Summary
A new method for recording both fluorescence and cryo-EM images of small bacterial cells was developed and used to identify chemoreceptor arrays in cryotomograms of intact Caulobacter crescentus cells. We show that in wild-type cells preserved in a near-native state, the chemoreceptors are hexagonally packed with a lattice spacing of 12 nm, just a few tens of nanometers away from the flagellar motor that they control. The arrays were always found on the convex side of the cell, further demonstrating that Caulobacter cells maintain dorsal/ventral as well as anterior/posterior asymmetry. Placing the known crystal structure of a trimer of receptor dimers at each vertex of the lattice accounts well for the density and agrees with other constraints. Based on this model for the arrangement of receptors, there are between one and two thousand receptors per array.
Introduction
Bacterial chemotaxis depends on two large macromolecular complexes: a chemosensory system that senses the environment and a motor that propels the cell (Bourret and Stock, 2002). The core of the chemosensory system is an array of transmembrane ‘chemoreceptors’ which binds to attractants or repellents in the surrounding media and regulates the activity of CheA histidine kinases attached to the receptors’ cytoplasmic domains. CheA phosphorylates CheY, which in turn diffuses to and binds the C-ring of flagellar motors. The relative ratio of phosphorylated and unphosphorylated CheY acts as a toggle switch, causing flagellar motors to switch the sense of their rotation (i.e. from clockwise to counter-clockwise or vice-versa), which in turn causes cells to either continue swimming forward (‘run’) or change their direction randomly (‘tumble’) in multiflagellated Escherichia coli cells (Berg, 2003). In Caulobacter crescentus, the switch in sense of rotation causes short reversals in swimming direction (Alley et al., 1991). The chemoreceptors are also called methyl-accepting chemotaxis proteins (MCPs) because their activity is regulated by the methyl-transferase CheR and the methyl-esterase CheB.
Chemoreceptors are linked at their proximal (cytoplasmic) tips by CheA and an additional protein CheW in such a way that they form tightly coupled arrays at the cell poles (Maddock and Shapiro, 1993; Bray et al., 1998; Lybarger and Maddock, 2001; Boukhvalova et al., 2002; Gestwicki and Kiessling, 2002). While high-resolution structures of all these components are available, including the cytoplasmic portion of MCPs (Kim et al., 1999; Park et al., 2006), CheA (Bilwes et al., 1999), CheW(Griswold et al., 2002), CheR (Djordjevic and Stock, 1997; 1998) and CheB (Djordjevic et al., 1998), the arrangement, size and position of the arrays with respect to the flagellar motors inside cells remains unclear. Understanding this ‘mesoscale’ architectural is critical, however, as it is vital to the arrays’ cooperative signal amplification and regulation (Ames et al., 2002; Gestwicki and Kiessling, 2002; Sourjik, 2004; Sourjik and Berg, 2004; Li and Hazelbauer, 2005; Parkinson et al., 2005). Based on crystal structures, two models of the arrangement of the MCPs, CheA and CheW in the array have been suggested (reviewed in Weis, 2006). In the ‘trimer of dimers’ model, the unit cell of a hexagonal lattice is formed by three receptor dimers (Shimizu et al., 2000). In the ‘hedgerows of dimers’ model, receptor dimers are arranged in rows (Park et al., 2006). Cryo-EM studies of soluble complexes of a partial receptor with CheA and CheW show an alternative ‘barrel’-like pattern (Francis et al., 2004; Wolanin et al., 2006).
In order to elucidate the architecture of the chemotaxis system, we visualized it directly within intact C. crescentus cells by electron crytomography (ECT; Lucic et al., 2005; Jensen and Briegel, 2007). C. crescentus is a Gram-negative bacteria characterized by a dimorphic life cycle. Motile ‘swarmer’ cells mature by releasing their single polar flagellum and developing a tube-like stalk and terminal holdfast which attaches the cells to surfaces (Wagner and Brun, 2007). These non-motile, ‘stalked’ cells then elongate and undergo multiple rounds of division, each time producing and releasing a swarmer progeny. In C. crescentus, the chemosensory and propulsion systems are coupled in space and time: chemoreceptor and flagellar gene expression is co-ordinated (Jensen et al., 2002), chemoreceptors localize to the flag-ellated pole in swarmer cells (Alley et al., 1992), and stalked cells contain neither chemoreceptors nor flagella (Shaw et al., 1983; Gomes and Shapiro, 1984; Alley et al., 1992). By recording both fluorescent light microscopical and electron tomographical images of particular C. crescentus cells, here we identify the chemoreceptor array in C. crescentus tomograms and describe its size, location and structure within intact cells. The results confirm previous electron microscopical studies that identified a similar structure in E. coli as the chemoreceptor array (Zhang et al., 2007), and support a modified ‘trimers of dimers’ network model for its architecture.
Results
Tomograms of wild-type cells
About 50 three-dimensional reconstructions (tomograms) of intact, frozen-hydrated, log-phase C. crescentus cells at various stages of the cell cycle were produced which revealed the typical ultrastructure of C. crescentus: an S-layer, outer membrane, peptidoglycan and inner membrane enclosing a cytoplasm containing ribosome-like particles, cytoskeletal filaments and occasional vesicles (Briegel et al., 2006). Most cells exhibited a stalk, a minority exhibited a flagellum, and still others exhibited no appendages at all, presumably because the cell was in the transitional state between shedding the flagella and forming a stalk. In 12 cells, we observed a plate-like cytoplasmic assembly (Fig. 1) that closely resembled the assemblies identified previously as chemoreceptor arrays in E. coli (Zhang et al., 2007). The ‘base’-plate was positioned 31 nm below (proximal to) the inner membrane (measured ‘peak-to-peak’), bent to follow the curvature of the cell wall. A series of thinner densities perpendicular to the base plate and membrane spanned the distance between them. These thin connecting densities passed through a second, fainter plate parallel to the cell wall about 10 nm below the inner membrane. In the nine cells where pili and/or flagella were obvious, the plate-like assembly colocalized to the convex side of the same pole (see Movie S1). The distance between the plate-like assemblies and the flagellar motors ranged from 17 to 130 nm. The assembly was not found in any of the stalked cells. The area of the inner membrane covered by these assemblies ranged from ~11 000 to 22 800 nm2. The assembly’s location, timing within the cell cycle, and resemblance to the chemoreceptor arrays of E. coli all suggested that it was the chemoreceptor array.
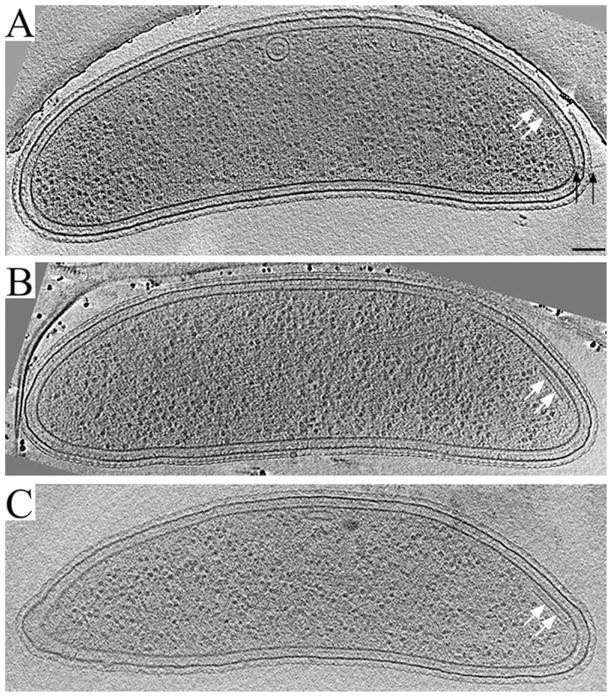
Fig. 1.
Reconstructions of wild-type C. crescentus swarmer cells. Representative ~12 nm thick slices through the centres of three C. crescentus swarmer cells are shown. At the convex side of the flagellated pole, a plate-like assembly (white arrows) is visible. The prominent base plate is connected to the inner membrane by thin, wall-like densities. A second, fainter layer of density parallel to the membrane can be discerned between the base plate and inner membrane. In most cases, pili (black arrows in A) and flagella (not shown) were also present at the same cell pole, but not in the same slice where the plate-like assembly was most visible (the case for the cells in B and C). Scale bar: 100 nm.
McpA mutants
To confirm whether this assembly was in fact the chemoreceptor array, we imaged a C. crescentus strain carrying a fusion of the red-fluorescent protein mCherry to the C-terminus of the mcpA gene on a xylose-inducible plasmid. Because expression of the mcpA–mCherry fusion was uncoupled from the cell cycle, after cells were incubated in the presence of xylose for a few hours every cell exhibited a bright fluorescent signal at one of the poles, regardless of its developmental stage. In the case of dividing cells, bright signals were seen at both poles. Such induced cells were plunge-frozen and tomograms of 13 poles were produced. Plate-like assemblies were now found in every cell, whether stalked or flagellated (Fig. 2), and always at the pole with the appendage, as had been seen previously in Caulobacter mutants where the carboxyl-terminal degradation domain of McpA was eliminated (Alley et al., 1993). The assemblies could also now be found on the concave as well as convex side of the cell or at the tip of the pole. Thus the pattern of mislocalization of the fluorescently tagged McpA matched that of the plate-like assemblies.
Fig. 2.
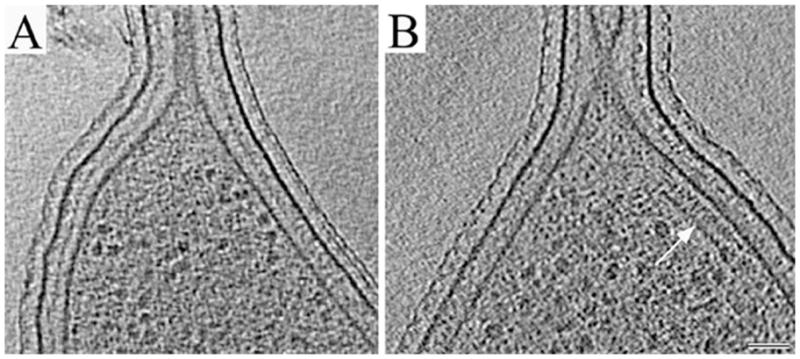
Mislocalization of the plate-like assembly to the stalked poles of McpA mutant cells.
A. 14 nm thick slice through a typical reconstruction of a wild-type stalked pole showing that in wild-type cells, the plate-like assembly was not seen in stalked cells.
B. 12 nm thick slice through a typical stalked pole of the McpA mutant after xylose induction, showing a plate-like assembly (white arrow). Scale bar: 50 nm.
To further investigate these correlated mislocalizations, we developed a new method to obtain both a fluorescent image and a cryotomogram of the same cell. After being induced with xylose, mutant cells were concentrated, lightly fixed, immobilized with poly-L-lysine on gold TEM ‘finder’ grids, sandwiched between a glass slide and coverslip together with excess media, and imaged in a high-powered oil-immersion fluorescence light microscope (fLM) (Fig. 3A). The grids were then gently but quickly removed from the glass slide, plunge-frozen and transferred to the electron cryomicroscope. The same region observed by fLM was located in the TEM (Fig. 3B) and tilt-series of specific cells were recorded. Altogether, seven data sets from two different grids were matched containing a total of nine cell poles (two poles are shown in Fig. 3C and D). The presence and absence of the putative chemoreceptor arrays in the tomograms correlated perfectly with the presence or absence of fluorescent foci (seven poles exhibited both a fluorescent focus and a plate-like assembly in the same location and two poles showed neither). We conclude that the plate-like assemblies are in fact the chemoreceptor arrays of C. crescentus.
Fig. 3.
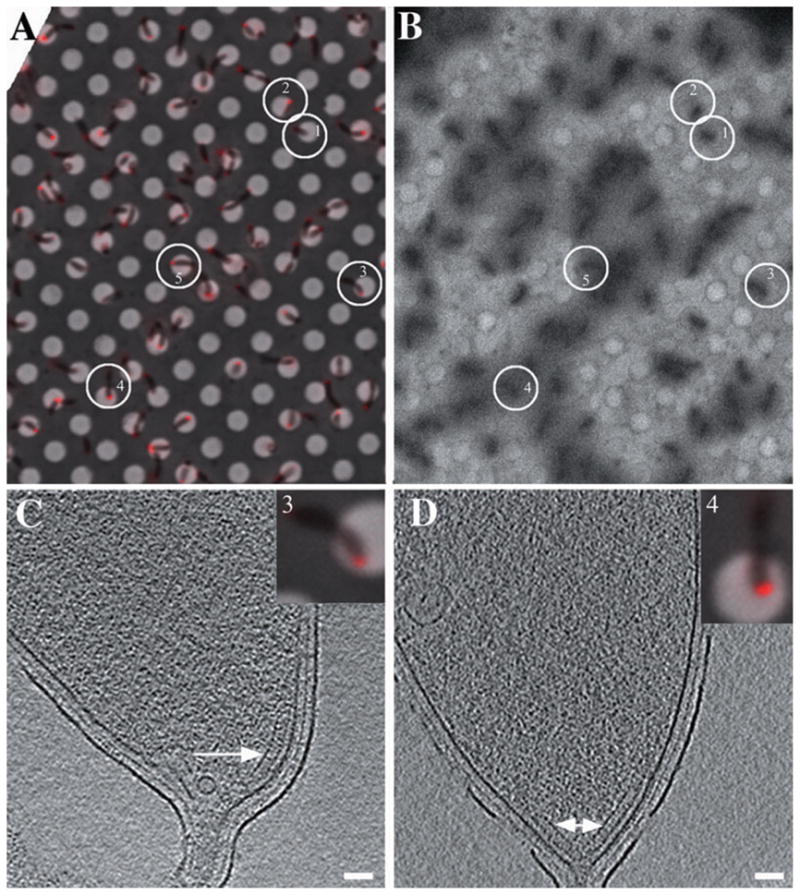
Correlated fLM and ECT of McpA mutant cells.
A. fLM image of slightly fixed, xylose-induced McpA–mCherry mutant cells immobilized on an EM finder grid with regularly spaced, 2 μm diameter holes in a thin carbon film. Most cells exhibit a single fluorescent focus at one pole, but elongated pre-divisional cells exhibit foci at both poles.
B. Low magnification EM image of the same region after the grid had been removed from the light microscope, plunge-frozen and inserted into the EM. Full tilt-series were recorded of the five circled cells, which maintained their positions well during the transfer.
C and D. 15 nm thick slices through the cell poles of cell #3 (C) and #4 (D). The presence, absence and positions of the plate-like assemblies (white arrow) correlated perfectly with fluorescence signals from McpA. Scale bars: 50 nm.
C. crescentus chemoreceptor arrays are hexagonally packed
In ‘side’ views such as that seen in Fig. 1, the chemoreceptor arrays were composed of the base plate and thin, regularly spaced, wall-like densities connecting the base with the inner membrane (an enlarged view of another example is shown in Fig. 4A). ‘Top’ views of the region right above the base layer (toward the inner membrane) showed a honeycomb-like arrangement of the wall-like densities (Fig. 4B). The power spectrum of this region revealed a hexagonal lattice with a spacing of 12 nm (Fig. 4B, inset). While applying a median filter to the tomogram reduced background noise (Fig. 4C and D), to further improve the signal-to-noise ratio we extracted and averaged seven subvolumes with obvious hexagonal ordering from one (unfiltered) tomogram and applied sixfold rotational symmetry to the resulting average. The side view of this averaged structure is shown in Fig. 5A, as well as several ‘top-views’ at indicated positions between the base plate and inner membrane (Fig. 5B–F). From the base plate (pink in Fig. 5G), a honeycomb-like hexagonal net was seen rising towards the membrane. Closer to the inner membrane the hexagonal order quickly deteriorated until it was no longer discernable. By measuring the surface area on the inner membrane covered by the arrays on several representative cells, the arrays were estimated to contain between 90 and 180 hexagons.
Fig. 4.
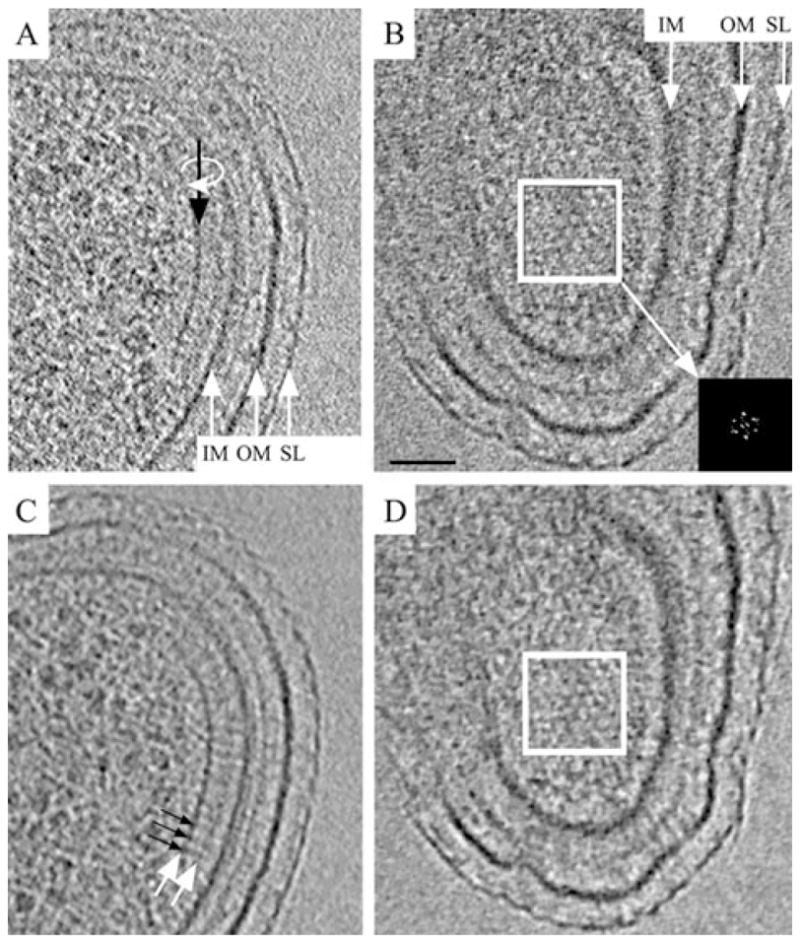
Structure of a wild-type chemoreceptor array.
A and B. 8 nm thick tomographic slices perpendicular (A) and parallel (B) to the membranes (the slice shown in B is related to that shown in A by a 90° rotation around the line indicated by the black arrow).
C and D. The same slices after median filtering. The base plate (left white arrow in C) is positioned 31 nm proximal to the inner membrane (IM, inner membrane; OM, outer membrane; SL, surface layer). Thin pillars of density (black arrows in C) connect the base plate with the inner membrane, and a second, fainter, plate-like layer (right white arrow in C) is visible between the base plate and inner membrane. The hexagonal arrangement of the chemoreceptor array right above the base plate is apparent in (B) and (D) (white box). The power spectrum (inset, enlarged to enhance clarity) reveals a lattice with a centre-to-centre spacing of 12 nm. Scale bar: 50 nm.
Fig. 5.
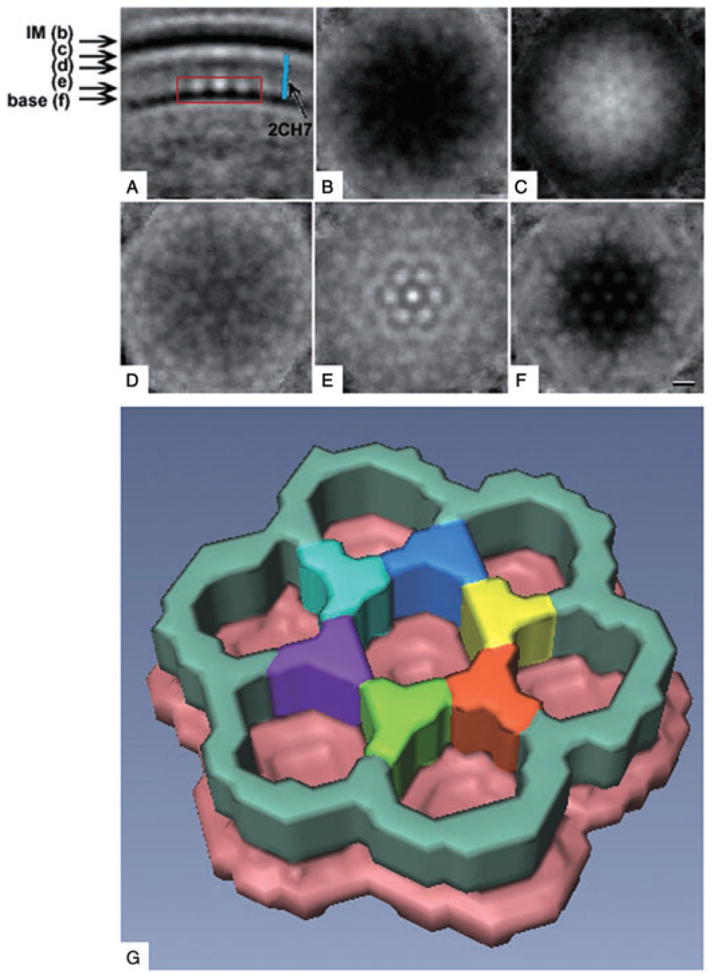
Average unit cell. The volumes surrounding seven well-ordered hexagonal centres were extracted from the tomogram shown in Fig. 4, aligned, averaged and sixfold rotationally symmetrized.
A. Slice through the resulting average perpendicular to the membrane (IM, inner membrane; base, base plate; blue rod, low resolution model of the cytoplasmic domain of receptor MCP1143 (pdb #2CH7), showing that its length and width match those of the thin wall-like connecting densities). [Note that the low-resolution model is of the receptor type 44H (41) and therefore slightly different than the receptors found in C. crescentus].
B–F. Slices parallel to the membrane, taken at the positions marked on the left in (A). The clear hexagonal order present in the base plate diminishes rapidly towards the inner membrane.
G. Manually segmented three-dimensional surface representation of the volume indicated by the red box in (A), with the six vertices of the central hexagon highlighted by different colours. Scale bar: 10 nm.
Discussion
Our understanding of many biological systems has progressed from knowledge of their basic functions to identification of the macromolecules involved to atomic models of each of their structures, but before accurate mechanistic computational models can be constructed, the details of how the components are arranged within living cells must also be elucidated. Work on bacterial chemotaxis has followed this pattern, resulting in a need for architectural details to constrain and refine already sophisticated computational models (Bray et al., 1998; Lipkow et al., 2005; Bray et al., 2007). Here we used a combination of fLM and ECT of wild-type and mutant cells to identify and describe the C. crescentus chemoreceptor array in intact cells.
A major technical challenge in ECT has been to unambiguously identify structures seen in tomograms. There are as yet no ‘GFP-like’, genetically encodable, electron-dense labels that would work in this context (ECT of intact bacterial cells). Instead, structures like cytoskeletal filaments have been identified by correlating their size, position or number with changes in their expression levels or stability (Li et al., 2007). Likewise, chemoreceptor arrays had previously been identified in images and tomograms of frozen-hydrated E. coli cells based on their polar localization, increase in number in cells overexpressing chemoreceptors, or dependence on CheA and CheW (Weis et al., 2003; Lefman et al., 2004; Zhang et al., 2004; 2007). Immunolabelling has also been used to help identify the chemoreceptor array (Weis et al., 2003; Lefman et al., 2004; Zhang et al., 2007). Our work here confirmed that identification through correlated fLM and ECT, a powerful combination of complementary microscopies.
To facilitate correlated light and electron cryomicroscopy, two groups recently developed cryostages that allow frozen-hydrated samples to be observed in light microscopes (Sartori et al., 2007; Schwartz et al., 2007). These stages allow near-native preservation of the sample while both fLM and cryo-EM images are recorded, without any changes in the sample except the unavoidable radiation damage. Unfortunately, because the cold temperatures prohibit the use of immersion oil and all but long-working-distance objective lenses, the resolution of these cryo-light microscopes (including the one we tested) is inadequate to discern localization patterns within small bacterial cells.
In order to obtain both high-resolution fLM and ECT images of the small C. crescentus cells of interest here, we immobilized them on EM finder grids, investigated their fluorescence patterns at room temperature and then plunge-froze them in preparation for electron cryomicroscopy. Minor changes in the sample did occur during the transfer from the light to the electron microscope, including positional shifts of some bacterial cells or even their complete detachment from the grid, and the structures of individual cells were slightly perturbed by the light fixation. We were nevertheless able to unambiguously identify the plate-like assemblies as chemoreceptor arrays by showing that they consistently and precisely colocalized with fluorescently tagged chemoreceptor subunits.
Once the arrays were identified, their architectural features were deduced from cryotomograms of unfixed, wild-type cells. Chemoreceptor arrays were seen to comprise a plate-like base parallel to and ~31 nm below the inner membrane; a series of wall-like densities that extended from the base to the inner membrane; and a second and weaker plate-like density ~10 nm below the inner membrane. The wall-like densities formed a hexagonal net with 12 nm spacing whose order deteriorated rapidly with distance from the base. Given that (i) CheA and CheW are known to bind each other and the cytoplasmic tips of MCPs (Falke and Hazelbauer, 2001; Boukhvalova et al., 2002); (ii) that at the other end of the receptors there is a hydrophobic region that must traverse the membrane to put the periplasmic domain in contact with extracellular stimuli (Maddock and Shapiro, 1993; Sourjik and Berg, 2004); and (iii) that the distance between the base layer and the inner membrane (31 nm) is consistent with the estimated length of MCPs (Kim et al., 1999; Park et al., 2006), we interpret the thin walls as the receptors and the ‘base plate’ their complex with CheA and CheW.
While the resolution in our tomograms was not high enough to reveal the exact packing of MCPs, trimers of receptor dimers, as seen in the crystal structure (Kim et al., 1999), fit compellingly well into the vertices of the hexagonal lattice to form linked rings of receptors (Fig. 6). Cross-linking assays had previously pointed to a ‘trimer-of-dimers’ organization of the receptors in vivo (Studdert and Parkinson, 2004), and a model derived from these studies predicted that the trimers would form the vertices of a hexagonal lattice (Studdert and Parkinson, 2005). Our model also agreed with genetic studies that found an important signalling role for trimers (Ames et al., 2002). Finally, recent studies using small particles of lipid bilayer containing one or a few chemoreceptors, called nanodiscs, further implicated ‘trimers-of-dimers’ as the core structure of kinase activation and control (Boldog et al., 2006). In contrast, other models based on ‘hedgerows’ (Park et al., 2006) or larger numbers of receptors per hexagon were incompatible with our reconstructions. Our data were also unreconcilable with models where trimers of receptor dimers stood alone in the centres of CheA/CheW hexagons (Shimizu et al., 2000), as the receptors formed apparently continuous walls which emerged from the thickest regions of the CheA/CheW base plate.
Fig. 6.
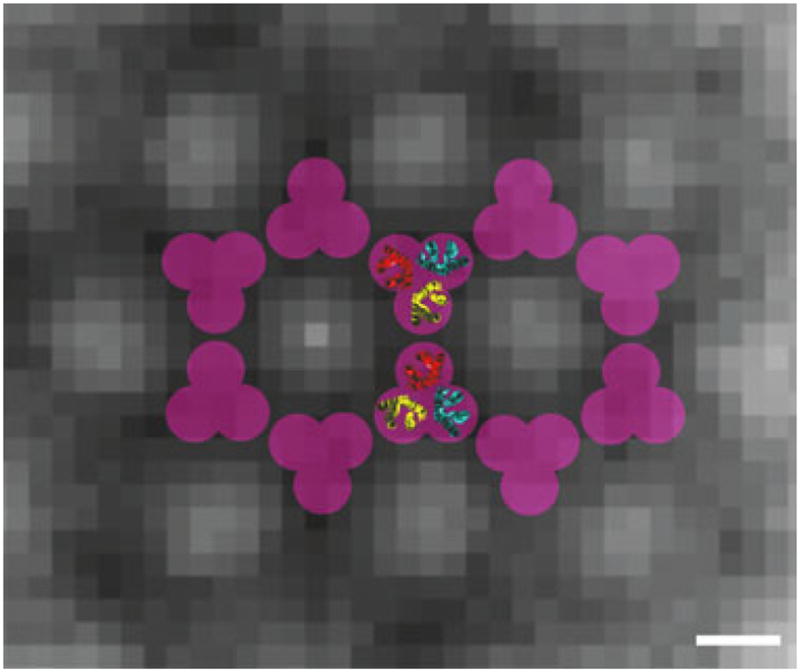
Model for the arrangement of the receptors in the chemotaxis array. The background is a slice of the symmetrized average unit cell parallel to the membrane and close to the CheA/CheW base plate. While the resolution of the average is not sufficient to dock the receptor structures precisely, the overall dimensions and symmetry (purple) of the bottom half of the trimer-of-receptor-dimers crystal structure (1QU7 pdb ID) (receptor dimers coloured in red, yellow and blue) fit well into the observed hexagonal lattice. Scale bar: 5 nm.
Seven major MCP classes have been identified by large-scale comparative genomic analyses based on the length and sequence conservation of the domains. C. crescentus contains 18 different MCPs. Sixteen of these are of type 36H, as are all five MCPs found in E. coli (Alexander and Zhulin, 2007). Of these 16 type 36H MCPs in C. crescentus, six contain the C-terminal pentapeptide tether to which CheR and CheB bind in E. coli (Alexander and Zhulin, 2007). The other two MCPs in C. crescentus are of type 38H and an un-aligned, ‘cytoplasmic’ type. Interestingly, despite these sequence similarities, the lattice spacing we observed in wild-type C. crescentus cells (12 nm) differed significantly from the lattice spacing reported in EM studies of E. coli overexpressing the Tsr receptor (7.5 nm) (Weis et al., 2003; Lefman et al., 2004). It is unclear whether the receptor packing in the E. coli study was altered owing to the elevated Tsr receptor concentrations, if the arrays were in different states, or if the architectures of E. coli and C. crescentus chemoreceptor arrays are simply different.
In any case, based on the surface area of the arrays observed here (11 000–22 800 nm2), typical C. crescentus chemoreceptor arrays contain ~90–180 hexagons. If in fact each intersection in the hexagonal lattice is composed of a trimer of receptor dimers, as we propose, the threefold axes of nearest-neighbour trimers of dimers would be 6.9 nm apart, and there would be ~1100–2200 individual receptors per array. When grown in rich media, the number of chemoreceptors per E. coli cell has been estimated biochemically to range from 3600 to 15 000, depending on the bacterial strain, the growth phase and the media (Li and Hazelbauer, 2004). Given that E. coli cells are roughly four times as voluminous as C. crescentus cells, the concentration of receptors appears to be similar.
The cytoplasmic base layer then must correspond to the complex formed by CheA, CheW and the cytoplasmic tips of the MCPs. Early models of the CheA-CheW layer were based on atomic models of CheA, CheW and the cytoplasmic portion of a receptor (Shimizu et al., 2000). Our results fail to support this model, as the predicted lattice spacing (20 nm) is almost twice as large as we observed. More recently, a structure of a CheA-CheW dimer was determined by crystallography and pulsed ESR of Thermotoga maritima proteins, from which a different model of the CheA-CheW-MCP1143C complex was proposed (Park et al., 2006). In this model, the CheA and CheW proteins are arranged in a lattice with a spacing of 13.7 nm, which is just slightly larger than the lattice spacing we observed here in C. crescentus, but the CheW dimer cleft in the T. maritima model is too small to accommodate the trimers of receptor dimers that appear to be present here (Park et al., 2006).
An even larger complex of CheA, CheW, and the cytoplasmic domain of an MCP has also been generated, first by fusing a leucine zipper dimerization domain to the aspartate receptor Tar (Francis et al., 2002; 2004), and later using the serine receptor Tsr without the dimerization domain (Wolanin et al., 2006). These complexes formed elongated ‘head-to-head’ barrel-like structures with two hollow cavities. The authors interpreted each side of the structure (one barrel) to be two sets of three receptor dimers, standing parallel in asymmetric rings to create each hollow cavity in the centre. The heads of the two barrels were linked by a double CheA/CheW plate in the middle. While a similar number of receptor monomers (24) would be present in each ring of our proposed hexagonal lattice, and the diameter of the barrel and our rings were roughly consistent, the two models differ substantially in other ways. First, the barrel-like structures seemed to be formed by two non-symmetric sides containing three uniquely arranged recepter dimers per side, whereas in our reconstruction the receptors appear to be hexagonally packed, at least near the CheA/CheW base. Second, in the barrel-like structures, two sets of receptors were joined ‘head-to-head’ by a doubly thick CheA/CheW layer. Because a similar arrangement had been seen in E. coli cells overproducing the chemotaxis receptor Tsr, where membrane invaginations allowed the signalling domains of two receptor arrays to lie flat against each other (as if a single array had folded over on itself; Lefman et al., 2004), it was proposed that this configuration might represent the activated conformation of the receptors (Wolanin et al., 2006). As in our tomograms of wild-type C. crescentus cells, neither membrane invaginations nor doubly thick receptor arrays were seen, and because CheA, CheW and the receptors are all highly conserved across species, it is unlikely that the signalling domain/signalling domain juxtaposition represents the natural configuration of activated receptors (Zhang et al., 2007), but instead occurs only under artificial conditions.
Thus, none of the existing specific models of the CheA/CheW/MCP array agree well with all our experimental findings, although other patterns such as the one proposed in Studdert and Parkinson (2005) could. Our data also suggest that the receptors are well organized near the CheA/CheW base, but lose that order as they rise towards the inner membrane. The region between the CheA/CheW base and the inner membrane has been called the ‘adaptation compartment’, where it is thought that CheR and CheB regulate the methylation of the receptors (Shimizu et al., 2000). In our tomograms there was a second, faint layer of density where the methylation sites are expected (Ames and Parkinson, 1994; Ames, 1996), ~10 nm below the inner membrane. This layer corresponds to the interaction sites of CheR and CheB with the MCPs, and is consistent with the proposed ‘brachiating mechanism’ of CheR (Levin et al., 2002). In this model, one of CheR’s two binding domains (either the one for the receptor’s flexible tether or the one for its methylation site) stays connected to a first receptor while the other domain binds to a second, neighbouring receptor, leading to a ‘hand-over-hand’ movement of the enzyme through the array. The small number of CheR and CheB molecules compared with receptors in an array (~400–15 000 in E. coli; Li and Hazelbauer, 2004), might not, however, be sufficient alone to explain the observed layer of density in that region.
Finally, the polar localization of the chemoreceptor array in swarmer cells had been shown previously (Alley et al., 1992), but our studies refined its location to the convex side of the flagellated cell pole, 17–130 nm away from the flagellar motor. While it is already known that the cytoskeletal protein Crescentin localizes to the concave side of C. crescentus cells, where it endows cells with their characteristic crescent shape (Ausmees et al., 2003), the presence of the chemotaxis array on only the convex side further highlights how C. crescentus cells maintain dorsal/ventral as well as anterior/posterior asymmetry. Whether this second dimension in intracellular differentiation is unique to C. crescentus or is widespread among bacteria has yet to be determined.
Experimental procedures
Specimen preparation
CB15N cells were grown at 30°C in peptone yeast extract (PYE) medium (Ely, 1991) to an optical density at 600 nm between 0.2 and 0.4. Two millilitres of the cell suspension was pelleted for 5 min at 1500 rcf. The pellet was rinsed in M2 Salt solution (Ely, 1991), then re-suspended in 30–50 μl of fresh M2 solution. The EM R2/2 copper/rhodium grids were glow-discharged and coated with a 3×-concentrated, 10 nm colloidal gold solution (Ted Pella, Redlands, CA). A5×-concentrated solution of 10 nm colloidal gold was added to the cells immediately before plunge freezing. A 4 μl droplet of the sample solution was applied to the EM grid, then automatically blotted and plunge-frozen in liquid ethane using a Vitrobot (FEI Company, Hillsboro, OR) (Iancu et al., 2007). The grids were stored under liquid nitrogen until data collection.
Synchronized cells
CB15N cells were synchronized according to published protocols (Tsay and Alley, 2001). One millilitre of synchronized cells was pelleted for 5 min at 1500 rcf, re-suspended in 30–50 μl M2 salt solution and plunge-frozen as described above.
Caulobacter McpA mutant
A fusion of the C-terminus of the Caulobacter McpA protein to the red-fluorescent protein, mCherry, was expressed in Caulobacter cells from a xylose-inducible promoter. Cells were grown overnight in PYE media containing 5 μg ml−1 kanamycin. One hundred microlitres of the cell suspension was added to 5 ml fresh PYE/kanamycin medium and induced with 0.03% xylose for at least 2 h prior to imaging. For plunge-freezing, 2 ml of the cell suspension was pelleted for 5 min at 1500 rcf and re-suspended in ~100 μl supernatant. Concentrated 10 nm colloidal gold solution treated with BSA was added to the concentrated cells directly before freezing. A 4 μl droplet of the sample solution was manually applied to the grid, automatically blotted, then plunged into a mixture of ethane (37%) and propane (63%) (W.F. Tivol, A. Briegel and G.J. Jensen, unpublished), using a Vitrobot (FEI Company, Hillsboro, OR).
Correlated fLM-ECT of McpA mutants
A 2 ml aliquot of cell culture was pelleted for 5 min at 1500 rcf. The pellet was re-suspended in 1 ml fixing solution containing 0.1% glutaraldehyde, 3% paraformaldehyde and 20 mM sodium phosphate (pH 7) and incubated for 30 min on ice. The cells were then pelleted, washed three times in washing buffer (140 mM NaCl, 3 mM potassium KCl, 8 mM Na2HPO4, 1.5 mM KH2PO4 and 0.05% Tween) and re-suspended in the imaging buffer containing 50 mM glucose, 10 mM EDTA and 20 mM Tris-HCl (pH 7.6). A 4 μl droplet of this solution was applied to an H2 gold finder TEM grid covered with R2/2 Quantifoil and treated with 4 μl of 0.05% poly-L-lysine in ddH2O. After blotting the grid from the side, the grid was gently rinsed with buffer. The grid was transferred onto a droplet of imaging buffer on a glass slide and covered with a coverslip. Imaging was carried out in a Nikon 90i fluorescence microscope, then the grid was carefully taken from the slide, 3 μl droplet of BSA-treated, 5×-concentrated, 10 nm gold solution was added to it, and then it was plunge-frozen into liquid ethane as described above. Tilt-series were recorded of the same cell clusters imaged in the fLM.
EM data collection
EM images were collected using a FEI Polara™ (FEI Company, Hillsboro, OR, USA), 300 kV FEG transmission electron microscope equipped with a Gatan energy filter (slit width 20 eV) on a 2 × 2 k Gatan Ultrascan CCD camera or later a lens-coupled 4 × 4 k Ultracam (Gatan, Pleasanton, CA). Pixels on the CCD represented between 0.67 and 1.2 nm on the specimen. Tilt-series from −60° to 60° with an increment between 0.5° and 1° were recorded semi-automatically around one or two axis (Iancu et al., 2005) at 10 and 12 μm underfocus using the predictive UCSF-Tomo package (Zheng et al., 2004). A cumulative dose of 200 e−/Å2 or less was used for the directly plunge frozen samples and 100 e−/Å2 or less for the correlated fLM-ECT tomograms.
Image processing
Three-dimensional reconstructions were calculated using the IMOD software package (Mastronarde, 1997). In order to obtain a higher signal-to-noise ratio map of the chemoreceptor array, we manually segmented an array with Amira (Mercury Computer Systems). Using a similar approach as described in Wright et al. (2007), we selected a layer just above the CheA/CheW base plate (where the hexagonal lattice was most visible) and projected that layer onto the surface of the array. Using a semi-automatic peak detection tool, we picked seven hexagonal centres from the best-ordered regions of the surface projection. Noting their positions, the normal vectors to the surface, and the directions to the closest neighbouring hexagonal centres, we extracted subvolumes surrounding each chosen centre, superimposed them by rotation and translation, averaged the set and finally applied sixfold rotational symmetry.
Supplementary Material
Acknowledgments
We thank Brian Crane for helpful discussions. This work was supported in part by NIH grants R01 AI067548 and P50 G082545, and DOE grant DE-FG02-04ER63785 to G.J. J., the Beckman Institute at Caltech, and gifts to Caltech from the Agouron Institute and the Gordon and Betty Moore Foundation. Zemer Gitai and John Werner were supported by Grant No. DE-FG02-05ER64136 from the Office of Science (BER), US Department of Energy. Rasmus B. Jensen thanks the ‘The Danish Natural Science Research Council’ for funding.
Footnotes
This material is available as part of the online article from: http://www.blackwell-synergy.com/doi/abs/10.1111/j.1365-2958.2008.06219.x
(This link will take you to the article abstract).
Please note: Blackwell Publishing is not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Alexander RP, Zhulin IB. Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc Natl Acad Sci USA. 2007;104:2885–2890. doi: 10.1073/pnas.0609359104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alley MRK, Gomes SL, Alexander W, Shapiro L. Genetic analysis of a temporally transcribed chemotaxis gene cluster in Caulobacter crescentus. Genetics. 1991;129:333–342. doi: 10.1093/genetics/129.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alley MRK, Maddock JR, Shapiro L. Polar localization of a bacterial chemoreceptor. Genes Dev. 1992;6:825–836. doi: 10.1101/gad.6.5.825. [DOI] [PubMed] [Google Scholar]
- Alley MRK, Maddock JR, Shapiro L. Requirement of the carboxyl terminus of a bacterial chemoreceptor for its targeted proteolysis. Science. 1993;259:1754–1757. doi: 10.1126/science.8456303. [DOI] [PubMed] [Google Scholar]
- Ames P, Parkinson JS. Constitutively Signaling fragments of Tsr, the Escherichiacoli serine chemoreceptor. J Bacteriol. 1994;176:6340–6348. doi: 10.1128/jb.176.20.6340-6348.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames P, Yu YA, Parkinson JS. Methylation segments are not required for chemotactic signalling by cytoplasmic fragments of Tsr, the methyl-accepting serine chemoreceptor of Escherichia coli. Mol Microbiol. 1996;1996:737–746. doi: 10.1046/j.1365-2958.1996.408930.x. [DOI] [PubMed] [Google Scholar]
- Ames P, Studdert CA, Reiser RH, Parkinson JS. Collaborative signaling by mixed chemoreceptor teams in Escherichia coli. Proc Natl Acad Sci USA. 2002;99:7060–7065. doi: 10.1073/pnas.092071899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausmees N, Kuhn JR, Jacobs-Wagner C. The bacterial cytosceleton: an intermediate filament-like function in cell shape. Cell. 2003;115:705–713. doi: 10.1016/s0092-8674(03)00935-8. [DOI] [PubMed] [Google Scholar]
- Berg HC. The rotary motor of bacterial flagella. Annu Rev Biochem. 2003;72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- Bilwes AM, Alex LA, Crane BR, Simon MI. Structure of CheA, a signal-transducing histidine kinase. Cell. 1999;96:131–141. doi: 10.1016/s0092-8674(00)80966-6. [DOI] [PubMed] [Google Scholar]
- Boldog T, Grimme S, Mingshan L, Sligar SG, Hazelbauer GL. Nanodiscs seperate chemoreceptor oligomeric states and reveal their signaling properties. Proc Natl Acad Sci USA. 2006;103:11509–11514. doi: 10.1073/pnas.0604988103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukhvalova MS, Dahlquist FW, Stewart RC. CheW binding interactions with CheA and Tar: importance for chemotaxis signaling in E. coli. J Biol Chem. 2002;277:22251–22259. doi: 10.1074/jbc.M110908200. [DOI] [PubMed] [Google Scholar]
- Bourret RB, Stock AM. Molecular information processing: lessons from bacterial chemotaxis. J Biol Chem. 2002;277:9625–9628. doi: 10.1074/jbc.R100066200. [DOI] [PubMed] [Google Scholar]
- Bray D, Levin MD, Morton-Firth CJ. Receptor clustering as a cellular mechanism to control sensitivity. Nature. 1998;393:85–88. doi: 10.1038/30018. [DOI] [PubMed] [Google Scholar]
- Bray D, Levin MD, Lipkow K. The chemotactic behavior of computer-based surrogate bacteria. Curr Biol. 2007;17:12–19. doi: 10.1016/j.cub.2006.11.027. [DOI] [PubMed] [Google Scholar]
- Briegel A, Dias DP, Li Z, Jensen RB, Frangakis AS, Jensen GJ. Multiple large filament bundles observed in Caulobacter crescentus by electron cryotomography. Mol Microbiol. 2006;62:5–14. doi: 10.1111/j.1365-2958.2006.05355.x. [DOI] [PubMed] [Google Scholar]
- Djordjevic S, Stock AM. Crystal structure of the chemotaxis receptor methyltransferase CheR suggests a conserved structural motiv for binding S-adensoylmethionine. Structure. 1997;5:545–558. doi: 10.1016/s0969-2126(97)00210-4. [DOI] [PubMed] [Google Scholar]
- Djordjevic S, Stock AM. Chemotaxis receptor recognition by protein methyltransferase CheR. Nat Struct Mol Biol. 1998;5:446–450. doi: 10.1038/nsb0698-446. [DOI] [PubMed] [Google Scholar]
- Djordjevic S, Goudreau PN, Xu Q, Stock AM, West AH. Sructural basis for methylesterase CchB regulation by a phosphorylation-activated domain. Proc Natl Acad Sci USA. 1998;95:1381–1386. doi: 10.1073/pnas.95.4.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B. Genetics of Caulobacter crescentus. Methods Enzymol. 1991;204:372–384. doi: 10.1016/0076-6879(91)04019-k. [DOI] [PubMed] [Google Scholar]
- Falke JJ, Hazelbauer GL. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem Sci. 2001;26:257–265. doi: 10.1016/s0968-0004(00)01770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis NR, Levit MN, Shaikh TR, Melanson LA, Stock JB, DeRosier DJ. Subunit organization in a soluble complex of Tar, CheW, and CheA by electron microscopy. J Biol Chem. 2002;277:36755–36759. doi: 10.1074/jbc.M204324200. [DOI] [PubMed] [Google Scholar]
- Francis NR, Wolanin PM, Stock JB, DeRosier DJ, Thomas DR. Three-dimensional structure and organization of a receptor/signalling complex. Proc Natl Acad Sci USA. 2004;101:17480–17485. doi: 10.1073/pnas.0407826101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gestwicki JE, Kiessling LL. Inter-receptor communication through arrays of bacterial chemoreceptors. Nature. 2002;415:81–84. doi: 10.1038/415081a. [DOI] [PubMed] [Google Scholar]
- Gomes SL, Shapiro L. Differential expression and positioning of chemotaxis methylation proteins in Caulobacter. J Mol Biol. 1984;178:551–568. doi: 10.1016/0022-2836(84)90238-9. [DOI] [PubMed] [Google Scholar]
- Griswold IJ, Zhou HJ, Matison M, Swanson RV, McIntosh LP, Simon MI, Dahlquist FW. The solution structure and interactions of CheW from Thermotoga maritima. Nat Struct Mol Biol. 2002;9:121–125. doi: 10.1038/nsb753. [DOI] [PubMed] [Google Scholar]
- Iancu C, Wright ER, Benjamin J, Tivol W, Dias DP, Murphy GE, et al. A ‘Flip-flop’ rotation stage for routine dual-axis electron cryotomography. J Struct Biol. 2005;151:288–297. doi: 10.1016/j.jsb.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Iancu CV, Tivol WF, Schooler JB, Dias DP, Henderson GP, Murphy GE, et al. Electron cryotomography sample preparation using the vitrobot. Nat Protoc. 2007;1:2813–2819. doi: 10.1038/nprot.2006.432. [DOI] [PubMed] [Google Scholar]
- Jensen GJ, Briegel A. How electron cryotomography is opening a new window onto prokaryotic ultrastructure. Curr Op Struct Biol. 2007;17:260–267. doi: 10.1016/j.sbi.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Jensen RB, Wang SC, Shapiro L. Dynamic localisation of proteins and DNA during a bacterial cell cycle. Nat Rev. 2002;3:167–176. doi: 10.1038/nrm758. [DOI] [PubMed] [Google Scholar]
- Kim KK, Yokota H, Kim SH. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature. 1999;400:787–792. doi: 10.1038/23512. [DOI] [PubMed] [Google Scholar]
- Lefman J, Zhang P, Hirai T, Weis RM, Juliani J, Bliss D, et al. Three-dimensional electron microscopic imaging of membrane invaginations in Escherichia coli overproducing the chemotaxis receptor Tsr. J Bacteriol. 2004;186:5052–5061. doi: 10.1128/JB.186.15.5052-5061.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin MD, Shimizu TS, Bray D. Binding and diffusion of CheR molecules within a cluster of membrane receptors. Biophs J. 2002;82:1809–1817. doi: 10.1016/S0006-3495(02)75531-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Hazelbauer GL. Cellular stoichiometry of the components of the chemotaxis signalling complex. J Bacteriol. 2004;186:3687–3694. doi: 10.1128/JB.186.12.3687-3694.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Hazelbauer GL. Adaptional assistance in clusters of bacterial chemoreceptors. Mol Microbiol. 2005;56:1617–1626. doi: 10.1111/j.1365-2958.2005.04641.x. [DOI] [PubMed] [Google Scholar]
- Li Z, Trimble MJ, Brun YV, Jensen GJ. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J. 2007;26:4694–4708. doi: 10.1038/sj.emboj.7601895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipkow K, Andrews SS, Bray D. Simulated diffusion of phosphorylated CheY through the cytoplasm of Escherichia coli. J Bacteriol. 2005;187:45–53. doi: 10.1128/JB.187.1.45-53.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucic V, Förster F, Baumeister W. Structural studies by electron tomography: from cells to molecules. Annu Rev Biochem. 2005;74:833–865. doi: 10.1146/annurev.biochem.73.011303.074112. [DOI] [PubMed] [Google Scholar]
- Lybarger SR, Maddock J. Polarity in action: asymmetric protein lokalization in bacteria. J Bacteriol. 2001;183:3261–3267. doi: 10.1128/JB.183.11.3261-3267.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddock JR, Shapiro L. Polar location of the chemoreceptor complex in the Escherichiacoli cell. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- Mastronarde DA. Dual-axis tomography: an approach with alignment methods that preserve resolution. J Struct Biol. 1997;120:343–352. doi: 10.1006/jsbi.1997.3919. [DOI] [PubMed] [Google Scholar]
- Park SY, Borbat PP, Gonzalez-Bonet G, Bhatnagar J, Pollard AM, Freed JH, et al. Reconstruction of the chemotaxis receptor-kinase assembly. Nat Struct Mol Biol. 2006;13:400–407. doi: 10.1038/nsmb1085. [DOI] [PubMed] [Google Scholar]
- Parkinson JS, Ames P, Studdert CA. Collaborative signalling by bacterial chemoreceptors. Cur Op Microbiol. 2005;8:116–121. doi: 10.1016/j.mib.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Sartori A, Gatz R, Beck F, Rigort A, Baumeister W, Plitzko JM. Correlative microscopy: bridging the gap between fluorescence light microscopy and cryo-electron tomography. J Struct Biol. 2007;160:135–145. doi: 10.1016/j.jsb.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Schwartz CL, Sarbash VI, Ataullakhanov FI, McIntosh JR, Nicastro D. Cryo-fluorescence microscopy facilitates correlations between light and cryo-electron microscopy and reduces the rate of photobleaching. J Microsc. 2007;227:98–109. doi: 10.1111/j.1365-2818.2007.01794.x. [DOI] [PubMed] [Google Scholar]
- Shaw P, Gomes SL, Sweeney K, Ely B, Shapiro L. Methylation involved in chemotaxis is regulated during Caulobacter differentiation. Proc Natl Acad Sci USA. 1983;80:5261–5265. [PMC free article] [PubMed] [Google Scholar]
- Shimizu TS, Le Novere N, Levin MD, Beavil AJ, Sutton BJ, Bray D. Molecular model of a lattice of signalling proteins involved in bacterial chemotaxis. Nat Cell Biol. 2000;2:792–796. doi: 10.1038/35041030. [DOI] [PubMed] [Google Scholar]
- Sourjik V. Receptor clustering and signal processing in E. coli chemotaxis. Trends Microbiol. 2004;12:569–576. doi: 10.1016/j.tim.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Sourjik V, Berg HC. Functional interactions between receptors in bacterial chemotaxis. Nature. 2004;428:437–441. doi: 10.1038/nature02406. [DOI] [PubMed] [Google Scholar]
- Studdert CA, Parkinson JS. Crosslinking snapshots of bacterial chemoreceptor squads. Proc Natl Acad Sci USA. 2004;101:2117–2122. doi: 10.1073/pnas.0308622100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studdert CA, Parkinson JS. Insights into the organization and dynamics of bacterial chemoreceptor clusters through in vivo crosslinking studies. Proc Natl Acad Sci USA. 2005;102:15623–15628. doi: 10.1073/pnas.0506040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsay JW, Alley MRK. Proteolysis of the Caulobacter McpA chemoreceptor is cell cycle regulated by a ClpX-dependent pathway. J Bacteriol. 2001;183:5001–5007. doi: 10.1128/JB.183.17.5001-5007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner JK, Brun YV. Out on a limb: how the Caulobacter stalk can boost the study of bacterial shape. Mol Microbiol. 2007;64:28–33. doi: 10.1111/j.1365-2958.2007.05633.x. [DOI] [PubMed] [Google Scholar]
- Weis RM. Inch by inch, row by row. Nat Struct Mol Biol. 2006;13:382–384. doi: 10.1038/nsmb0506-382. [DOI] [PubMed] [Google Scholar]
- Weis RM, Hirai T, Chalah A, Kessel M, Peters PJ, Subramaniam S. Electron microscopic analysis of membrane assemblies formed by the bacterial chemotaxis receptor Tsr. J Bacteriol. 2003;185:3636–3643. doi: 10.1128/JB.185.12.3636-3643.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolanin PM, Baker MD, Thomas DR, DeRosier DJ, Stock AM. Self-assembly of receptor/signaling complexes in bacterial chemotaxis. Proc Natl Acad Sci USA. 2006;103:14313–14318. doi: 10.1073/pnas.0606350103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright ER, Schooler JB, Ding HJ, Kieffer C, Fillmore C, Sundquist WI, Jensen GJ. Electron cryotomography of imature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. EMBO J. 2007;26:2218–2226. doi: 10.1038/sj.emboj.7601664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Boos W, Heymann J, Gnaegi H, Kessel M, Subramaniam S. Receptor arrays in frozen-hydrated E. coli direct visualization of receptor arrays in frozen-hydrated sections and plunge-frozen specimens of E. coli engineered to overproduce the chemotaxis receptor Tsr. J Microsc. 2004;216:76–83. doi: 10.1111/j.0022-2720.2004.01395.x. [DOI] [PubMed] [Google Scholar]
- Zhang P, Khursigara CM, Hartnell LM, Subramaniam S. Direct visualization of Escherichia coli chemotaxis receptor arrays using cryo-electron microscopy. Proc Natl Acad Sci USA. 2007;104:3777–3781. doi: 10.1073/pnas.0610106104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng QS, Braunfeld MB, Sedat JW, Agard DA. An improved strategy for automated electron microscopy. J Struct Biol. 2004;147:91–101. doi: 10.1016/j.jsb.2004.02.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.