Abstract
Paracoccidioides brasiliensis isolates from 10 nine-banded armadillos (Dasypus novemcinctus) were comparable with 19 clinical isolates by sequence analysis of the PbGP43 gene and ribosomal internal transcribed spacer 1 (ITS1) and ITS2 and by random amplified polymorphic DNA. In this original ITS study, eight isolates differed by one or three sites among five total substitution sites.
In humans, the thermally dimorphic fungus Paracoccidioides brasiliensis causes paracoccidioidomycosis (PCM), a systemic granulomatous mycosis prevalent in rural areas of Latin American countries. Infection generally occurs by inhalation of conidia, which transform into pathogenic yeasts in the pulmonary alveoli (18). Nine-banded armadillos (Dasypus novemcinctus) have recently been considered a natural reservoir of P. brasiliensis (2, 9, 21, 24, 29). Apparently, different organs of individual armadillos can be infected with P. brasiliensis bearing distinct genotypes and virulence capacities, but the data on genetic polymorphism have so far been restricted to samples isolated from a few animals (25, 26).
Recently, P. brasiliensis strains with typical morphology have been isolated from the spleen, liver, and mesenteric lymph nodes of 10 armadillos captured in the counties of Botucatu, PratÂnia, and Manduri (1, 11) (Fig. 1), located in the area of Botucatu, São Paulo state, Brazil, where PCM is endemic (17). We have shown that these isolates are able to cause PCM infection of various degrees in hamsters (11).
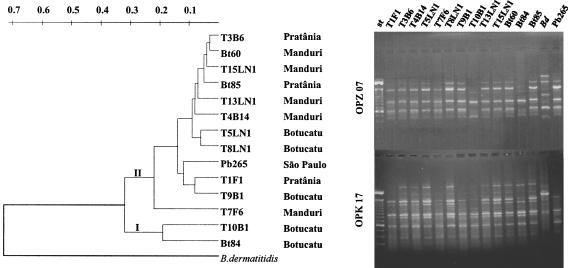
FIG. 1.
(Left) dendrogram of P. brasiliensis isolated from humans (Bt60, Bt84, Bt85, and Pb265) and armadillos (T-named), including T1F1, T3B6, and T4B14 studied before (26), distributed according to a matrix of genetic similarity generated from the analysis of RAPD profiles with 24 primers. The data were assembled by using the neighbor-joining method, with B. dermatitidis (ATCC 26199) as an outgroup, which shared 27% genetic similarity. Main clades I and II and the county of origin are indicated. (Right) representative RAPD profiles (with primers OPZ07 and OPK17). st, 100-pb ladder (strongest band, 600 bp). The distance between the armadillos' burrows has been mapped by using the global positioning system (1). The approximate average distances were 33 km (Botucatu and PratÂnia), 86 km (Botucatu and Manduri), and 62 km (Manduri and PratÂnia). Animals from the same county lived between 0.35 and 9.88 km apart.
These samples have been compared at the DNA sequencing level with clinical isolates Bt60, Bt84, Bt85 (from the same area as shown in Fig. 1), Pb265, and Pb1 to Pb16 (detailed in reference 19) (Pb16 was isolated from soil). Our aim was to distinguish between human and armadillo isolates based on the polymorphism of two loci: the internal transcribed spacer 1 (ITS1) and ITS2 of the ribosomal DNA complex and the PbGP43 gene (8), both already used in the identification of P. brasiliensis by PCR (4, 10, 12, 27, 28). PbGP43 (1,329-bp long, with one 78-bp intron) encodes the major gp43 fungal antigen (8, 23, 30), and its polymorphism has been previously characterized by using Pb1 to Pb16 (19). The ITS region has been successfully used for typing of pathogenic fungi (13), including Histoplasma capsulatum (14), which is genetically related to P. brasiliensis, as inferred from 18S and ITS analysis (3, 22). Our interest was to verify its usefulness in P. brasiliensis intraspecific differentiation.
Fungal isolates were maintained as yeasts at 35°C (11, 19); DNA extraction was carried out by using a glass beads protocol (31) or as previously described (7, 19) for Pb1 to Pb16. PbGP43 exon 2 was PCR-amplified according to standard protocols with specific primers 5′-TCATCTCACGTCGCATCTCACATT-3′ (sense) and 5′-GGCTCCTCAAAGTCTGCCATGAGGAAG-3′ (antisense), which extend from nucleotide 733 to nucleotide 1,213 (8). Universal primers ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) and ITS5 (5′-GGAAGTAAAAGTCGTAACAAGG-3′) were used to generate a 634-bp PCR product that included the ITS1, 5.8S, and ITS2. The fragments were purified through MicroSpin S-400 HR columns (Amersham Pharmacia) and used as templates in sequencing reactions (DYEnamic Terminator Cycle Sequencing kit; Amersham Pharmacia). Both strands were sequenced in an ABI model 373A automated sequencer. For Pb1 to Pb16, the procedures for DNA amplification and sequencing of cloned fragments are detailed elsewhere (19).
A previous study (19) defined the existence of six PbGP43 genotypes based on the distribution of 21 substitution sites, which occurred mostly in exon 2 (nucleotides 578 to 1166), generating predominantly nonsynonymous amino acid changes. The partial PbGP43 sequences obtained in this work matched three of these genotypes, and most of them belonged in groups E and F (Table 1). Lymph node (LN-numbered) isolates seemed to fit preferentially in group E, while those from spleen (B-numbered) and liver (F-numbered) were mostly in group F. In a previous study (25), PbGP43 sequences of three isolates from organs of the same armadillo were similar to those of either group F (spleen) or B (liver and lymph node).
TABLE 1.
P. brasiliensis isolates grouped according to nucleotide substitution sites in a consensus PbGP43 sequencea
| Group | PbGP43 substitution sitesb | Isolate(s) from indicated study
|
|
|---|---|---|---|
| Previousc | Present | ||
| A | 268, 578, 617, 628, 751, 763, 799, 830, 856, 872, 981, 1082, 1086, 1157, 1166 | Pb2, Pb3, Pb4 | T10F1, Bt84 |
| B | 617, 799, 821, 852 | Pb15, Pb16 | None |
| C | 617, 799 | Pb1 | None |
| D | 589 | Pb5, Pb10, Pb11 | None |
| E | 874, 965 | Pb6, Pb7, Pb8, Pb14 | T5LN1, T9B1, T13LN1, T15LN1 |
| F | 874, 965, 1143 | Pb9, Pb12, Pb13 | T1F1, T3B6, T4B14, T7F6, T8LN1, Bt60 |
The PbGP43 sequence was reported by Morais et al. (19). The sequences obtained in the present work covered only the sites shown in bold (part of exon 2). See reference 19 for details about the nucleotide substitutions, corresponding amino acid changes, and extensive discussion about the gp43 isoforms.
The nucleotide numbers are the same as those used by Cisalpino et al. (8).
We found two polymorphic sites in ITS1 and three in ITS2 (Table 2), but 73% of the sequences were identical to the consensus, which matched that previously deposited in GenBank (accession no. AF38360). The 5.8S subunit was conserved. Site 518 (A to G) was mutated in five isolates and could be defining two P. brasiliensis genetic groups. However, a maximum-likelihood phylogenetic tree had weak bootstrap support and is not presented. Note that the ITS primer pair previously suggested for amplification of a fragment specific for P. brasiliensis (12) does not contain any polymorphic sites.
TABLE 2.
Polymorphic nucleotides found in the ribosomal ITS1 and ITS2 sequences from 30 P. brasiliensis isolatesa
| Siteb | Consensusc | Isolate
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Pb10 | Pb8 | Pb12 | Pb13 | Pb4 | T10B1 | Bt84 | Pb2 | ||
| 113 | G | A | A | ||||||
| 156 | G | T | T | ||||||
| 384 | C | T | T | ||||||
| 518 | A | G | G | G | G | G | |||
| 554 | T | C | |||||||
Twenty-two sequences corresponded to the consensus. Sequence polymorphisms were analyzed using the Megalign program of the Lasergene System (DNAstar Inc.).
Site 1 corresponds to the first base of ITS1.
The isolates with sequence consensus (same as GenBank accession no. AF38360) were Pb1, Pb3, Pb5, Pb6, Pb7, Pb9, Pb11, Pb14, Pb15, Pb16, T1F1, T3B6, T4B14, T5LN1, T7F6, T8LN1, T9B1, T13LN1, T15LN1, Bt60, Bt85, and Pb265.
The PbGP43 sequences of T10B1 and Bt84 were similar to that of Pb4, which together with those of Pb2 and Pb3 were phylogenetically distant from the others in a maximum-likelihood tree (19). These sequences are highly polymorphic and translate peculiarly basic gp43 isoforms, which contain some differential antibody epitopes (19, 20). Among the five isolates presently known to encode basic gp43 (Table 1), four (Bt84/Pb2, Pb4/T10B1) had polymorphic ITS of two different patterns (Table 2). Moreover, T10B1 and Bt84, both from Botucatu, shared the same clade I in a dendrogram resulting from random amplified polymorphic DNA (RAPD) analysis (Fig. 1). T10B1 was significantly more aggressive than the other armadillo isolates in the hamster experimental model, killing the animals after 2 weeks of intratesticular infection (11).
The biggest clade II of the RAPD tree assembled most of the armadillo isolates tested (Fig. 1), which did not carry any mutation in ITS (Table 1) and had similar PbGP43 genotypes (Table 2). Our RAPD analysis, carried out as previously described (6) in a thermocycler (MJ Research, Inc.,Waltham, Mass.) with 24 random primers (Operon Technology), originally aimed at distinguishing the isolates geographically, as previously reported for P. brasiliensis (6). Indeed, their distribution into branches seemed to correlate with the county of origin.
In this communication, we showed the first genetic analysis of P. brasiliensis from a large number of armadillos and confirmed their similarity with clinical isolates by DNA sequencing. We showed the first sequence comparison of the ITS1 and ITS2 regions from many isolates, among which eight differed by one or three sites among five total substitution sites. Our results suggest the existence of two genetic groups, since those defined by PbGP43 and RAPD analyses did not necessarily coincide with the ITS groups. We believe that both PbGP43 and ITS will be useful in further genetic studies of P. brasiliensis similar to those that revealed intraspecific genetic groups and cryptic sex in Coccidioides immitis and H. capsulatum (5, 15, 16).
Nucleotide sequence accession numbers.
The GenBank accession numbers for the new ITS sequences are AY374336 to AY374339.
Acknowledgments
This work was supported by FAPESP, PRONEX/CNPq, CAPES, FIRCA, and CODI.
We thank J. W. Taylor for sequencing facilities and suggestions; J. B. Pesquero for accessibility to automated sequencing; R. C. Fenille for technical support; M. F. Sugizaki, A. Sano, Z. P. Camargo, A. Restrepo, and T. I. E. Svidzinski for providing fungal isolates; T. Kasuga for discussion; and A. Restrepo for critically reviewing the manuscript.
REFERENCES
- 1.Bagagli, E., M. Franco, S. M. G. Bosco, F. Hebeler-Barbosa, L. A. Trinca, and M. R. Montenegro. 2003. High frequency of Paracoccidioides brasiliensis infection in armadillos (Dasypus novemcinctus): an ecological study. Med. Mycol. 41:217-223. [DOI] [PubMed] [Google Scholar]
- 2.Bagagli, E., A. Sano, K. I. Coelho, S. Alquati, M. Miyaji, Z. P. de Camargo, G. M. Gomes, M. Franco, and M. R. Montenegro. 1998. Isolation of Paracoccidioides brasiliensis from armadillos (Dasypus novemcinctus) captured in an endemic area of paracoccidioidomycosis. Am. J. Trop. Med. Hyg. 58:505-512. [DOI] [PubMed] [Google Scholar]
- 3.Bialek, R., A. Ibricevic, A. Fothergill, and D. Begerow. 2000. Small subunit ribosomal DNA sequence shows Paracoccidioides brasiliensis closely related to Blastomyces dermatitidis. J. Clin. Microbiol. 38:3190-3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bialek, R., A. Ibricevic, C. Aepinus, L. K. Najvar, A. W. Fothergill, J. Knobloch, and J. R. Graybill. 2000. Detection of Paracoccidioides brasiliensis in tissue samples by a nested PCR assay. J. Clin. Microbiol. 38:2940-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burt, A., D. A. Carter, G. L. Koenig, T. J. White, and J. W. Taylor. 1996. Molecular markers reveal cryptic sex in the human pathogen Coccidioides immitis. Proc. Natl. Acad. Sci. USA 93:770-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calcagno, A. M., G. Niño-Vega, F. San Blas, and G. San Blas. 1998. Geographic discrimination of Paracoccidioides brasiliensis strains by randomly amplified polymorphic DNA analysis. J. Clin. Microbiol. 36:1733-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cisalpino, P. S., J. F. Silveira, and L. R. Travassos. 1994. RNA and DNA isolation from Paracoccidioides brasiliensis yeast cells: establishment of cDNA genomic libraries and PCR amplification, p. 461-467. In B. Maresca and G. S. Kobayashi (ed.), Molecular biology of pathogenic fungi, a laboratory manual. Telos Press, New York, N.Y.
- 8.Cisalpino, P. S., R. Puccia, L. M. Yamauchi, M. I. Cano, J. F. da Silveira, and L. R. Travassos. 1996. Cloning, characterization, and epitope expression of the major diagnostic antigen of Paracoccidioides brasiliensis. J. Biol. Chem. 271:4553-4560. [DOI] [PubMed] [Google Scholar]
- 9.Corredor, G. G., J. H. Castano, A. Peralta, S. Díez, M. Arango, J. McEwen, and A. Restrepo. 1999. Isolation of Paracoccidioides brasiliensis from the nine-banded armadillo Dayspus novemcinctus in an endemic area for paracoccidioidomycosis in Colombia. Rev. Iberoam. Micol. 19:216-220. [PubMed] [Google Scholar]
- 10.Gomes, G. M., P. S. Cisalpino, C. P. Taborda, and Z. P. de Camargo. 2000. PCR for diagnosis of paracoccidioidomycosis. J. Clin. Microbiol. 38:3478-3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hebeler-Barbosa, F., M. R. Montenegro, and E. Bagagli. 2003. Virulence profiles of ten Paracoccidioides brasiliensis isolates obtained from armadillos (Dasypus novemcinctus). Med. Mycol. 41:89-96. [DOI] [PubMed] [Google Scholar]
- 12.Imai, T., A. Sano, Y. Mikami, K. Watanabe, F. H. Aoki, M. L. Branchini, R. Negroni, K. Nishimura, and M. Miyaji. 2000. A new PCR primer for the identification of Paracoccidioides brasiliensis based on rRNA sequences coding the internal transcribed spacers (ITS) and 5.8S regions. Med. Mycol. 38:323-326. [DOI] [PubMed] [Google Scholar]
- 13.Iwen, P. C., S. H. Hinrichs, and M. E. Rupp. 2002. Utilization of the internal transcribed spacer regions as molecular targets to detect and identify human fungal pathogens. Med. Mycol. 40:87-109. [DOI] [PubMed] [Google Scholar]
- 14.Jiang, B., M. S. Bartlett, S. D. Allen, J. W. Smith, L. J. Wheat, P. A. Connolly, and C. H. Lee. 2000. Typing of Histoplasma capsulatum isolates based on nucleotide sequence variation in the internal transcribed spacer regions of rRNA genes. J. Clin. Microbiol. 38:241-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kasuga, T., J. W. Taylor, and T. J. White. 1999. Phylogenetic relationships of varieties and geographical groups of the human pathogenic fungus Histoplasma capsulatum Darling. J. Clin. Microbiol. 37:653-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koufopanou, V., A. Burt, and J. W. Taylor. 1997. Concordance of gene genealogies reveals reproductive isolation in the pathogenic fungus Coccidioides immitis. Proc. Natl. Acad. Sci. USA 94:5478-5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marques, S. A., M. F. Franco, R. P. Mendes, N. C. Silva, C. Baccili, E. D. Curcelli, A. C. Feracin, C. S. Oliveira, J. V. Tagliarini, and N. L. Dillon. 1983. Epidemiologic aspects of paracoccidioidomycosis in the endemic area of Botucatu (São Paulo-Brazil). Rev. Inst. Med. Trop. São Paulo 25:87-92. [PubMed] [Google Scholar]
- 18.McEwen, J. G., B. I. Restrepo, M. E. Salazar, and A. Restrepo. 1987. Nuclear staining of Paracoccidioides brasiliensis conidia. J. Med. Vet. Mycol. 25:343-345. [PubMed] [Google Scholar]
- 19.Morais, F. V., T. F. Barros, M. K. Fukada, P. S. Cisalpino, and R. Puccia. 2000. Polymorphism in the gene coding for the immunodominant antigen gp43 from the pathogenic fungus Paracoccidioides brasiliensis. J. Clin. Microbiol. 38:3960-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moura-Campos, C., J. L. Gesztesi, A. P. Vincentini, J. D. Lopes, and Z. P. Camargo. 1995. Expression and isoforms of gp43 in different strains of Paracoccidioides brasiliensis. J. Med. Vet. Mycol. 33:223-227. [DOI] [PubMed] [Google Scholar]
- 21.Naiff, R. D., L. C. Ferreira, T. V. Barrett, M. F. Naiff, and J. R. Arias. 1986. Enzootic paracoccidioidomycosis in armadillos (Dasypus novemcinctus) in the State of Pará. Rev. Inst. Med. Trop. São Paulo 28:19-27. [DOI] [PubMed] [Google Scholar]
- 22.Peterson, S. W., and L. Sigler. 1998. Molecular genetic variation in Emmonsia crescens and Emmonsia parva, etiologic agents of adiaspiromycosis, and their phylogenetic relationship to Blastomyces dermatitidis (Ajellomyces dermatitidis) and other systemic fungal pathogens. J. Clin. Microbiol. 36:2918-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Puccia, R., and L. R. Travassos. 1991. 43-kilodalton glycoprotein from Paracoccidioides brasiliensis: immunochemical reactions with sera from patients with paracoccidioidomycosis, histoplasmosis, or Jorge Lobo's disease. J. Clin. Microbiol. 29:1610-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Restrepo, A., J. G. McEwen, and E. Castañeda. 2001. The habitat of Paracoccidioides brasiliensis: how far from solving the riddle? Med. Mycol. 39:233-241. [DOI] [PubMed] [Google Scholar]
- 25.Sano, A., J. Defaveri, R. Tanaka, K. Yokoyama, N. Kurita, M. Franco, K. I. Coelho, E. Bagagli, M. R. Montenegro, M. Miyaji, and K. Nishimura. 1999. Pathogenicities and GP43kDa gene of three Paracoccidioides brasiliensis isolates originated from a nine-banded armadillo (Dasypus novemcinctus). Mycopathologia 144:61-65. [DOI] [PubMed] [Google Scholar]
- 26.Sano, A., R. Tanaka, K. Yokoyama, M. Franco, E. Bagagli, M. R. Montenegro, Y. Mikami, M. Miyaji, and K. Nishimura. 1999. Comparison between human and armadillo Paracoccidioides brasiliensis isolates by random amplified polymorphic DNA analysis. Mycopathologia 143:165-169. [DOI] [PubMed] [Google Scholar]
- 27.Sano, A., K. Yokoyama, M. Tamura, Y. Mikami, I. Takahashi, K. Fukushima, M. Miyaji, and K. Nishimura. 2001. Detection of gp43 and ITS1-5.8S-ITS2 ribosomal RNA genes of Paracoccidioides brasiliensis in paraffin-embedded tissue. Nippon Ishinkin. Gakkai Zasshi. 42:23-27. [DOI] [PubMed] [Google Scholar]
- 28.Semighini, C. P., Z. P. de Camargo, R. Puccia, M. H. Goldman, and G. H. Goldman. 2002. Molecular identification of Paracoccidioides brasiliensis by 5′ nuclease assay. Diagn. Microbiol. Infect. Dis. 44:383-386. [DOI] [PubMed] [Google Scholar]
- 29.Silva-Vergara, M. L., R. Martinez, Z. P. Camargo, M. H. Malta, C. M. Maffei, and J. B. Chadu. 2000. Isolation of Paracoccidioides brasiliensis from armadillos (Dasypus novemcinctus) in an area where the fungus was recently isolated from soil. Med. Mycol. 38:193-199. [DOI] [PubMed] [Google Scholar]
- 30.Travassos, L. R., C. Taborda, L. K. Iwai, E. Cunha-Neto, and R. Puccia. 2003. The gp43 from Paracoccidioides brasiliensis: a major diagnostic antigen and vaccine candidate, p. 279-296. In J. E. Domer and G. S. Kobayashi (ed.), Mycota XII, human fungal pathogens. Springer-Verlag, Berlin, Germany.
- 31.van Burik, J. A., R. W. Schreckhise, T. C. White, R. A. Bowden, and D. Myerson. 1998. Comparison of six extraction techniques for isolation of DNA from filamentous fungi. Med. Mycol. 36:299-303. [PubMed] [Google Scholar]