Abstract
Serotyping is a universally accepted subtyping method for Listeria monocytogenes. Identification of the strain serotype permits differentiation between important food-borne strains (1/2a, 1/2b, and 4b) and provides a “gold standard” for comparing isolates analyzed in different labs and with different techniques. Although an efficient enzyme-linked immunosorbent assay serotyping protocol was described recently, identification of PCR serotyping primers would further increase the ease and accessibility of this classification system. Serotyping PCR primers were designed from variable regions of the L. monocytogenes genome. Three primer sets were used in conjunction with a previously described Division III primer set in order to classify 122 L. monocytogenes strains into five serotype groups [1/2a(3a), 1/2b, 1/2c(3c), 4b(d,e), and 4a/c]. Results of the PCR method agreed with those of the conventional slide agglutination method for 97, 100, 94, and 91% of strains belonging to serotypes 1/2a, 1/2b, 1/2c, and 4b, respectively.
Listeria monocytogenes is a gram-positive, facultative, intracellular bacterial pathogen that causes morbidity and mortality in humans and livestock. It is a significant food-borne pathogen due its widespread distribution in nature, its ability to survive in a wide range of environmental conditions, and its ability to grow at refrigeration temperatures. Although human listeriosis has a low rate of incidence, L. monocytogenes causes severe illness and mortality in susceptible individuals. Neonates, the elderly, and the immunosuppressed are particularly at risk. Approximately 2,500 human listeriosis cases occur annually in the United States, resulting in 500 deaths (20).
Numerous molecular subtyping techniques have identified two major phylogenetic divisions within the species. Division I consists of serotypes 1/2b, 3b, 4b, 4d, and 4e, and Division II consists of serotypes 1/2a, 1/2c, 3a, and 3c (1-3, 5, 13, 24). A third division consisting of less common serotypes 4a and 4c has also been identified (25). L. monocytogenes has a strongly clonal population structure (8), and virulence gene evolution has paralleled that of somatic and flagellar antigens (11, 30).
L. monocytogenes strains are serotyped according to variation in the somatic (O) and flagellar (H) antigens (28). Although more than 14 serotypes of L. monocytogenes have been described (14), only three serotypes (1/2a, 1/2b, and 4b) cause the vast majority of clinical cases (29). Interestingly, although serotype 1/2a is the most frequently isolated from food, serotype 4b causes the majority of human epidemics (12). Therefore, it is likely that serotype designation is associated with virulence potential.
Because of the importance of L. monocytogenes epidemiology to human health, a number of discriminatory subtyping methods have been described for this organism (2, 6, 9, 15, 16, 19, 21, 22, 27, 31). Although pulsed-field gel electrophoresis is the most commonly employed subtyping technique, new subtyping technologies are constantly introduced and tested in hopes of increasing the resolution, speed, and reproducibility of L. monocytogenes subtyping. The most recent examples of this are multilocus sequence subtyping (26) and microarray genomic analysis (4, 8). It is important that the strains included in these studies are initially characterized by using a universally accepted method such as serotyping. Serotyping makes it possible to compare results from different studies while providing a biological context for phylogenetic or phenetic relationships that are described by a new method.
The major drawbacks to routine serotyping include cost, availability and standardization of reagents, as well as the technical expertise needed to perform the assay. Recently, Palumbo et al. (23) described an enzyme-linked immunosorbent assay serotyping format used in conjunction with a commercially available kit that makes serotyping much less expensive, more efficient, and therefore more accessible to research and clinical laboratories. Nevertheless, identification of serotype-specific PCR primers would further increase the ease of identifying strain serotype. Jinneman and Hill (17) described mismatch amplification mutation assay (MAMA) PCR primers that allow strains to be easily classified into phylogenetic divisions, and here we describe PCR primers that allow the four major serotypes (1/2a, 1/2b, 1/2c, and 4b) to be identified with greater than 90% accuracy.
MATERIALS AND METHODS
Bacterial strains.
A panel of 122 L. monocytogenes strains was assembled from human (n = 46), veterinary (n = 20), and environmental (n = 41) samples, as well as from samples from undetermined sources (n = 15). A majority of the strains were from the United States or Canada, and 10 were of European origin. Ten serotypes were included in the analysis, although the majority of the strains tested belonged to the four most common serotypes: 1/2a (n = 30), 1/2b (n = 20), 1/2c (n = 18), and 4b (n = 35).
Serotyping.
Samples were serotyped with a commercially available serotyping kit (Denka Seiken Co., Tokyo, Japan) according to the manufacturer's instructions, or they had been previously serotyped by a reference laboratory.
PCR primer design.
Division-specific multiplex primers (Table 1) were designed on the basis of a divergent region of the L. monocytogenes genome that was previously identified by means of microarray analysis (4, 8). Microarray probe 302 (GenBank accession no. BH170518) has high sequence similarity to an iron transport protein and was shown by microarray analysis to hybridize only to strains belonging to phylogenetic Divisions I and III; therefore, primers were designed on the basis of sequence differences that distinguish the two major divisions.
TABLE 1.
PCR primers used to serotype L. monocytogenes strains
| Primer set | Forward primer sequence | Reverse primer sequence | Product size (bp) | Anneal temp (°C) | Specificity |
|---|---|---|---|---|---|
| D1a | CGATATTTTATCTACTTTGTCA | TTGCTCCAAAGCAGGGCAT | 214 | 59 | Division I or III |
| D2a | GCGGAGAAAGCTATCGCA | TTGTTCAAACATAGGGCTA | 140 | 59 | Division II |
| FlaAb | TTACTAGATCAAACTGCTCCc | AAGAAAAGCCCCTCGTCC | 538 | 54 | Serotypes 1/2a and 3a |
| GLTb | AAAGTGAGTTCTTACGAGATTT | AATTAGGAAATCGACCTTCT | 483 | 45 | Serotypes 1/2b and 3b |
PCR products run on a 2% agarose gel.
PCR products run on a 1.2% agarose gel.
The underlined nucleotide is a mismatch that was introduced to increase primer specificity.
Strains identified as belonging in Division I or III were further subtyped by using primers designed to differentiate serotypes 4 and 1/2b (Table 1). These primers (hereinafter called GLT primers) were designed from a 1/2b serotype-specific region flanking the gltA-gltB cassette described by Lei et al. (18) (Table 1). Strains identified as serotype 4 were further serotyped for identification of Division III strains using the MAMA-C PCR primers as described by Jinneman and Hill (17). If strains tested positive with these primers, then the serotype was identified as 4a/c. Serovar 4 strains that tested negative were considered serotype 4b, although it is possible that they belong to the relatively rare serotype 4d or 4e.
Strains identified as belonging in Division II were serotyped by using primers designed from the flaA gene, which encodes the L. monocytogenes flagellin protein (Table 1) (10, 25). Sequencing primers were designed from regions of the flaA gene that are conserved between serotypes 4b (strain 12067; GenBank accession no. X65624) and 1/2a (strain EGD; GenBank accession no. AL591824). These primers amplify a region corresponding to nucleotides 738 to 1276 of strain 12067. The primers were used to amplify and sequence a 1.2-kbp region of the flaA gene of seven 1/2a strains and six 1/2c strains. These 13 strains were chosen because microarray analysis had shown them to be genetically diverse (4, 8). A mismatch-containing forward primer was designed on the basis of sequence differences that distinguish serotypes 1/2a and 1/2c and was paired with the reverse sequencing primer. Strains that tested positive with the FlaA primers were considered serotype 1/2a or 3a. Strains testing negative were considered serotype 1/2c or 3c.
DNA extraction.
L. monocytogenes strains were stored long term in brain heart infusion medium with 10% sterile glycerol at −80°C. Strains were recovered by streaking on brain heart infusion agar plates and were grown overnight at 37°C. Cells were scraped and resuspended in 500 μl of 1× Tris-EDTA, vortexed, and boiled for 10 min. The samples were centrifuged, and the supernatant was aliquoted into new microcentrifuge tubes. Samples were then stored at −20°C until used.
PCR amplification.
Amplification reaction mistures contained primers at a concentration of 50 pmol/μl, 1 U of Taq polymerase, 1× reaction buffer (Fisher Scientific, Pittsburgh, Pa.), 0.2 mM each deoxynucleoside triphosphate (Promega, Madison, Wis.), 2.5 mM MgCl2, and 2 μl of template DNA in a 25-μl reaction volume. PCR cycling conditions were as follows: 95°C for 3 min followed by 25 cycles (with D1, D2, and GLT primers) or 30 cycles (FlaA primers) of 95°C for 30 s, 45 to 59°C for 30 s (see Table 1 for primer-specific annealing temperatures), and 72°C for 1 min, followed by a final step of 72°C for 10 min after cycling was completed. The insert size was determined by gel electrophoresis (1.2 to 2% agarose) (see Table 1 for product sizes). MAMA-C PCR amplifications were performed as described previously by Jinneman and Hill (17).
Statistical analysis.
NCSS 2001 software (NCSS Statistical Software, Kaysville, Utah) was used for statistical analysis. Sensitivity and specificity were defined by using standard serotyping as the “gold standard” and were based only on the PCR results from strains that were correctly classified by other primers. For example, only PCR results from 1/2a, 1/2c, 3a, and 3c strains were included in the sensitivity and specificity calculations for the FlaA primer set. Therefore, sensitivity was defined as the number of strains that tested positive for the serotype(s) that the primers were designed to detect (and that were also classified as that serotype by slide agglutination) divided by the number of strains that were classified as this serotype by slide agglutination.
RESULTS
The major phylogenetic division of 122 L. monocytogenes strains was determined by multiplex PCR carried out with primers designed from a region of an iron transport protein (GenBank accession no. BH170518) (Table 1) (Fig. 1). Ninety-nine percent (121 of 122) of strains tested with primer pairs D1 and D2 were typeable, and 98% (118 of 121) of the strains were placed in the correct phylogenetic division (Table 2). Overall, the sensitivities were 100 and 95%, and the specificities were 96 and 100% for the D1 and D2 primer sets, respectively. Listeria innocua (ATCC 51742) and Listeria ivanovii (ATCC 700402) isolates were also tested with the D1 and D2 multiplex primers, with L. innocua testing positive for Division I and L. ivanovii testing negative with both primer sets.
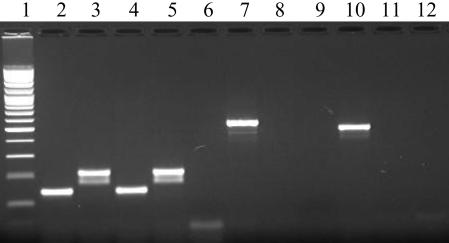
FIG. 1.
PCR amplification results. Lane 1, 100-bp ladder; lanes 2 to 6, D1 and D2 multiplex primers, strains ILSI 3 (1/2a), ILSI 6 (1/2b), H9333 (1/2c), ILSI 1 (4b), and no template control, respectively; lanes 7 to 9, FlaA primers, strains ILSI 3, H9333, and no template control, respectively; lanes 10 to 12, GLT primers, strains ILSI 6, ILSI 1, and no template control, respectively.
TABLE 2.
Summary of PCR resultsa
| Primer | Division II
|
Division I
|
Division III
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1/2a | 1/2c | 3a | 3c | 1/2b | 3b | 4b | 4d | 4a | 4c | |
| D1 | 1/30 | 0/18 | 0/5 | 1/2 | 20/20 | 2/2 | 35/35 | 1/1 | 3/3 | 6/6 |
| D2 | 29/30 | 18/18 | 4/5 | 1/2 | 0/20 | 0/2 | 0/35 | 0/1 | 0/3 | 0/6 |
| FlaAb | 29/29 | 1/18 | 4/4 | 0/1 | NA | NA | NA | NA | NA | NA |
| GLTc | 0/1 | NA | NA | 1/1 | 20/20 | 2/2 | 0/35 | 0/1 | 0/3 | 0/6 |
| MAMA-Cd | NA | NA | NA | NA | NA | NA | 3/35 | 0/1 | 3/3 | 6/6 |
Values indicate the number of positive reactions out of the total number of strains tested.
Only strains testing positive with D2 primers were tested with FlaA primers.
Only strains testing positive with D1 primers were tested with GLT primers.
Only strains testing negative with GLT primers were tested using MAMA-C primers.
Strains testing positive with D1 primers (n = 69) were further subtyped by using GLT primers designed from a 1/2b serotype-specific region flanking the gltA-gltB cassette (Table 1) (18). All 4b strains were negative (n = 35), and all 1/2b strains were positive (n = 20) when tested with these primers (Table 2, Fig. 1). The only 1/2a strain misclassified by the D1 and D2 primers was negative using the GLT primers, as were the 4a (n = 3), 4c (n = 6), and 4d (n = 1) strains. The 3c strain misclassified by the D1 and D2 primers tested positive when the GLT primers were used, as did the two 3b strains. Overall, 100% of strains correctly classified by the D1 and D2 primers as belonging in Division I or III were classified correctly with this primer set. Consequently, the sensitivity was 100% and the specificity was 100% for this primer set.
Samples testing negative with the GLT primers were further subtyped by using the previously described MAMA-C primers (17) for inclusion in Division III. All serotype 4a samples (n = 3), 4c samples (n = 6), 91% (32 of 35) of 4b samples, and the 4d sample were serotyped correctly with these primers (Table 2). The 1/2a strain misclassified by the D1 and D2 primers tested positive. Overall, the sensitivity was 100%, and the specificity was 92% for this primer set.
Samples classified by multiplex PCR as belonging in Division II (n = 52) were further subtyped with primers designed to be specific for serotype 1/2a based on the flaA gene (10, 25). One hundred percent of serotype 1/2a (29 of 29) and 3a (4 of 4) strains tested positive with these primers, as did 6% (1 of 18) of the 1/2c strains (Table 2). Ninety-four percent of the 1/2c strains and the single 3c strain tested negative. The sensitivity was 100% and the specificity was 95% for this primer set.
A concordance analysis was conducted given the null hypothesis that serotype and PCR assays produce similar results. In this case, it was assumed that slide agglutination serotype classification represented the gold standard so that discrepancies between the two methods were attributable to the PCR assay. A McNemar test for paired observations with a discontinuity adjustment was applied for each category (e.g., Division I or II) or serotype (e.g., 1/2b) relevant to the primer set being tested. In all cases, classification of strains into division or serotype using PCR primers was concordant with classification based on conventional serotyping (McNemar test; P ≥ 0.25).
To verify that the assay results were reproducible, all 122 strains were tested in three separate experiments with similar results (range, 98 to 100% reproducibility). A subset of 10 strains was retested independently by a different technician not associated with the project, and reproducibility was 100%.
DISCUSSION
Although the genetic basis for serotype identification is not well defined, genetic analyses indicate that the evolution of somatic and flagellar antigens has paralleled that of many other genes (8, 11, 30). Therefore, we hypothesized that serotyping PCR primers may be designed as an alternative to the slide agglutination method of serotyping. Primers were designed by using genomic regions that had been previously identified as diverse by sequence analysis, monoclonal antibody binding, and/or microarray analysis (4, 8, 18, 25). In particular, microarray subtyping has been previously used to identify regions of the genome that vary between phylogenetic division and/or serotype (4, 8). One probe with sequence similarity to an iron transport protein was identified as being unique to Division I, and this information has allowed the design of division-specific multiplex primers.
Three other probes encoding different proteins involved in cell wall synthesis were identified by microarray (8; M. K. Borucki, unpublished data) and sequenced. Four sets of primers were tested for the ability to distinguish between serotypes within Division I. Results from all four primer sets were comparable to those obtained with the GLT primers (data not shown). However, due to the fact that the gltA-gltB region has been shown to be serotype specific by the use of monoclonal antibodies, we felt that these primers were the most biologically relevant.
In conclusion, PCR primers accurately identified both phylogenetic divisions and serotypes [1/2a(3a), 1/2b, 1/2c(3c), 4b(d,e), and 4a/c]. A panel of 122 L. monocytogenes strains was tested; the sensitivity of the primer sets ranged from 95 to 100%, and specificity ranged from 92 to 100%. For isolates of the four major serotypes (1/2a, 1/2b, 1/2c and 4b), results of the PCR method agreed with those of the slide agglutination method for 97 (29 of 30), 100 (20 of 20), 94 (17 of 18), and 91% (32 of 35) of strains, respectively. Of the primers sets, the MAMA-C primer set was the least accurate (Table 2). This is not surprising, as these primers were designed to differentiate according to division rather than serotype. Additionally, one of the three 4b strains (strain FSL-J1-158) identified using the MAMA-C primers as belonging to Division III has also been classified as such by sequence analysis (7).
PCR is routinely performed in most diagnostic and research laboratories, and these primers should greatly increase the ease of L. monocytogenes serotyping. Future research will focus on the development of PCR primers that distinguish the rare serotypes as well as refining the assay into a more efficient multiplex format.
Acknowledgments
We gratefully acknowledge the excellent technical assistance provided by Edith Orozco, James Reynolds, So Hyun Kim, Marlene Bakko, and Melissa Krug. L. monocytogenes isolates were kindly provided by Peggy Hayes and Lewis Graves (Centers for Disease Control and Prevention), Jinxin Hu (Washington State Department of Health), Karen Jinneman (U.S. Food and Drug Administration), Irene Wesley (USDA Agricultural Research Service-National Animal Disease Center), Franco Pagotto (Health Canada), Lisa Gorski (USDA Agricultural Research Service), and Martin Wiedmann (Cornell University). We thank Don Knowles for his critical review of the manuscript and Todd Ward (USDA Agricultural Research Service) for helpful discussion.
Funding for this work was provided by the USDA Agricultural Research Service (grant no. CWU 5348-32000-017-00D) and the Agricultural Animal Health Program (College of Veterinary Medicine, Washington State University).
REFERENCES
- 1.Aarts, H. J., L. E. Hakemulder, and A. M. Van Hoef. 1999. Genomic typing of Listeria monocytogenes strains by automated laser fluorescence analysis of amplified fragment length polymorphism fingerprint patterns. Int. J. Food Microbiol. 49:95-102. [DOI] [PubMed] [Google Scholar]
- 2.Bibb, W. F., B. Schwartz, B. G. Gellin, B. D. Plikaytis, and R. E. Weaver. 1989. Analysis of Listeria monocytogenes by multilocus enzyme electrophoresis and application of the method to epidemiologic investigations. Int. J. Food Microbiol. 8:233-239. [DOI] [PubMed] [Google Scholar]
- 3.Bibb, W. F., B. G. Gellin, R. Weaver, B. Schwartz, B. D. Plikaytis, M. W. Reeves, R. W. Pinner, and C. V. Broome. 1990. Analysis of clinical and food-borne isolates of Listeria monocytogenes in the United States by multilocus enzyme electrophoresis and application of the method to epidemiologic investigations. Appl. Environ. Microbiol. 56:2133-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borucki, M. K., M. J. Krug, W. T. Muraoka, and D. R. Call. 2003. Discrimination among Listeria monocytogenes isolates using a mixed genome DNA microarray. Vet. Microbiol. 92:351-362. [DOI] [PubMed] [Google Scholar]
- 5.Brosch, R., J. Chen, and J. B. Luchansky. 1994. Pulsed-field fingerprinting of listeriae: identification of genomic divisions for Listeria monocytogenes and their correlation with serovar. Appl. Environ. Microbiol. 60:2584-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchrieser, C., R. Brosch, B. Catimel, and J. Rocourt. 1993. Pulsed-field gel electrophoresis applied for comparing Listeria monocytogenes strains involved in outbreaks. Can. J. Microbiol. 39:395-401. [DOI] [PubMed] [Google Scholar]
- 7.Cai, S., D. Y. Kabuki, A. Y. Kuaye, T. G. Cargioli, M. S. Chung, R. Nielsen, and M. Wiedmann. 2002. Rational design of DNA sequence-based strategies for subtyping Listeria monocytogenes. J. Clin. Microbiol. 40:3319-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Call, D. R., M. K. Borucki, and T. E. Besser. 2003. Mixed-genome microarrays reveal multiple serotype and lineage-specific differences among strains of Listeria monocytogenes. J. Clin. Microbiol. 41:632-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Cesare, A., J. L. Bruce, T. R. Dambaugh, M. E. Guerzoni, and M. Wiedmann. 2001. Automated ribotyping using different enzymes to improve discrimination of Listeria monocytogenes isolates, with a particular focus on serotype 4b strains. J. Clin. Microbiol. 39:3002-3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dons, L., O. F. Rasmussen, and J. E. Olsen. 1992. Cloning and characterization of a gene encoding flagellin of Listeria monocytogenes. Mol. Microbiol. 6:2919-2929. [DOI] [PubMed] [Google Scholar]
- 11.Ericsson, H., H. Unnerstad, J. G. Mattsson, M. L. Danielsson-Tham, and W. Tham. 2000. Molecular grouping of Listeria monocytogenes based on the sequence of the inIB gene. J. Med. Microbiol. 49:73-80. [DOI] [PubMed] [Google Scholar]
- 12.Gilot, P., A. Genicot, and P. Andre. 1996. Serotyping and esterase typing for analysis of Listeria monocytogenes populations recovered from foodstuffs and from human patients with listeriosis in Belgium. J. Clin. Microbiol. 34:1007-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graves, L. M., B. Swaminathan, M. W. Reeves, S. B. Hunter, R. E. Weaver, B. D. Plikaytis, and A. Schuchat. 1994. Comparison of ribotyping and multilocus enzyme electrophoresis for subtyping of Listeria monocytogenes isolates. J. Clin. Microbiol. 32:2936-2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graves, L. M., B. Swaminathan, and S. Hunter. 1999. Subtyping Listeria monocytogenes, p. 279-298. In E. T. Ryser and E. H. Marth (ed.), Listeria, listeriosis, and food safety. Marcel Dekker, Inc., New York, N.Y.
- 15.Graves, L. M., and B. Swaminathan. 2001. PulseNet standardized protocol for subtyping Listeria monocytogenes by macrorestriction and pulsed-field gel electrophoresis. Int. J. Food Microbiol. 65:55-62. [DOI] [PubMed] [Google Scholar]
- 16.Jersek, B., E. Tcherneva, N. Rijpens, and L. Herman. 1996. Repetitive element sequence-based PCR for species and strain discrimination in the genus Listeria. Lett. Appl. Microbiol. 23:55-60. [DOI] [PubMed] [Google Scholar]
- 17.Jinneman, K. C., and W. E. Hill. 2001. Listeria monocytogenes lineage group classification by MAMA-PCR of the listeriolysin gene. Curr. Microbiol. 43:129-133. [DOI] [PubMed] [Google Scholar]
- 18.Lei, X. H., F. Fiedler, Z. Lan, and S. Kathariou. 2001. A novel serotype-specific gene cassette (gltA-gltB) is required for expression of teichoic acid-associated surface antigens in Listeria monocytogenes of serotype 4b. J. Bacteriol. 183:1133-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mazurier, S. I., and K. Wernars. 1992. Typing of Listeria strains by random amplification of polymorphic DNA. Res. Microbiol. 143:499-505. [DOI] [PubMed] [Google Scholar]
- 20.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Norrung, B., and N. Skovgaard. 1993. Application of multilocus enzyme electrophoresis in studies of the epidemiology of Listeria monocytogenes in Denmark. Appl. Environ. Microbiol. 59:2817-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Donoghue, K., K. Bowker, J. McLauchlin, D. S. Reeves, P. M. Bennett, and A. P. MacGowan. 1995. Typing of Listeria monocytogenes by random amplified polymorphic DNA (RAPD) analysis. Int. J. Food Microbiol. 27:245-252. [DOI] [PubMed] [Google Scholar]
- 23.Palumbo, J. D., M. K. Borucki, R. E. Mandrell, and L. Gorski. 2003. Serotyping of Listeria monocytogenes by enzyme-linked immunosorbent assay and identification of mixed-serotype cultures by colony immunoblotting. J. Clin. Microbiol. 41:564-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piffaretti, J. C., H. Kressebuch, M. Aeschbacher, J. Bille, E. Bannerman, J. M. Musser, R. K. Selander, and J. Rocourt. 1989. Genetic characterization of clones of the bacterium Listeria monocytogenes causing epidemic disease. Proc. Natl. Acad. Sci. USA 86:3818-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rasmussen, O. F., P. Skouboe, L. Dons, L. Rossen, and J. E. Olsen. 1995. Listeria monocytogenes exists in at least three evolutionary lines: evidence from flagellin, invasive associated protein and listeriolysin O genes. Microbiology 141:2053-2061. [DOI] [PubMed] [Google Scholar]
- 26.Salcedo, C., L. Arreaza, B. Alcala, L. de la Fuente, and J. A. Vazquez. 2003. Development of a multilocus sequence typing method for analysis of Listeria monocytogenes clones. J. Clin. Microbiol. 41:757-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sciacchitano, C. J. 1998. DNA fingerprinting of Listeria monocytogenes using enterobacterial repetitive intergenic consensus (ERIC) motifs-polymerase chain reaction/capillary electrophoresis. Electrophoresis 19:66-70. [DOI] [PubMed] [Google Scholar]
- 28.Seeliger, H. P., and K. Hohne. 1979. Serotyping of Listeria monocytogenes and related species. Methods Microbiol. 13:31-49. [Google Scholar]
- 29.Tappero, J. W., A. Schuchat, K. A. Deaver, L. Mascola, J. D. Wenger, et al. 1995. Reduction in the incidence of human listeriosis in the United States. Effectiveness of prevention efforts? JAMA 273:1118-1122. [DOI] [PubMed] [Google Scholar]
- 30.Vines, A., M. W. Reeves, S. Hunter, and B. Swaminathan. 1992. Restriction fragment length polymorphism in four virulence-associated genes of Listeria monocytogenes. Res. Microbiol. 143:281-294. [DOI] [PubMed] [Google Scholar]
- 31.Wiedmann, M., J. L. Bruce, C. Keating, A. E. Johnson, P. L. McDonough, and C. A. Batt. 1997. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect. Immun. 65:2707-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]