discussion
3.1. Construction of the Expression Plasmids Carrying the Wild Type and Mutant Alleles of PSEN1 Gene
The wild type and 1174 C→G, L392V mutant of PSEN1 gene were selected as a model system to assess the allele-specificity of the siRNAs and to select the duplexes, that discriminate between two target RNAs differing by the single nucleotide. Plasmids coding the wild type and mutant type genes were prepared as described in Section 2. At first, the full length (1404 bp) wild type and mutant PSEN1 genes were cloned in ORF (open reading frame) at the 3′-UTR of EGFP gene coded in the pEGFP-C1 plasmid. The expression products were designed to contain the EGFP protein fused by five amino acids linker (-Ser-Gly-Leu-Arg-Ser-) with intact Wt-PSEN1 or Mut-PSEN1 protein. The resulting plasmids pEGFP-Wt-PSEN1(1400) or pEGFP-Mut-PSEN1(1400) were cotransfected with the reporter plasmid pDsRed-N1 coding RFP, into HeLa cells. After 48 hour incubation, only very low levels of the fluorescence of EGFP fusion proteins were observed. Probably, attaching the 473-aa long polypeptide chain at the C-termini of EGFP resulted in abnormal folding of the fusion protein and, in consequence, in the low level of the fluorescence. Therefore these plasmids were excluded from further experiments and new, shorten fusion genes were prepared, containing a 344 bp 3′-terminal fragment of PSEN1 gene, in the wild-type and mutated versions, attached to the 3′-UTR of EGFP gene in the pEGFP-C1 expression plasmid. Additionally, the translation stop codons TAG and TAA were introduced into EGFP upstream of the coding sequence of fragment of the PSEN1 gene. The length of the obtained plasmids was confirmed by 0.5% agarose gel electrophoresis (Figure 1). We expected the high value of the fluorescence of EGFP in the case of intact fusion transcript EGFP-PSEN1 and low or no fluorescence if the fusion transcript is degraded by RNAi. Results of the dual fluorescence assay for all reporter plasmids (pEGFP-Wt-PSEN1(1400); pEGFP-Mut-PSEN1(1400); pEGFP-Wt-PSEN1(400) and pEGFP-Mut-PSEN1(400)) are shown in Figure 2. Plasmids pEGFP-Wt-PSEN1(400) and pEGFPMut-PSEN1(400) were used in further studies of the siRNA activity toward the Wt- and Mut-PSEN1 genes both expressed in HeLa cells.
Figure 1.
Electrophoretic analysis of the plasmids carrying the wild type (Wt) and mutant (Mut) alleles of the PSEN-1 gene and original, commercially available plasmids: (1) pEGFP-C1 (4731 bp), (2) pDsRed-N1 (4689 bp) (3) pEGFP-Wt-PSEN1(400) (5075 bp), (4) pEGFP-Mut-PSEN1(400) (5075 bp) (0.5% agarose gel).
Figure 2.
Comparison of the fluorescence values of EGFP-PSEN1 fusion proteins expressed from the EGFP-PSEN1 gene coding plasmids. Plasmids pEGFP-Wt PSEN1(1400) and pEGFP-Mut PSEN1(1400) contain full length (1404 bp) wild type (Wt) and mutein (Mut) (C→G 1174) PSEN1 gene introduced at the 3′-UTR of EGFP gene; plasmids pEGFP-Wt-PSEN1(400) and pEGFP-Mut-PSEN1(400) contain insert with shorter version (344 bp) of PSEN1 (Wt or Mut) introduced at the 3′-UTR of the EGFP gene.
3.2. Design, Synthesis, and Evaluation of the Activity of siRNAs Targeting Mutant Allele of PSEN1
3.2.1. siRNA Molecules
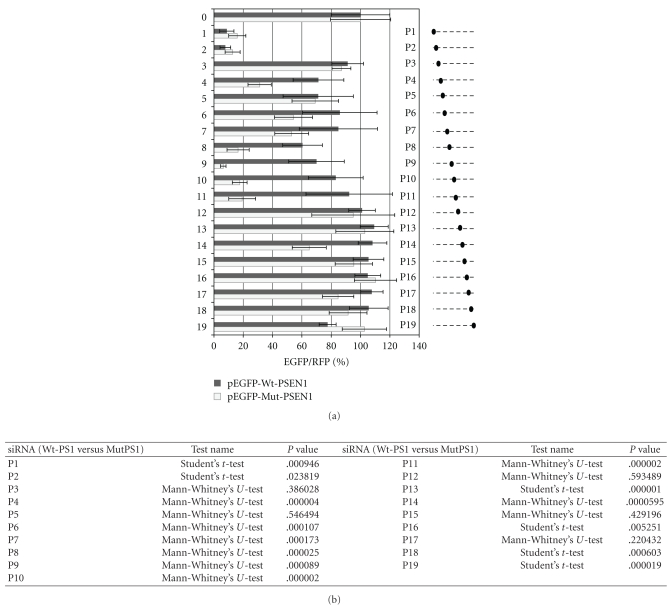
We synthesized a set of 19 siRNAs, designed across the L392V point mutation in the human PSEN1 gene. The L392V mutant contains a guanosine at the position 1174 of mRNA, whereas the wild-type mRNA bears a cytidine at that position. Each siRNA antisense strand was fully complementary to the mutant PSEN1 mRNA, but in relation to the wild type mRNA contained a C:C mismatch (Figure 3). The structure of all tested oligonucleotides was confirmed by mass spectrometry (MALDI-TOF MS, Table 1), and their purity was assessed on 20% denaturing PAGE (data not shown). All siRNA duplexes used in these experiments had the typical structure of 19 bp fully complementary duplex with 2-nt overhangs at each 3′-end with typically used two thymidine units (TT). This approach was intended for the proper interaction of RNA duplex with proteins of RNAi machinery [49]. Ago2 protein, the main catalytic core of the RISC (RNA-induced silencing complex) binds the guide strand, which leads the complex to the complementary mRNA strand. Ago2-mediated RNA cleavage requires perfect Watson-Crick base pairing between the guide strand of siRNA and the target mRNA, spanning both, the seed region (positions from 2nd to 8th nucleotide) and the cleavage site (phosphate group opposite to the internucleotide linkage located between 10th and 11th nucleotide, counted from the 5′-end of the guide strand [50].
Figure 3.
The sequences of the antisense strands of siRNAs used in studies. The guide strand of each siRNA is fully complementary to the mutant (C→G) PSEN1 transcript and it has one mismatch (C:C) in respect to the wild type gene. The region between 2nd to 8th nucleotide, counted from the 5′-end of the antisense strand of siRNA is known as the seed region.
3.2.2. Silencing Activity Measured in the Dual Fluorescence Assay
In order to evaluate the silencing activity of siRNAs, we used the already described dual fluorescence reporter system [51–55]. This assay is based on the measurement of the relative fluorescence intensity of EGFP and RFP proteins, expressed from exogenously delivered plasmids cotransfected together with siRNA. In our case, the HeLa cells were transfected with pEGFP-Wt-PSEN1(400) or pEGFP-Mut-PSEN1(400) and pDsRed-N1 as well as with P1–P19 siRNAs listed in Table 1 and Figure 3. The results of the fluorescence measurements are shown in Figure 4. The P8–P11 siRNAs, with the perfect complementarity to the mutant allele, but in the same time with one mismatch (cytidine:cytidine, C:C) to the wild type gene placed at the central positions of the duplex, efficiently silenced the mutant gene (up to 90%, P < .0001), while only slightly reducing the wild type gene expression (silencing ca. 10–30%, P < .0001). Interestingly, duplexes P1 and P2 showed a very strong silencing property towards both—the wild type and the mutated genes. This high silencing activity of the latter duplexes may be related to the high thermodynamic asymmetry of the duplex ends (discussed in Section 3.4) in comparison to other siRNA duplexes or to the better access of siRNA to the target region of the mRNA. The remaining P3, P5, P6, P7, and P12–P19 duplexes were almost inactive in relation to both the wild type as well as the mutant gene.
Figure 4.
(a) Comparison of the fluorescence level obtained from analysis of HeLa cells cotransfected with pEGFP-Wt-PSEN1(400) or pEGFP-Mut-PSEN1(400), pDsRRed-N1 and indicated siRNAs (0—control nonsilencing siRNA and siRNAs targeting the mutated form of PSEN1 transcript (denoted as P1–P19) (1 nM). Dotted lines at the right panel represent the antisense strands of the used siRNAs. Positions of the C:C mismatch between the guide strand of siRNA and the wild-type PSEN1 mRNA are indicated by the black dots. The level of the relative EGFP/RFP fluorescence of the cells transfected with control non-silencig siRNA was used as 100%. The results are mean values from three independent experiments. (b) Statistical analysis of differences between two groups of data (Wt-PSEN1 versus Mut-PSEN1) calculated by the use the Student's t-test (for data with normal distribution) or by the nonparametric Mann-Whitney's U-test. The differences with P < .05 were considered as statistically significant.
3.2.3. Silencing Activity Measured by Flow Cytometry
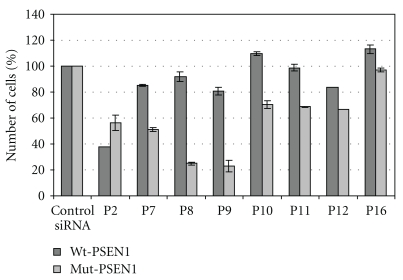
HeLa cells cotransfected with pEGFP-Wt-PSEN1(400) or pEGFP-Mut-PSEN1(400), pDsRed-N1 plasmids, and with selected duplexes P2, P7, P8, P9, P10, P11, P12, and P16 siRNAs were analyzed by FACS flow cytometry. The amount of cells containing the EGFP and RFP fluorescence in the tested samples (cells transfected with siRNAs) was determined and compared with the amount of cells in control samples (cells treated with transfection reagent only or treated with nonsilencing, control siRNA). The duplexes perfectly complementary to the L392V PSEN1 mRNA but with one mismatch (C:C) towards the wild type gene, lockated at the central position of the antisense strand (as P8 and P9), exhibited the highest allele-specificity and silencing efficiency exclusively towards the mutant gene (Figure 5). Thus, the data obtained in this analysis confirmed the above described results.
Figure 5.
Results of flow cytometry assay. HeLa cells were cotransfected by pEGFP-Wt-PSEN1(400) or pEGFP-Mut-PSEN1(400), pDsRed-N1, and indicated siRNAs, using Lipofectamine 2000 as a transfection reagent (details described in Section 2). HeLa cells containing both, the EGFP and RFP fluorescence, were counted in the BD FACS Calibur Flow Cytometry System (Becton Dickinson) using Ar-ion laser (488 nm). Fluorescence dot plots were generated using Cell Quest software.
3.2.4. Evaluation of the Level of Aβ42 by ELISA
The activity of siRNAs towards endogenous target genes was evaluated by the measurement of the level of extracellular Aβ42 released from the transfected cells. For this purpose human fibroblasts derived from the patients with FAD, carrying L392V mutantion in PSEN1 (NOV4), were used [56, 57]. Human fibroblasts, carrying the wild type of PSEN1 (CELMA), derived from healthy individuals were used as a control [56, 57]. The amount of the Aβ42 peptide was determined by the sandwich ELISA test. The normalized results obtained for this assay for both types of cells transfected with control, nonsilencing, and P10 siRNA are shown in Figure 6. The amount of the Aβ42 peptide released from NOV4 cells (NOV4Aβ42) transfected with P10 siRNA was reduced by ca. 40% (P = .000021) compared to fibroblasts NOV4 treated only with Lipofectamine 2000 or transfected with control nonsilencing siRNA. Application of siRNAs directed towards the mutated PSEN1 gene did not influence the level of Aβ42 released from control fibroblasts (CELMAAβ42), which was much lower than in NOV4 cells (both NOV4Aβ42 and CELMAAβ42 levels are showed at the graph as 100%). The results obtained may constitute a proof-of-concept that silencing of the mutant allele of PSEN1, exerted by fully complementary siRNA, led to the reduced expression of the mutant Presenilin1, which in consequence gave less of the toxic Aβ42 product.
3.3. Modulation of Allele-Specific Activity of siRNAs by Chemical Modifications
3.3.1. Modification of siRNA with 2-Thiouridine
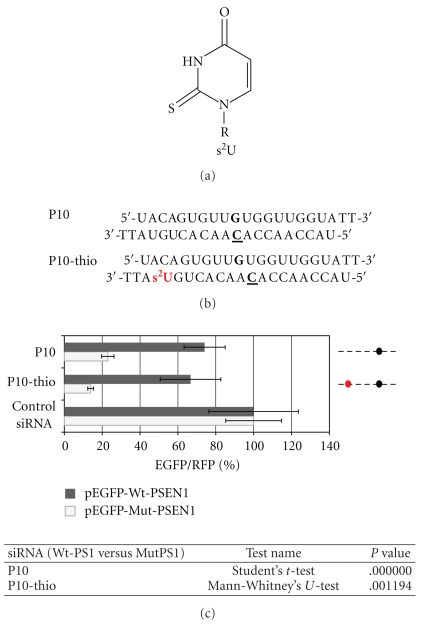
The allele-specific activity was evaluated for siRNA modified with naturally occurring 2-thiouridine (s2U) nucleoside. As it has been already demonstrated, s2U, due to preferred C3′-endo conformation of the ribose ring, improves the thermodynamic stability of s2U-containing double stranded RNA helices [52, 58, 59]. Additionally, s2U unit, when present in the anticodon region of transfer RNA (tRNA), enhances its specificity of Watson-Crick base pairing with A and restricts wobble base pairing with G [58, 59]. We choose P10 duplex, which previously has shown satisfactory discrimination between wild type and mutant alleles, and introduced the s2U unit at the position 18 of the antisense strand (P10-thio) (Figure 7(b)). The s2U modification at this position of the RNA duplex (the 3′-end of duplex) improves its thermodynamic asymmetry and facilitates incorporation of the antisense strand into the RISC complex [52]. Moreover, s2U introduction can impact positively on the uptake of siRNA into cells, due to the presence of sulfur moiety, which increases the hydrophobic properties of the molecule. Results of the dual fluorescence assay performed for HeLa cells transfected with P10-thio siRNA are presented in Figure 7(c). It is clearly shown that introduction of the s2U unit at the 3′-end of the antisense strand of P10 siRNA further improved allele discrimination (~10%) due to the increased thermodynamic asymmetry of the duplex.
Figure 7.
(a) 2-Thiouridine (s2U) structure, R = β-D-ribofuranoside residue. (b) Sequences of the P10 and P10-thio modified siRNAs. (c) Comparison of the relative (EGFP/RFP) fluorescence in HeLa cells transfected with reporter plasmids: pEGFP-Wt-PSEN1(400) or pEGFP-Mut-PSEN1(400), pDsRed-N1 and siRNA: P10 and P10-thio (1 nM). Dotted lines represent the antisense strands of the used siRNAs. Positions of the C-C mismatch between the guide strand of siRNA and the wild-type PSEN1 mRNA are indicated by the black dots. Red dot represents the site of the s2U modification at the antisense strand. The level of the relative EGFP/RFP fluorescence in cells transfected with control nonsilencing siRNA was used as 100%. Statistical analysis of differences between two groups of data (Wt-PSEN1 versus Mut-PSEN1) were calculated by the use the Student's t-test (for data with normal distribution) or by the nonparametric Mann-Whitney's U-test. The differences with P < .05 were considered as statistically significant.
3.3.2. Silencing Activity of Sulfur-Containing siRNA Evaluated by Fluorescence Microscopy
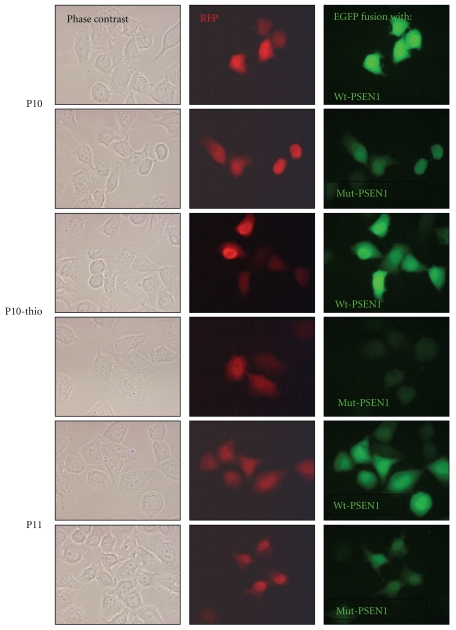
The activity of the selected siRNAs was also observed by the fluorescence microscopy. Figure 8 shows fluorescence images of the fixed samples obtained from HeLa cells transfected with P10, P10-thio, and P11 siRNA duplexes. Here again, the cells carrying the wild type gene (EGFP-Wt-PSEN1(400)) expressed green fluorescent protein at the high level and independently of the siRNA sequence. In contrast, slides containing cells expressing the mutant gene (EGFP-Mut-PSEN1(400)) have shown less fluorescently labeled cells, and the most pronounced result was obtained for cells transfected with the P10-thio siRNA duplex, confirming high discrimination ratio of siRNA against the wild type and mutant alleles of the PSEN1 gene.
Figure 8.
Silencing effects of siRNAs directed towards the Mut-PSEN1 (C→G1174). Fluorescence microscopy image of cells expressing the wild type and mutant fusion protein in the presence of siRNA: P10, P10-thio, and P11.
3.3.3. Modification of siRNA with Wobble Base Pair
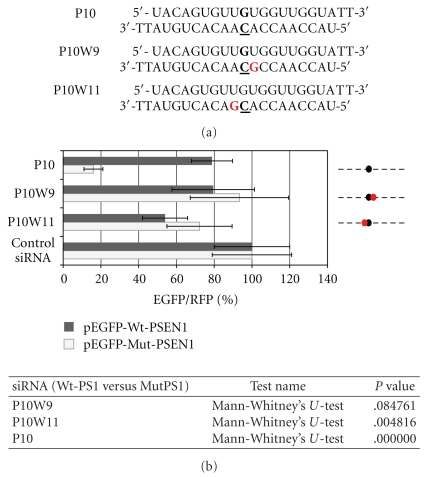
The region near the central positions at the antisense strand of siRNA (10th-11th) is interesting, primarily from the perspective of a nucleolytic reaction catalyzed by Ago2 [49, 50]. To investigate the influence of correct base pairing in this region on allele-specificity and the overall activity of RNA duplex, the wobble type modification was introduced at the two positions of the antisense strand, namely position 9 or 11 counting from the 5′end, adjacent to the mutation site (Figure 9(a)). According to our earlier assumption, introduction of such structural disorder into RNA duplex reduces the thermodynamic stability of the duplex and weakens the interaction between guide strand and the target mRNA. We supposed that the wobble modification could increase the allele-specificity of siRNAs targeted towards the mutant gene. Instead, we observed significant deterioration of the silencing activity of both modified siRNAs (P10W9 and P10W11) (Figure 9(b)). Most probably, introduced disorder in the base pairing like wobble G:U considerably disturbs the recognition of the double stranded RNA helix (containing mRNA and the antisense strand of siRNA) by Ago2, what results in inhibition of the silencing process.
Figure 9.
(a) Sequences of the P10W9 and P10W11 siRNAs modified in the central domain at the position 9 or 11; A is replaced by G (in red) to generate the wobble base pair with complementary mRNA strand. (b) Comparison of the florescence levels obtained after co-transfection of HeLa cells with pEGFP-Wt-PSEN1(400) or pEGFP-Mut-PSEN1(400), pDsRed-N1 and indicated siRNAs (P10, P10W9, or P10W11). Dotted lines represent the antisense strands of the used siRNAs. Positions of the C-C mismatch between the guide strand of siRNA and the wild-type PSEN1 mRNA are indicated by the black dots. Red dots represent the sites of the wobble base pairs between the guide strand of siRNA and the wild- or mutated-type of PSEN1 mRNA. The level of the relative EGFP/RFP fluorescence in cells transfected with control nonsilencing siRNA was used as 100%. The results are mean values from three independent experiments. Statistical analysis of differences between two groups of data (Wt-PSEN1 versus Mut-PSEN1) were calculated by the use by the nonparametric Mann-Whitney's U-test. The differences with P < .05 were considered as statistically significant.
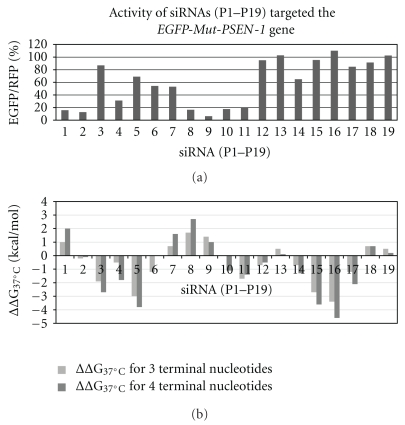
3.4. Thermodynamic Properties of siRNA Duplexes (P1–P19) versus Their Silencing Activity
Thermodynamic stability of RNA duplexes and secondary structure of RNA are often predicted by using free-energy parameters from a near-neighbor model [60]. To assess the relative thermodynamic stability of the duplex ends, we counted up the theoretical values of Gibbs free energy, ΔG37°C, of three and of four base pairs at the 5′-end and the 3′-end of siRNA (numbering according to the guide strand polarity) for each duplex used in our studies, by the use of the method recommended by Freier et al. [60]. The standard-state free energy of helix formation was counted as the sum of the following terms: (i) a free-energy change for helix initiation associated with forming the first base pair in the duplex, (ii) the sum of propagation free energies for forming each subsequent base pair, and (iii) free-energy increments for unpaired terminal nucleotides. Then, differences between the free Gibbs energy of the 5′-end and the 3′-end for each duplex were calculated independently for three and four consecutive base pairs according to the following equation: ΔΔG37°C = ΔG37°C (5′-end) − ΔG37°C (3′-end). The results of this analysis are shown in Table 2 and in Figure 10. The positive ΔΔG37°C values indicate the higher thermodynamic stability of the 3′-end of the duplex, thus the antisense strand should be selected as the guide strand and the siRNA duplex should be more active. The negative values of ΔΔG37°C mean the higher thermodynamic stability of the 5′-end of the duplex, thus the antisense strand is not preferred to be selected as a guide strand and, in consequence, the siRNA duplex would be expected to be less active towards the target gene. Comparison of the two graphs representing the silencing activity (Figure 10(a)) and the thermodynamic features of the duplex ends (Figure 10(b)) can explain observed differences in the activity of the used siRNAs, for example, duplexes P3–P6 and P14–P17 are characterized by the negative values of Gibbs free energy and are neglectably active. On the contrary, duplexes P1, P2, and P7–P10 showing positive or close to zero values of Gibbs free energy are more active as RNAi triggers. In some cases, silencing efficacy of siRNA duplexes depends on other factors as the secondary structure of target mRNA or availability of target sequence for RNAi proteins [61]. Also the other, not identified factors may govern the silencing process of given mRNA targets with short interfering RNAs.
Table 2.
Differences between the free Gibbs energy of 5′-end and 3′-end of the duplex (resp. to the polarity of the antisense strand), calculated independently for three and for four consecutive base pairs shown for each duplex (ΔΔG37°C = ΔG37°C(5′-end) − ΔG37°C (3′-end)). The values of ΔG37°C were calculated according to [60].
| siRNA | Sequence | ΔΔG37°C (kcal/mol) | ΔΔG37°C (kcal/mol) |
|---|---|---|---|
| (for 3 bp) | (for 4 bp) | ||
| P1 | 5′-GUGG—UCAGTT-3′ | 1 | 1.6 |
| 3′-TTCACC—AGUC-5′ | |||
| P2 | 5′-UGUG—CUCATT-3′ | −0.2 | −0.1 |
| 3′-TTACAC—GAGU-5′ | |||
| P3 | 5′-UUGU—CCUCTT-3′ | −1.9 | −2.7 |
| 3′-TTAACA—GGAG-5′ | |||
| P4 | 5′-GUUG—GCCUTT-3′ | −0.5 | −1.8 |
| 3′-TTCAAC—CGGA-5′ | |||
| P5 | 5′-UGUU—AGCCTT-3′ | −3 | −3.8 |
| 3′-TTACAA—UCGG-5′ | |||
| P6 | 5′-GUGU—AAGCTT-3′ | −1.2 | 0 |
| 3′-TTCACA—UUCG-5′ | |||
| P7 | 5′-AGUG—AAAGTT-3′ | 0.7 | 1.6 |
| 3′-TTUCAC—UUUC-5′ | |||
| P8 | 5′-CAGU—UAAATT-3′ | 1.7 | 2.7 |
| 3′-TTGUCA—AUUU-5′ | |||
| P9 | 5′-ACAG—GUAATT-3′ | 1.4 | 1 |
| 3′-TTUGUC—CAUU-5′ | |||
| P10 | 5′-UACA—GGUATT-3′ | 0 | −1.1 |
| 3′-TTAUGU—CCAU-5′ | |||
| P11 | 5′-CUAC—UGGUTT-3′ | −1.7 | −1.4 |
| 3′-TTGAUG—ACCA-5′ | |||
| P12 | 5′-UCUA—UUGGTT-3′ | −0.7 | −0.5 |
| 3′-TTAGAU—AACC-5′ | |||
| P13 | 5′-UUCU—GUUGTT-3′ | 0.5 | 0.1 |
| 3′-TTAAGA—CAAC-5′ | |||
| P14 | 5′-UUUC—GGUUTT-3′ | −0.7 | −1.3 |
| 3′-TTAAAG—CCAA-5′ | |||
| P15 | 5′-UUUU—UGGUTT-3′ | −2.7 | −3.6 |
| 3′-TTAAAA—ACCA-5′ | |||
| P16 | 5′-AUUU—GUGGTT-3′ | −3.4 | −4.6 |
| 3′-TTUAAA—CACC-5′ | |||
| P17 | 5′-CAUU—UGUGTT-3′ | −1.2 | −2.1 |
| 3′-TTGUAA—ACAC-5′ | |||
| P18 | 5′-UCAU—UUGUTT-3′ | 0.7 | 0.7 |
| 3′-TTAGUA—AACA-5′ | |||
| P19 | 5′-UUCA—GUUGTT-3′ | 0.5 | 0.2 |
| 3′-TTAAGU—CAAC-5′ |
Figure 10.
Comparison of the activity of siRNAs targeted Mut-PSEN1 gene (a) with their thermodynamic data (b). siRNAs were fully complementary to the Mut-PSEN1 transcript. Differences between the free Gibbs energy of 5′-end and 3′-end of the duplex (resp. to the polarity of the antisense strand) were calculated for three and for four consecutive base pairs: (ΔΔG37°C = ΔG37°C (5′-end) − ΔG37°C (3′-end)) (compare with Table 2).
4. Conclusions
Dominantly inherited disorders, caused by the presence of the toxic mutant allele besides the wild type gene, constitute attractive targets for allele-specific gene silencing. Gene therapy aimed at the elimination of the mutant allele causing the disease can be achieved by an RNA interference technology. Clinical success of such therapies depends mostly on the ability of RNAi to discriminate between the wild type and mutant alleles. In this paper, we demonstrate the allele-specific silencing activity of the short interfering RNAs directed towards mutant allele of PSEN1 gene. L392V PSEN1 mutant gene contains 1174 C→G mutation, which results in amino acid substitution, Leu to Val, at the position 392 of polypeptide chain of the protein. This mutation was found in patients with early onset FAD. At first, we synthesized a set of siRNAs, designed across the point mutation and determined their activity towards the mutant and wild type genes by a dual fluorescence assay, flow cytometry assay, and fluorescence microscopy images. The most active and allele-specific were the siRNA duplexes fully complementary to the target gene, which paired with the wild type allele with the C:C mismatch located in the central part of the mRNA-siRNA duplex (P8–P11). These duplexes silenced the mutant gene up to 90% and only slightly reduced the wild type gene, up to 10–30%. Introduction of the 2-thiouridine nucleoside into the 3′-end of the siRNA duplex (in respect to the polarity of the antisense strand) increased silencing activity of the duplex and discrimination ratio between the wild type and mutant allele (P10-thio). The wobble base pair modification introduced at the 9 or 11 positions of the antisense strand (P10W9 and P10W11), adjacent to the mutation site, reduced the thermodynamic stability of the duplex. We observed significant deterioration of the silencing activity of both modified siRNAs. Most probably, introduced disorder in the base pairing (G:U) considerably disturbed the recognition of the double stranded RNA helix (containing mRNA and the antisense strand of siRNA) by Ago2, what resulted in inhibition of the silencing process. The activity of the screened siRNAs was also evaluated by measurement of the level of extracellular Aβ42 released from the primary fibroblasts, carrying the (L392V) PSEN1 mutant (NOV4) in comparison to control fibroblasts (CELMA), carrying the wild type PSEN1, determined by ELISA. The normalized results obtained for NOV4 cells transfected with P10 and P10-thio siRNA showed reduced amount of Aβ42 by ca. 40% compared to the control fibroblasts treated only with transfection reagent. This result is a proof-of-concept that silencing of the mutant allele of PSEN1, exerted by fully complementary siRNA, led to reduced expression of the mutant Presenilin 1, which in consequence gave less of the toxic Aβ42 product.
Acknowledgments
This study was supported by the Ministry of Science and Higher Education, projects PBZ-MNiSW-07/I/2007 for years 2008–2010 and ERA-NET-NEURON/01/2009 for years 2009–2012 to B. Nawrot, MNiSW N N204 540039 for years 2010–2013 to M. Sierant and by Grant from the Cassa di Risparmio di Pistoia e Pescia 2009.0220 to B. Nacmias. Authors thank Dr Milena Sobczak for the chemical synthesis of the siRNA strands and Dr Lukasz Peczek for statistical analysis of the resulted data.
References
- 1.Sjögren T, Sjögren H, Lindgren AG. Morbus Alzheimer and morbus Pick; a genetic, clinical and pathoanatomical study. Acta Psychiatrica et Neurologica Scandinavica. Supplementum. 1952;82:1–152. [PubMed] [Google Scholar]
- 2.Goldgaber D, Lerman MI, McBride OW, Saffiotti U, Gajdusek DC. Characterization and chromosomal localization of a cDNA encodingbrain amyloid of Alzheimer's disease. Science. 1987;235(4791):877–880. doi: 10.1126/science.3810169. [DOI] [PubMed] [Google Scholar]
- 3.Tanzi RE, Gusella JF, Watkins PC, et al. Amyloid β protein gene: cDNA, mRNA distribution, and genetic linkage near the Alzheimer locus. Science. 1987;235(4791):880–884. doi: 10.1126/science.2949367. [DOI] [PubMed] [Google Scholar]
- 4.St George-Hyslop PH, Tanzi RE, Polinsky RJ, et al. The genetic defect causing familial Alzheimer's disease maps on chromosome 21. Science. 1987;235(4791):885–890. doi: 10.1126/science.2880399. [DOI] [PubMed] [Google Scholar]
- 5.Goate AM, Owen MJ, James LA, et al. Predisposing locus for Alzheimer’s disease on chromosome 21. The Lancet. 1989;1(8634):352–355. doi: 10.1016/s0140-6736(89)91725-x. [DOI] [PubMed] [Google Scholar]
- 6.Goate A, Chartier-Harlin MC, Mullan M, et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature. 1991;349(6311):704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- 7.Sherrington R, Rogaev EI, Liang Y, et al. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer's disease. Nature. 1995;375(6534):754–760. doi: 10.1038/375754a0. [DOI] [PubMed] [Google Scholar]
- 8.Levy-Lahad E, Wijsman EM, Nemens E, et al. A familial Alzheimer’s disease locus on chromosome I. Science. 1995;269(5226):970–973. doi: 10.1126/science.7638621. [DOI] [PubMed] [Google Scholar]
- 9.Rogaev EI, Sherrington R, Rogaeva EA, et al. Familial Alzheimer's disease in kindreds with missense mutations in a gene on chromosome 1 related to the Alzheimer's disease type 3 gene. Nature. 1995;376(6543):775–778. doi: 10.1038/376775a0. [DOI] [PubMed] [Google Scholar]
- 10.Alzheimer Disease and Frontotemporal Dementia Mutation Database. http://www.molgen.ua.ac.be/ADMutations/
- 11.Laudon H, Hansson EM, Melén K, et al. A nine-transmembrane domain topology for presenilin 1. Journal of Biological Chemistry. 2005;280(42):35352–35360. doi: 10.1074/jbc.M507217200. [DOI] [PubMed] [Google Scholar]
- 12.Spasic D, Tolia A, Dillen K, et al. Presenilin-1 maintains a nine-transmembrane topology throughout the secretory pathway. Journal of Biological Chemistry. 2006;281(36):26569–26577. doi: 10.1074/jbc.M600592200. [DOI] [PubMed] [Google Scholar]
- 13.Wolfe MS. γ-secretase in biology and medicine. Seminars in Cell and Developmental Biology. 2009;20(2):219–224. doi: 10.1016/j.semcdb.2008.12.011. [DOI] [PubMed] [Google Scholar]
- 14.Li H, Wolfe MS, Selkoe DJ. Toward structural elucidation of the γ-secretase complex. Structure. 2009;17(3):326–334. doi: 10.1016/j.str.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Strooper B, Annaert W. Novel research horizons for presenilins and γ-secretases in cell biology and disease. Annual Review of Cell and Developmental Biology. 2010;26:235–260. doi: 10.1146/annurev-cellbio-100109-104117. [DOI] [PubMed] [Google Scholar]
- 16.Lai MT, Chen E, Crouthamel MC, et al. Presenilin-1 and presenilin-2 exhibit distinct yet overlapping γ-secretase activities. Journal of Biological Chemistry. 2003;278(25):22475–22481. doi: 10.1074/jbc.M300974200. [DOI] [PubMed] [Google Scholar]
- 17.Smith IF, Green KN, LaFerla FM. Calcium dysregulation in Alzheimer’s disease: recent advances gained from genetically modified animals. Cell Calcium. 2005;38(3-4):427–437. doi: 10.1016/j.ceca.2005.06.021. [DOI] [PubMed] [Google Scholar]
- 18.Serban G, Kouchi Z, Baki L, et al. Cadherins mediate both the association between PS1 and β-catenin and the effects of PS1 on β-catenin stability. Journal of Biological Chemistry. 2005;280(43):36007–36012. doi: 10.1074/jbc.M507503200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Z, Hartmann H, Do VM, et al. Destabilization of β-catenin by mutations in presenilin-1 potentiates neuronal apoptosis. Nature. 1998;395(6703):698–702. doi: 10.1038/27208. [DOI] [PubMed] [Google Scholar]
- 20.Ikeuchi T, Kaneko H, Miyashita A, et al. Mutational analysis in early-onset familial dementia in the Japanese population: the role of PSEN1 and MAPT R406W mutations. Dementia and Geriatric Cognitive Disorders. 2008;26(1):43–49. doi: 10.1159/000141483. [DOI] [PubMed] [Google Scholar]
- 21.Pfister EL, Kennington L, Straubhaar J, et al. Five siRNAs targeting three SNPs may provide therapy for three-quarters of Huntington’s disease patients. Current Biology. 2009;19(9):774–778. doi: 10.1016/j.cub.2009.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Engelman J, Friedlander RM. Allele-specific silencing of mutant Huntington’s disease gene. Journal of Neurochemistry. 2009;108(1):82–90. doi: 10.1111/j.1471-4159.2008.05734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lombardi MS, Jaspers L, Spronkmans C, et al. A majority of Huntington’s disease patients may be treatable by individualized allele-specific RNA interference. Experimental Neurology. 2009;217(2):312–319. doi: 10.1016/j.expneurol.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 24.Rodriguez-Lebron E, Denovan-Wright EM, Nash K, Lewin AS, Mandel RJ. Intrastriatal rAAV-mediated delivery of anti-huntingtin shRNAs induces partial reversal of disease progression in R6/1 Huntington’s disease transgenic mice. Molecular Therapy. 2005;12(4):618–633. doi: 10.1016/j.ymthe.2005.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harper SQ, Staber PD, He X, et al. RNA interference improves motor and neuropathological abnormalities in a Huntington’s disease mouse model. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(16):5820–5825. doi: 10.1073/pnas.0501507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Warby SC, Montpetit A, Hayden AR, et al. CAG expansion in the Huntington disease gene is associated with a specific and targetable predisposing haplogroup. American Journal of Human Genetics. 2009;84(3):351–366. doi: 10.1016/j.ajhg.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Bilsen PHJ, Jaspers L, Lombardi MS, Odekerken JCE, Burright EN, Kaemmerer WF. Identification and allele-specific silencing of the mutant huntingtin allele in Huntington’s disease patient-derived fibroblasts. Human Gene Therapy. 2008;19(7):710–718. doi: 10.1089/hum.2007.116. [DOI] [PubMed] [Google Scholar]
- 28.Sapru MK, Yates JW, Hogan S, Jiang L, Halter J, Bohn MC. Silencing of human α-synuclein in vitro and in rat brain using lentiviral-mediated RNAi. Experimental Neurology. 2006;198(2):382–390. doi: 10.1016/j.expneurol.2005.12.024. [DOI] [PubMed] [Google Scholar]
- 29.Schwarz DS, Ding H, Kennington L, et al. Designing siRNA that distinguish between genes that differ by a single nucleotide. PLoS Genetics. 2006;2(9, article e140) doi: 10.1371/journal.pgen.0020140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maxwell MM, Pasinelli P, Kazantsev AG, Brown RH., Jr. RNA interference-mediated silencing of mutant superoxide dismutase rescues cyclosporin A-induced death in cultured neuroblastoma cells. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(9):3178–3183. doi: 10.1073/pnas.0308726100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raoul C, Abbas-Terki T, Bensadoun JC, et al. Lentiviral-mediated silencing of SOD1 through RNA interference retards disease onset and progression in a mouse model of ALS. Nature Medicine. 2005;11(4):423–428. doi: 10.1038/nm1207. [DOI] [PubMed] [Google Scholar]
- 32.Ralph GS, Radcliffe PA, Day DM, et al. Silencing mutant SOD1 using RNAi protects against neurodegeneration and extends survival in an ALS model. Nature Medicine. 2005;11(4):429–433. doi: 10.1038/nm1205. [DOI] [PubMed] [Google Scholar]
- 33.Yokota T, Miyagishi M, Hino T, et al. siRNA-based inhibition specific for mutant SOD1 with single nucleotide alternation in familial ALS, compared with ribozyme and DNA enzyme. Biochemical and Biophysical Research Communications. 2004;314(1):283–291. doi: 10.1016/j.bbrc.2003.12.098. [DOI] [PubMed] [Google Scholar]
- 34.Xia H, Mao Q, Eliason SL, et al. RNAi suppresses polyglutamine-induced neurodegeneration in a model of spinocerebellar ataxia. Nature Medicine. 2004;10(8):816–820. doi: 10.1038/nm1076. [DOI] [PubMed] [Google Scholar]
- 35.Miller VM, Xia H, Marrs GL, et al. Allele-specific silencing of dominant disease genes. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(12):7195–7200. doi: 10.1073/pnas.1231012100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alves S, Nascimento-Ferreira I, Auregan G, et al. Allele-specific RNA silencing of mutant ataxin-3 mediates neuroprotection in a rat model of Machado-Joseph disease. PLoS One. 2008;3(10) doi: 10.1371/journal.pone.0003341. Article ID e3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abdelgany A, Wood M, Beeson D. Allele-specific silencing of a pathogenic mutant acetylcholine receptor subunit by RNA interference. Human Molecular Genetics. 2003;12(20):2637–2644. doi: 10.1093/hmg/ddg280. [DOI] [PubMed] [Google Scholar]
- 38.Ohnishi Y, Tamura Y, Yoshida M, Tokunaga K, Hohjoh H. Enhancement of allele discrimination by introduction of nucleotide mismatches into siRNA in allele-specific gene silencing by RNAi. PLoS One. 2008;3(5) doi: 10.1371/journal.pone.0002248. Article ID e2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie Z, Romano DM, Kovacs DM, Tanzi RE. Effects of RNA interference-mediated silencing of γ-secretase complex components on cell sensitivity to caspase-3 activation. Journal of Biological Chemistry. 2004;279(33):34130–34137. doi: 10.1074/jbc.M401094200. [DOI] [PubMed] [Google Scholar]
- 40.Xie Z, Romano DM, Tanzi RE. Effects of RNAi-mediated silencing of PEN-2, APH-1a, and nicastrin on wild-type vs FAD mutant forms of presenilin. Journal of Molecular Neuroscience. 2005;25(1):67–77. doi: 10.1385/JMN:25:1:067. [DOI] [PubMed] [Google Scholar]
- 41.Campion D, Brice A, Hannequin D, et al. A large pedigree with early-onset Alzheimer’s disease: clinical, neuropathologic, and genetic characterization. Neurology. 1995;45(1):80–85. doi: 10.1212/wnl.45.1.80. [DOI] [PubMed] [Google Scholar]
- 42.Campion D, Flaman JM, Brice A, et al. Mutations of the presenilin I gene in families with early-onset Alzheimer’s disease. Human Molecular Genetics. 1995;4(12):2373–2377. doi: 10.1093/hmg/4.12.2373. [DOI] [PubMed] [Google Scholar]
- 43.Rogaev EI, Sherrington R, Rogaeva EA, et al. Familial Alzheimer’s disease in kindreds with missense mutations in a gene on chromosome 1 related to the Alzheimer’s disease type 3 gene. Nature. 1995;376(6543):775–778. doi: 10.1038/376775a0. [DOI] [PubMed] [Google Scholar]
- 44.Campion D, Dumanchin C, Hannequin D, et al. Early-onset autosomal dominant Alzheimer disease: prevalence, genetic heterogeneity, and mutation spectrum. American Journal of Human Genetics. 1999;65(3):664–670. doi: 10.1086/302553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raux G, Guyant-Maréchal L, Martin C, et al. Molecular diagnosis of autosomal dominant early onset Alzheimer’s disease: an update. Journal of Medical Genetics. 2005;42(10):793–795. doi: 10.1136/jmg.2005.033456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ikeuchi T, Kaneko H, Miyashita A, et al. Mutational analysis in early-onset familial dementia in the Japanese population: the role of PSEN1 and MAPT R406W mutations. Dementia and Geriatric Cognitive Disorders. 2008;26(1):43–49. doi: 10.1159/000141483. [DOI] [PubMed] [Google Scholar]
- 47.Caruthers MH. Gene synthesis machines: DNA chemistry and its uses. Science. 1985;230(4723):281–285. doi: 10.1126/science.3863253. [DOI] [PubMed] [Google Scholar]
- 48.Nawrot B, Sochacka E. Preparation of short interfering RNA containing the modified nucleosides 2-thiouridine, pseudouridine, or dihydrouridine. Current Protocols in Nucleic Acid Chemistry. 2009;(37):16.2.1–16.2.16. doi: 10.1002/0471142700.nc1602s37. [DOI] [PubMed] [Google Scholar]
- 49.Kim DH, Rossi JJ. Strategies for silencing human disease using RNA interference. Nature Reviews Genetics. 2007;8(3):173–184. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- 50.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431(7006):343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 51.Chiu YL, Rana TM. RNAi in human cells: basic structural and functional features of small interfering RNA. Molecular Cell. 2002;10(3):549–561. doi: 10.1016/s1097-2765(02)00652-4. [DOI] [PubMed] [Google Scholar]
- 52.Sipa K, Sochacka E, Kazmierczak-Baranska J, et al. Effect of base modifications on structure, thermodynamic stability, and gene silencing activity of short interfering RNA. RNA. 2007;13(8):1301–1316. doi: 10.1261/rna.538907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nawrot B, Sipa K. Chemical and structural diversity of siRNA molecules. Current Topics in Medicinal Chemistry. 2006;6(9):913–925. doi: 10.2174/156802606777303658. [DOI] [PubMed] [Google Scholar]
- 54.Sierant M, Kubiak K, Kazmierczak-Baranska J, Warashina M, Kuwabara T, Nawrot B. Evaluation of BACE1 silencing in cellular models. International Journal of Alzheimer’s Disease. 2009;2009:10 pages. doi: 10.4061/2009/257403. Article ID 257403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nawrot B, Sierant M, Paduszynska A. Emerging drugs and targets for Alzheimer’s disease. In: Martinez A, editor. RNA Interference of Genes Related to Alzheimer’s Disease. chapter 26. Vol. 2. RSC Publishing; 2009. pp. 230–266. [Google Scholar]
- 56.Cecchi C, Fiorillo C, Sorbi S, et al. Oxidative stress and reduced antioxidant defenses in peripheral cells from familial Alzheimer’s patients. Free Radical Biology and Medicine. 2002;33(10):1372–1379. doi: 10.1016/s0891-5849(02)01049-3. [DOI] [PubMed] [Google Scholar]
- 57.Sierant M, Kubiak K, Kazmierczak-Baranska J, et al. RNA interference in silencing of genes of Alzheimer's disease in cellular and rat brain models. Nucleic Acids Symposium Series. 2008;(52):41–42. doi: 10.1093/nass/nrn021. [DOI] [PubMed] [Google Scholar]
- 58.Agris PF, Sierzputowska-Gracz H, Smith W, Malkiewicz A, Sochacka E, Nawrot B. Thiolation of uridine carbon-2 restricts the motional dynamics of the transfer RNA wobble position nucleoside. Journal of the American Chemical Society. 1992;114(7):2652–2656. [Google Scholar]
- 59.Davis D. Biophysical and conformational properties of modified nucleotides in RNA (magnetic resonance studies) In: Grosjean H, Benne B, editors. Modification and Editing of RNA. Washington, DC, USA: ASM Press; 1998. pp. 85–102. [Google Scholar]
- 60.Freier SM, Kierzek R, Jaeger JA. Improved free-energy parameters for predictions of RNA duplex stability. Proceedings of the National Academy of Sciences of the United States of America. 1986;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Westerhout EM, Berkhout B. A systematic analysis of the effect of target RNA structure on RNA interference. Nucleic Acids Research. 2007;35(13):4322–4330. doi: 10.1093/nar/gkm437. [DOI] [PMC free article] [PubMed] [Google Scholar]