Abstract
T lymphocytes are a key regulatory component of the adaptive immune system. Understanding how the micro- and nano-scale details of the extracellular environment influence T cell activation may have wide impact on the use of T cells for therapeutic purposes. In this article, we examine how the micro- and nano-scale presentation of ligands to cell surface receptors, including microscale organization and nanoscale mobility, influences the activation of T cells. We extend these studies to include the role of cell-generated forces, and the rigidity of the microenvironment, on T cell activation. These approaches enable delivery of defined signals to T cells, a step toward understanding the cell-cell communication in the immune system, and developing micro/nano- and material- engineered systems for tailoring immune responses for adoptive T cell therapies.
INTRODUCTION
The immune system protects against pathogens and other agents. The adaptive immune system, a recently evolved part of the vertebrate immune system, forms a strong line of defense against biological challenges through its ability to recognize new antigens, develop an appropriate response, and rapidly recall this action on subsequent re-exposure. T lymphocytes are a key regulatory component of this system, coordinating the activity of other cells and directly carrying out specific immune functions. Given these roles, T cell manipulation has been proposed as a therapeutic treatment for many diseases, most notably cancer [1–5]. A key step in many of these treatments, collectively referred to as adoptive immunotherapy, is the ex vivo stimulation and expansion of T cells, with subsequent re-introduction of these cells into the patient. This process is most often carried out by presenting T cells with ligands to specific receptors present on the T cell surface. The T Cell Receptor (TCR) complex which provides the primary antigenic signal conferring specificity and the CD28, and LFA-1 receptors which provide costimulatory and adhesive signals have been the most intensively used. Additional co-receptors assist TCR signaling in responding to different types of major histocompatibility complex (MHC) antigen presenting molecules. CD4 assists with recognition of extracellular peptides on MHC class II and CD8 assists with intracellular peptides presented on MHC class I. Mature T cells express CD4 or CD8 and both types of cells are valued adoptive immunotherapy. In vivo, these ligands are presented to T cells on the surface of specialized Antigen Presenting Cells (APCs), and this association with the plasma membrane is required for optimal effect. While ideal for use in stimulating T cells in vivo, culture and growth of native APCs is difficult and inconsistent. As such, the growth of T cells for contemporary adoptive immunotherapy is often initiated by ligands presenting on engineered beads or artificial APCs [6–9] (Fig. 1B). These approaches allow production of sufficient quantities of T cells for clinical use, but have severe limitations including lack of control over the phenotype or performance of the resultant cells [4].
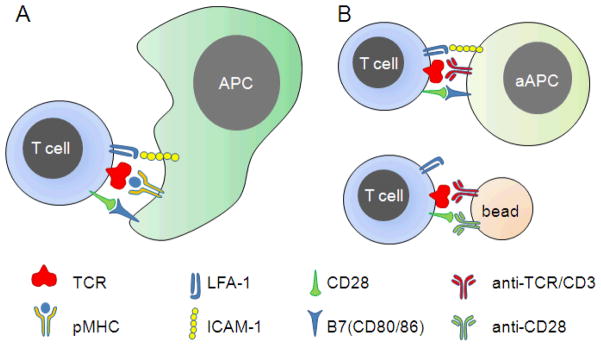
Figure 1. T cell activation.
(A) Activation of T cells in vivo is mediated in large part by contact-mediated communication with Antigen Presenting Cells (APCs). (B) For therapeutic ex vivo expansion, activation is commonly carried out by replacing the APC with either engineered cells or beads that engage the same receptors involved in T cell/APC interaction.
We propose that the process of T cell expansion can be improved by designing biomaterials to better capture the native T cell/APC interface, a small (70 μm2) region termed the immunological synapse (IS), which serves as a major point of communication. The binding of TCR to peptide-loaded MHCs (pMHC), CD28 by B7, and LFA-1 by ICAM-1, three major receptor-ligand complexes, occurs in this interface, directing a range of subsequent cell functions including polarization, cytokine secretion, signal integration, and asymmetric cell division [10–13]. An emerging picture is that the micro- and nano-scale details of this structure play important roles in coordinating the function of these cells. This report describes recent progress towards understanding these aspects, which may lead to new design rules for biomaterials that when used in T cell expansion will improve adoptive immunotherapy.
1. MICRO-/NANO-SCALE PATTERNING
A distinguishing characteristic of the immune synapse is the presence of highly complex, micrometer-scale organizations of signaling complexes within this structure. The mature synapse was initially described as a microscale, annular disc of LFA-1/ICAM-1 supramolecular activation clusters (SMACs) surrounding centralized clusters of TCR/pMHC [14]. However, additional patterns of these and other receptor pairs, including CD28/B7, have been identified (Figure 2A) [15–18], raising the possibility that the different motifs may convey specific instructions to the T cells, and conversely be used to control cellular activation.
Figure 2. Micropatterned activation of T cells.
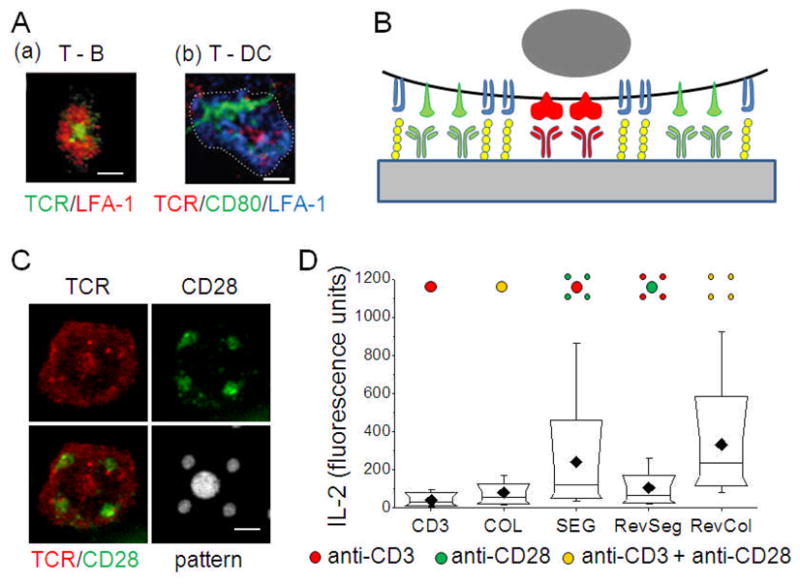
(A) Signaling complexes for distinct patterns of organization within the immune synapse. Adapted from [14] and [18]. (B) Micropattering of activating ligands to cell surface receptors allows the study of how cells respond to specific organizations of signaling complexes. (C) Demonstration of the ability to control receptor organization using activating antibodies to CD3 (central 2 μm dot) and CD28 (satellite 1 μm features). (D) Comparison of IL-2 secretion by mouse naïve CD4+ T cells on specific microscale patterns of CD3 and CD28. Scale bars: 2 μm.
Recent years have seen the application of micro- and nano-patterning methods to understanding the immune synapse [19–22]. Most notably, Doh and Irvine [19] introduced a lithographic approach to create complementary patterns of ICAM-1 and an activating antibody to CD3 (a key signaling component of the TCR) on material surfaces, and demonstrated that preactivated T cells recognize different microscale patterns of these ligands. Our group built upon this concept [21], introducing techniques to combine higher numbers of patterns on a single surface (Figure 2B and C). By applying multiple rounds of microcontact printing to a single surface, we created independently-defined patterns of activating antibodies to CD3 and CD28, surrounded and separated by ICAM-1. These immobilized proteins were highly effective in patterning the engagement of T cell receptors (Figure 2C).
Naïve CD4+ T cells isolated from mice were able to recognize different patterns of activating antibodies to CD3 and CD28, as measured by secretion of IL-2, a key cytokine involved in T cell activation. Figure 2D compares IL-2 secretion across a select set of patterns indicated along the bottom axis of that graph, revealing a set of design rules: 1) activation of naïve CD4+ T cells is enhanced by engaging CD28 in the periphery of the T cell-substrate interface, as opposed to the center, 2) a similar change in CD3 engagement had a minor influence on IL-2 secretion, and 3) segregation/colocalization of the CD3 and CD28 signaling had a very small impact on T cell function.
Together, these studies point to a powerful role of spatial organization in directing T cell activation and, potentially, subsequent differentiation and function. Incorporation of these patterning concepts into biomaterial design may provide a similar, new level of control over T cell expansion.
2. MODULATING T CELL ACTIVATION THROUGH SUBSTRATE RIGIDITY
The mechanical rigidity of the extracellular environment is increasingly recognized as a modulator of numerous cell functions, including formation/disassembly of adhesion complexes (i.e., focal adhesions), cell spreading, gene transcription, and cell fate [23–25]. This rigidity-sensing is coupled with force generation by the acto-myosin cytoskeleton in the cell, which may alter the phosphorylation of key molecules and switch integrins between resting and active states [26, 27].
In T cells, formation of an IS also involves a high level of acto-myosin activity, which is initiated by TCR microclusters [28] and driven by continuous centripetal transportation of these microclusters [28–31]. Inhibiting actin polymerization abrogates the formation of TCR microclusters [28] and the activation processes [32, 33]. Myosin IIA was found to be the motor protein for the centripetal actin flow and microcluster movement [34]. Actin polymerization and myosin II-induced retraction at the periphery of the IS result in “contractile oscillation” [35], which may help stabilize integrin-mediated adhesion through force-mediated signaling [36]. Inhibition of myosin IIA activity significantly reduced phosphorylation of Zap70 and LAT, two pivotal signaling events in T cell activation [34], implying that the mechanical force generated inside T cell may play a role in elevating activation signals.
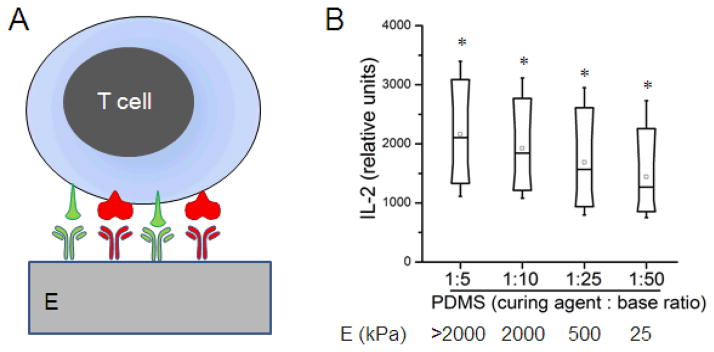
We tested this hypothesis by intervening with the mechanical contraction process using antibody-coated substrates of varied rigidities. The bulk rigidity of planar polydimethylsiloxane (PDMS, Sylgard 184) substrates, which were coated with activating antibodies to CD3 and CD28 (Figure 3A) was controlled by varying the ratio of elastomer base to curing agent, resulting in substrates ranging in Young’s modulus from tens of kiloPascals to several MegaPascals (Figure 3B) [37]. PDMS was prepared as a thin (hundreds of micrometers) layer backed by glass. Cells can probe the first few micrometers of elastomer depth [23], so these preparations appear as a half-infinite slab to the cells, yet are thin enough to allow microscopy and handling. The amount of antibody adsorbed to each surface was measured using an ELISA-based approach, and was found to be similar across this range of compositions/rigidities (P < 0.005), varying by approximately 5%.
Figure 3. Rigidity sensing by T cells.
(A) Mouse CD4+ T cells were activated using antibodies to CD3 and CD28, adsorbed onto planar substrates of varying rigidity. (B) Box plots comparing IL-2 secretion by cells as a function of substrate rigidity. * Each condition was statistically different from all others, α = 0.05, n > 2000 cells per surface.
Figure 3B compares IL-2 secretion by naïve mouse CD4+ T cells as a function of PDMS formulation; the Box plots in this figure illustrate one representative experiment of three repetitions. T cell costimulation was sensitive to the rigidity of the underlying substrate, where cells on the stiffer substrate had higher IL-2 secretion. All samples were statistically different from each other by Kruskal-Wallis and ANOVA tests (α = 0.05, n > 2000 cells per surface).
Control over the mechanical properties of an activating surface may similarly modulate long-term T cell function, including division, proliferation, and differentiation, providing a new level of control over ex vivo T cell expansion.
3. LATERAL MOBILITY OF SIGNALING CLUSTERS
Unlike extracellular matrix proteins, ligands to proteins involved in cell-cell communication, such as those presented by an APC, exhibit lateral mobility across the cell surface, owing to its association with the plasma membrane. Supported lipid bilayers (SLBs) presenting membrane proteins capture the lateral mobility of the natural cell surface, and have emerged as a powerful model for investigating cell signaling [38–46]. Using this system, Chan et al. [47] demonstrated that laterally mobile, GPI-tethered CD58 was much more potent than an immobile counterpart in promoting cell interaction. Subsequent implementations of this model provide a rare look into the organization of the immunological synapse [39, 40]. T cells cluster and reorganize various components of antigen-presenting cells that have been isolated and tethered to planar supported lipid bilayer, a phenomenon not possible if the proteins were immobilized to the experimental surface.
It is increasingly recognized that the plasma membrane exhibits considerable micro- and nano-scale order. Sub-cellular patterning of a single type of SLB has been studied using techniques of e-beam lithography [41, 48], micro-contact printing [49], photolithography and parylene peel-off [50]. The ability to pattern multiple bilayers of different composition on a single surface is important for bridging between studies using uniform SLBs and micro-patterned immobilized proteins [19, 22]. However, patterning at sub-cellular resolution has been elusive, owing to the limitations of non-mixing laminar flow [51–54]. More recently, researchers have patterned SLBs with sub-micrometer precision and multiple compositions using AFM-related techniques [55–57], but these are not well-suited for covering the relatively large areas intended for cell-based experiments (millimeters to centimeters on a side).
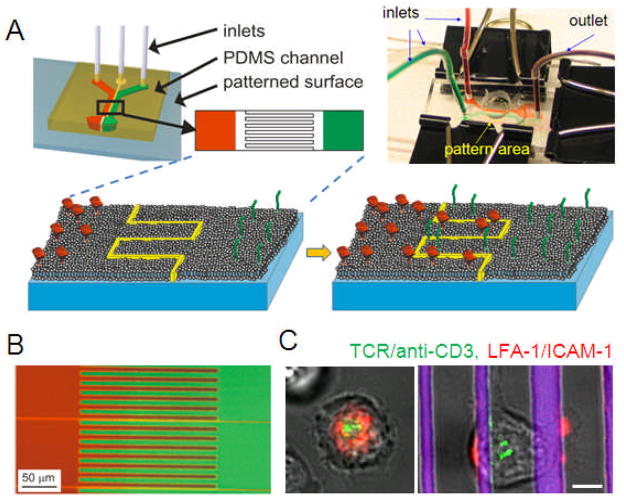
As shown in Figure 4A, we introduced a SLB patterning strategy that takes advantage of diffusive transport in supported membranes after formation on a substrate [58]. A bilayer-compatible substrate is divided into multiple regions, consisting of two large open regions separated by a middle region which contains a continuous barrier that divides the entire surface into two topologically distinct regions. A three-flow chamber system is used to form three different types of lipid bilayer on this surface; bilayers containing two different tethered proteins are deposited on the outer regions, while a plain bilayer is formed on the middle zone. Over time, tethered biomolecules will diffuse from the outer regions into the interdigitated middle region, leading to an interlacing of regions each separately containing different components (Figure 4B). Importantly, the spatial resolution of the resultant bilayers is determined by the barrier, providing finer resolution than that provided by laminar flow, reaching potentially into the realm of tens of nanometers [59]. With this approach, multiple ligands can be presented to cells, each confined to separate regions of the cell-surface interface while retaining the mobility required for effective membrane protein function. Figure 4C shows the use of this platform in presenting spatially segregated, micropatterned ligands to the T cell surface proteins TCR and LFA-1.
Figure 4. Multicomponent supported lipid bilayers.
(A) Microfluidic approaches allow patterning of supported lipid bilayers at subcellular levels. (B) Example of a two-component lipid bilayer system. (C) Comparison of T cell receptor organization on unpatterned (left) and segregated (right) lipid bilayers presenting ligands to TCR and LFA-1; scale bar = 5 μm. Adapted from [58].
The supported lipid bilayer model has been extremely useful for contemporary investigations into the impact of membrane protein mobility on cell signaling. Adaptation of these design rules into new biomaterials, however, poses several challenges, including the fragility and limited lifetime of supported lipid bilayer. Continued advances in artificial amphiphilic molecules capturing key properties of lipids may yield to more effective techniques for including lateral mobility into material systems. Conversely continued design of polymer structure may allow the capture of the nanoscale behavior of natural lipids into these systems.
CONCLUSIONS
This article reviews recent advances in understanding the role of micro- and nano-scale organization and cellular biomechanics in directing T cell function. Together, these principles may lead to biomaterials that provide significantly enhanced control over T cell expansion, leading to improved implementation of adoptive immunotherapy.
Acknowledgments
This work is supported in part by the National Institutes of Health, EY016586 and EB008199.
References
- 1.Finn OJ. Cancer immunology. New England Journal of Medicine. 2008;358:2704–15. doi: 10.1056/NEJMra072739. [DOI] [PubMed] [Google Scholar]
- 2.Hunder NN, Wallen H, Cao J, Hendricks DW, Reilly JZ, Rodmyre R, Jungbluth A, Gnjatic S, Thompson JA, Yee C. Treatment of metastatic melanoma with autologous CD4+ T cells against NY-ESO-1. New England Journal of Medicine. 2008;358:2698–703. doi: 10.1056/NEJMoa0800251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.June CH. Adoptive T cell therapy for cancer in the clinic. Journal of Clinical Investigation. 2007;117:1466–76. doi: 10.1172/JCI32446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.June CH. Principles of adoptive T cell cancer therapy. Journal of Clinical Investigation. 2007;117:1204–12. doi: 10.1172/JCI31446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weiner LM. Cancer immunotherapy--the endgame begins. New England Journal of Medicine. 2008;358:2664–5. doi: 10.1056/NEJMp0803663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dudley ME, Wunderlich JR, Yang JC, Sherry RM, Topalian SL, Restifo NP, Royal RE, Kammula U, White DE, Mavroukakis SA, Rogers LJ, Gracia GJ, Jones SA, Mangiameli DP, Pelletier MM, Gea-Banacloche J, Robinson MR, Berman DM, Filie AC, Abati A, Rosenberg SA. Adoptive cell transfer therapy following non-myeloablative but lymphodepleting chemotherapy for the treatment of patients with refractory metastatic melanoma. Journal of Clinical Oncology. 2005;23:2346–57. doi: 10.1200/JCO.2005.00.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riddell SR, Greenberg PD. The use of anti-CD3 and anti-CD28 monoclonal antibodies to clone and expand human antigen-specific T cells. Journal of Immunoogicall Methods. 1990;128:189–201. doi: 10.1016/0022-1759(90)90210-m. [DOI] [PubMed] [Google Scholar]
- 8.Rosenberg SA, Restifo NP, Yang JC, Morgan RA, Dudley ME. Adoptive cell transfer: a clinical path to effective cancer immunotherapy. Nature Reviews Cancer. 2008;8:299–308. doi: 10.1038/nrc2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suhoski MM, Golovina TN, Aqui NA, Tai VC, Varela-Rohena A, Milone MC, Carroll RG, Riley JL, June CH. Engineering Artificial Antigen-presenting Cells to Express a Diverse Array of Co-stimulatory Molecules. Molecular Therapeutics. 2007;15:981–988. doi: 10.1038/mt.sj.6300134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang JT, Palanivel VR, Kinjyo I, Schambach F, Intlekofer AM, Banerjee A, Longworth SA, Vinup KE, Mrass P, Oliaro J, Killeen N, Orange JS, Russell SM, Weninger W, Reiner SL. Asymmetric T lymphocyte division in the initiation of adaptive immune responses. Science. 2007;315:1687–91. doi: 10.1126/science.1139393. [DOI] [PubMed] [Google Scholar]
- 11.Dustin ML. Hunter to gatherer and back: immunological synapses and kinapses as variations on the theme of amoeboid locomotion. Annual Review of Cell and Developmental Biology. 2008;24:577–96. doi: 10.1146/annurev.cellbio.24.110707.175226. [DOI] [PubMed] [Google Scholar]
- 12.Dustin ML, Colman DR. Neural and immunological synaptic relations. Science. 2002;298:785–9. doi: 10.1126/science.1076386. [DOI] [PubMed] [Google Scholar]
- 13.Dustin ML, Olszowy MW, Holdorf AD, Li J, Bromley S, Desai N, Widder P, Rosenberger F, van der Merwe PA, Allen PM, Shaw AS. A novel adaptor protein orchestrates receptor patterning and cytoskeletal polarity in T-cell contacts. Cell. 1998;94:667–77. doi: 10.1016/s0092-8674(00)81608-6. [DOI] [PubMed] [Google Scholar]
- 14.Monks CR, Freiberg BA, Kupfer H, Sciaky N, Kupfer A. Three-dimensional segregation of supramolecular activation clusters in T cells. Nature. 1998;395:82–6. doi: 10.1038/25764. [DOI] [PubMed] [Google Scholar]
- 15.Thauland TJ, Koguchi Y, Wetzel SA, Dustin ML, Parker DC. Th1 and Th2 cells form morphologically distinct immunological synapses. Journal of Immunology. 2008;181:393–9. doi: 10.4049/jimmunol.181.1.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trautmann A, Valitutti S. The diversity of immunological synapses. Current Opinion in Immunology. 2003;15:249–54. doi: 10.1016/s0952-7915(03)00040-2. [DOI] [PubMed] [Google Scholar]
- 17.Tseng SY, Liu M, Dustin ML. CD80 cytoplasmic domain controls localization of CD28, CTLA-4, and protein kinase Ctheta in the immunological synapse. Journal of Immunology. 2005;175:7829–36. doi: 10.4049/jimmunol.175.12.7829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tseng SY, Waite JC, Liu M, Vardhana S, Dustin ML. T cell-dendritic cell immunological synapses contain TCR-dependent CD28-CD80 clusters that recruit protein kinase C theta. Journal of Immunology. 2008;181:4852–63. doi: 10.4049/jimmunol.181.7.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doh J, Irvine DJ. Immunological synapse arrays: patterned protein surfaces that modulate immunological synapse structure formation in T cells. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:5700–5. doi: 10.1073/pnas.0509404103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Senaratne W, Sengupta P, Jakubek V, Holowka D, Ober CK, Baird B. Functionalized surface arrays for spatial targeting of immune cell signaling. Journal of the American Chemical Society. 2006;128:5594–5. doi: 10.1021/ja058701p. [DOI] [PubMed] [Google Scholar]
- 21.Shen K, Thomas VK, Dustin ML, Kam LC. Micropatterning of costimulatory ligands enhances CD4+ T cell function. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:7791–6. doi: 10.1073/pnas.0710295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen K, Qi J, Kam LC. Microcontact printing of proteins for cell biology. Journal of Visualized Experiments. 2008 doi: 10.3791/1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–89. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 24.Giannone G, Sheetz MP. Substrate rigidity and force define form through tyrosine phosphatase and kinase pathways. Trends in Cell Biology. 2006;16:213–23. doi: 10.1016/j.tcb.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 25.Pelham RJ, Jr, Wang Y. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:13661–5. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang G, Huang AH, Cai Y, Tanase M, Sheetz MP. Rigidity sensing at the leading edge through alphavbeta3 integrins and RPTPalpha. Biophysical Journal. 2006;90:1804–9. doi: 10.1529/biophysj.105.072462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Astrof NS, Salas A, Shimaoka M, Chen J, Springer TA. Importance of force linkage in mechanochemistry of adhesion receptors. Biochemistry. 2006;45:15020–8. doi: 10.1021/bi061566o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campi G, Varma R, Dustin ML. Actin and agonist MHC-peptide complex-dependent T cell receptor microclusters as scaffolds for signaling. Journal of Experimental Medicine. 2005;202:1031–1036. doi: 10.1084/jem.20051182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freiberg BA, Kupfer H, Maslanik W, Delli J, Kappler J, Zaller DM, Kupfer A. Staging and resetting T cell activation in SMACs. Nature Immunology. 2002;3:911–7. doi: 10.1038/ni836. [DOI] [PubMed] [Google Scholar]
- 30.Krummel MF, Sjaastad MD, Wulfing C, Davis MM. Differential clustering of CD4 and CD3zeta during T cell recognition. Science. 2000;289:1349–52. doi: 10.1126/science.289.5483.1349. [DOI] [PubMed] [Google Scholar]
- 31.Huppa JB, Gleimer M, Sumen C, Davis MM. Continuous T cell receptor signaling required for synapse maintenance and full effector potential. Nature Immunology. 2003;4:749–55. doi: 10.1038/ni951. [DOI] [PubMed] [Google Scholar]
- 32.Valitutti S, Dessing M, Aktories K, Gallati H, Lanzavecchia A. Sustained signaling leading to T cell activation results from prolonged T cell receptor occupancy. Role of T cell actin cytoskeleton. Journal of Experimental Medicine. 1995;181:577–84. doi: 10.1084/jem.181.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wulfing C, Davis MM. A receptor/cytoskeletal movement triggered by costimulation during T cell activation. Science. 1998;282:2266–9. doi: 10.1126/science.282.5397.2266. [DOI] [PubMed] [Google Scholar]
- 34.Ilani T, Vasiliver-Shamis G, Vardhana S, Bretscher A, Dustin ML. T cell antigen receptor signaling and immunological synapse stability require myosin IIA. Nature Immunology. 2009;10:531–9. doi: 10.1038/ni.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dobereiner HG, Dubin-Thaler BJ, Hofman JM, Xenias HS, Sims TN, Giannone G, Dustin ML, Wiggins CH, Sheetz MP. Lateral membrane waves constitute a universal dynamic pattern of motile cells. Physical Review Letters. 2006;97:038102. doi: 10.1103/PhysRevLett.97.038102. [DOI] [PubMed] [Google Scholar]
- 36.Dustin ML. Cell adhesion molecules and actin cytoskeleton at immune synapses and kinapses. Current Opinion in Cell Biology. 2007;19:529–33. doi: 10.1016/j.ceb.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsai J, Kam L. Rigidity-dependent cross talk between integrin and cadherin signaling. Biophysical Journal. 2009;96:L39–41. doi: 10.1016/j.bpj.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dori Y, Bianco-Peled H, Satija SK, Fields GB, McCarthy JB, Tirrell M. Ligand accessibility as means to control cell response to bioactive bilayer membranes. Journal of Biomedical Materials Research. 2000;50:75–81. doi: 10.1002/(sici)1097-4636(200004)50:1<75::aid-jbm11>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 39.Grakoui A, Bromley SK, Sumen C, Davis MM, Shaw AS, Allen PM, Dustin ML. The immunological synapse: a molecular machine controlling T cell activation. Science. 1999;285:221–7. doi: 10.1126/science.285.5425.221. [DOI] [PubMed] [Google Scholar]
- 40.Groves JT, Dustin ML. Supported planar bilayers in studies on immune cell adhesion and communication. Journal of Immunological Methods. 2003;278:19–32. doi: 10.1016/s0022-1759(03)00193-5. [DOI] [PubMed] [Google Scholar]
- 41.Mossman KD, Campi G, Groves JT, Dustin ML. Altered TCR signaling from geometrically repatterned immunological synapses. Science. 2005;310:1191–3. doi: 10.1126/science.1119238. [DOI] [PubMed] [Google Scholar]
- 42.Oliver AE, Ngassam V, Dang P, Sanii B, Wu H, Yee CK, Yeh Y, Parikh AN. Cell Attachment Behavior on Solid and Fluid Substrates Exhibiting Spatial Patterns of Physical Properties. Langmuir. 2009 doi: 10.1021/la900166u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pautot S, Lee H, Isacoff EY, Groves JT. Neuronal synapse interaction reconstituted between live cells and supported lipid bilayers. Nature Chemical Biology. 2005;1:283. doi: 10.1038/nchembio737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perez TD, Nelson WJ, Boxer SG, Kam L. E-Cadherin Tethered to Micropatterned Supported Lipid Bilayers as a Model for Cell Adhesion. Langmuir. 2005;21:11963–8. doi: 10.1021/la052264a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stroumpoulis D, Zhang H, Rubalcava L, Gliem J, Tirrell M. Cell adhesion and growth to Peptide-patterned supported lipid membranes. Langmuir. 2007;23:3849–56. doi: 10.1021/la062375p. [DOI] [PubMed] [Google Scholar]
- 46.Yokosuka T, Kobayashi W, Sakata-Sogawa K, Takamatsu M, Hashimoto-Tane A, Dustin ML, Tokunaga M, Saito T. Spatiotemporal regulation of T cell costimulation by TCR-CD28 microclusters and protein kinase C theta translocation. Immunity. 2008;29:589–601. doi: 10.1016/j.immuni.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chan PY, Lawrence MB, Dustin ML, Ferguson LM, Golan DE, Springer TA. Influence of Receptor Lateral Mobility On Adhesion Strengthening Between Membranes Containing Lfa-3 and Cd2. Journal of Cell Biology. 1991;115:245–255. doi: 10.1083/jcb.115.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DeMond AL, Groves JT. Interrogating the T cell synapse with patterned surfaces and photoactivated proteins. Current Opinion in Immunology. 2007;19:722–7. doi: 10.1016/j.coi.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 49.Kam L, Boxer SG. Cell adhesion to protein-micropatterned-supported lipid bilayer membranes. Journal of Biomedical Materials Research. 2001;55:487–95. doi: 10.1002/1097-4636(20010615)55:4<487::aid-jbm1041>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 50.Orth RN, Wu M, Holowka DA, Craighead HG, Baird BA. Mast Cell Activation on Patterned Lipid Bilayers of Subcellular Dimensions. Langmuir. 2003;19:1599–1605. [Google Scholar]
- 51.Kam L, Boxer SG. Formation of supported lipid bilayer composition arrays by controlled mixing and surface capture. Journal of the American Chemical Society. 2000;122:12901–2. [Google Scholar]
- 52.Kam L, Boxer SG. Spatially selective manipulation of supported lipid bilayers by laminar flow: steps towards biomembrane microfluidics. Langmuir. 2003;19:1624–31. [Google Scholar]
- 53.Yang T, Simanek EE, Cremer P. Creating addressable aqueous microcompartments above solid supported phospholipid bilayers using lithographically patterned poly(dimethylsiloxane) molds. Analytical Chemistry. 2000;72:2587–9. doi: 10.1021/ac000131i. [DOI] [PubMed] [Google Scholar]
- 54.Yoshina-Ishii C, Boxer SG. Arrays of mobile tethered vesicles on supported lipid bilayers. Journal of the American Chemical Society. 2003;125:3696–7. doi: 10.1021/ja029783+. [DOI] [PubMed] [Google Scholar]
- 55.Jackson BL, Groves JT. Scanning probe lithography on fluid lipid membranes. Journal of the American Chemical Society. 2004;126:13878–9. doi: 10.1021/ja046040a. [DOI] [PubMed] [Google Scholar]
- 56.Lenhert S, Sun P, Wang Y, Fuchs H, Mirkin CA. Massively parallel dip-pen nanolithography of heterogeneous supported phospholipid multilayer patterns. Small. 2007;3:71–5. doi: 10.1002/smll.200600431. [DOI] [PubMed] [Google Scholar]
- 57.Shi J, Chen J, Cremer PS. Sub-100 nm patterning of supported bilayers by nanoshaving lithography. Journal of the American Chemical Society. 2008;130:2718–9. doi: 10.1021/ja077730s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shen K, Tsai J, Shi P, Kam LC. Self-aligned supported lipid bilayers for patterning the cell-substrate interface. Journal of the American Chemical Society. 2009;131:13204–5. doi: 10.1021/ja904721h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsai J, Sun E, Gao Y, Hone JC, Kam LC. Non-Brownian diffusion of membrane molecules in nanopatterned supported lipid bilayers. Nano Letters. 2008;8:425–30. doi: 10.1021/nl072304q. [DOI] [PMC free article] [PubMed] [Google Scholar]