Abstract
Coxiella burnetii is a Gram-negative obligate intracellular pathogen and the causative agent of Q fever in humans. Q fever causes acute flu-like symptoms and may develop into a chronic disease leading to endocarditis. Its potential as a bioweapon has led to its classification as a category B select agent. An effective inactivated whole-cell vaccine (WCV) currently exists but causes severe granulomatous/necrotizing reactions in individuals with prior exposure, and is not licensed for use in most countries. Current efforts to reduce or eliminate the deleterious reactions associated with WCVs have focused on identifying potential subunit vaccine candidates. Both humoral and T cell-mediated responses are required for protection in animal models. In this study, nine novel immunogenic C. burnetii proteins were identified in extracted whole-cell lysates using 2D electrophoresis, immunoblotting with immune guinea pig sera, and tandem MS. The immunogenic C. burnetii proteins elicited antigen-specific IgG in guinea pigs vaccinated with whole-cell killed Nine Mile phase I vaccine, suggesting a T cell-dependent response. Eleven additional proteins previously shown to react with immune human sera were also antigenic in guinea pigs, showing the relevance of the guinea pig immunization model for antigen discovery. The antigens described here warrant further investigation to validate their potential use as subunit vaccine candidates.
INTRODUCTION
Coxiella burnetii is an obligate intracellular Gram-negative bacterium that resides and replicates within phagolysosomes (Voth & Heinzen, 2007). C. burnetii infections may be asymptomatic or present as an influenza-like illness in humans termed acute Q fever (Maurin & Raoult, 1999). Acute Q fever is generally self-limiting, but a chronic, latent infection may persist for years, leading to chronic Q fever or endocarditis in response to reactivation-triggering events such as immunosuppression (Maurin & Raoult, 1999). Mortality rates in the general population from Q fever are 1–2 % with a much higher incidence of death associated with pregnant women and individuals with preexisting valvulopathies (Maurin & Raoult, 1999). Domestic animals, arthropods and humans all serve as reservoirs for C. burnetii, which is shed in milk, urine, faeces and during parturition (Arricau Bouvery et al., 2003; Maurin & Raoult, 1999). The primary vector of transmission to humans appears to be contaminated sheep, cattle and goats. C. burnetii is highly infectious (infective dose <10), produces variants that are extremely stable in the environment, and is easily disseminated and transmitted by aerosols and dust particles. The United States Center for Disease Control has deemed C. burnetii a potential bioterrorism threat and classified it as a category B select agent.
C. burnetii has an LPS layer that undergoes phase variation upon repeated serial passage analogous to the smooth LPS to rough LPS transition of Gram-negative enteric bacteria (Hackstadt et al., 1985). The transition to truncated smooth phase II LPS has been attributed to an 18 kb genomic deletion (Vodkin & Williams 1986). Products of genes associated with the deleted region possess homology to proteins known to be involved in sugar conversions and the synthesis of LPS (Hoover et al., 2002). The difference in virulence between phase I (virulent) and phase II (avirulent) clonal variants appears to be due to the presence of terminal O-antigen sugars in phase I organisms that are absent in phase II clones (Hackstadt et al., 1985). Inactivated whole-cell C. burnetii phase I vaccine preparations are effective at protecting vaccinees from subsequent C. burnetii infection, while inactivated whole-cell phase II preparations are not. A formalin-inactivated whole-cell phase I vaccine is commercially available (Q-Vax), but no vaccine for Coxiella is currently licensed for use in the USA. Protective whole-cell vaccines (WCVs) may produce adverse reactions that include delayed-type hypersensitivity/necrotizing reactions at the site of injection in individuals with prior exposure to C. burnetii, and the risk of adverse reactions increases with subsequent immunizations. A chloroform : methanol residue vaccine (CMRV) of C. burnetii phase I cells has been found to confer protection against subsequent C. burnetii challenge in a number of animal models (Williams & Cantrell, 1982; Waag et al., 1997, 2002) and reduces the adverse reactions associated with the WCV (Ascher et al., 1983). The adverse reactions from this extracted preparation are still considered too severe for licensure in the USA. Since a CMRV reduces the severity of reactions yet still induces a protective immune response, it is plausible that an effective subunit vaccine can be developed to further reduce or eliminate these adverse reactions.
Vaccination with a WCV or CMRV induces T cell proliferation, seroconversion and protection against lethal challenge (Williams et al., 1986; Waag et al., 1997, 2002). Protein components and not LPS are responsible for T cell proliferation (Izzo et al., 1991). Protective immunity against Coxiella can be transferred to naïve animals by CD4+ T cells or serum from WCV- or CMRV-vaccinated animals (Andoh et al., 2007; Zhang et al., 2007). The mechanism of antibody-mediated immunity to C. burnetii infections is not clear but does not involve activating Fc receptors or complement (Shannon et al., 2009).
Guinea pigs infected with C. burnetii consistently mimic clinical and pathological symptoms seen in human acute Q fever, exhibiting enlarged spleen and lymph nodes, granulomas in liver, bone marrow and spleen, detectable bacteraemia, fever, and similar dose/response effects (Maurin & Raoult, 1999; Russell-Lodrigue et al., 2006). The robustness of the guinea pig model has also demonstrated differences in virulence between C. burnetii genogroups (Russell-Lodrigue et al., 2009). Importantly, guinea pigs display consistent lethal dose–responses to aerosol challenge (Waag et al., 1997) but are protected by WCV vaccination (Waag et al., 2002), which results in intense delayed-type hypersensitivity and dermal granulomatous reactions (Ascher et al., 1983).
This study was conducted to identify and further expand the number of C. burnetii Nine Mile proteins that generate a T cell-dependent IgG response in the guinea pig model. Newly identified immunogenic proteins may be of value in developing a subunit vaccine against C. burnetii infection as well as serodiagnostic targets.
METHODS
Growth and preparation of C. burnetii RSA 493 Nine Mile phase I whole-cell killed vaccine (WCKI).
C. burnetii NMI RSA 493 (phase I) was grown in embryonated chicken yolk sacs and purified by differential sedimentation using sucrose density gradients, as described by Hendrix & Mallavia (1984). Purified phase I cells were inactivated by electron beam irradiation with a minimum 10 kGy dose and tested for non-viability in a severe combined immunodeficiency (SCID) mouse model as described previously (Andoh et al., 2003), and used for 2D analysis and for WCKI vaccination of guinea pigs. C. burnetii RSA 493 Nine Mile phase I (clone 7) and its phase II clone RSA 439 (clone 4) were grown in mouse L929 fibroblast cell cultures and purified by gradient centrifugation, as reported by Williams et al. (1981). Phase I, but not phase II, cells were similarly inactivated by irradiation and tested for non-viability in SCID mice. For all sources of bacteria, bacterial concentrations were determined as dry weight of organisms, and bacteria were resuspended in PBS containing 0.25 M sucrose and used for 2D analysis.
Animals, immunization and serum collection.
Six female Hartley outbred guinea pigs were obtained from Charles River Laboratories. WCKI was emulsified in a 1 : 1 ratio of incomplete Freund's adjuvant (IFA) (Sigma) and 0.9 % sterile NaCl. Three animals were each immunized with 30 μg WCKI delivered subcutaneously into each rear flank at a concentration of 15 μg 125 μl−1. Three different animals received 125 μl emulsified IFA and 0.9 % sterile NaCl (1 : 1) delivered subcutaneously into each rear flank (sham-immunized). Animals were boosted twice more at 2 week intervals using their respective protocol. Three days after the final boost, animals were euthanized with 400 μl Beuthanasia-D (Schering-Plough) delivered intraperitoneally and exsanguinated to collect serum. Following centrifugation, individual sera were stored at −20 °C as frozen aliquots. Prior to immunoblotting, the sera from the three WCKI-immunized guinea pigs were pooled using equal volumes (immune sera). Similarly, the sera from the three sham-immunized guinea pigs were pooled using equal volumes (sham sera). The same pooled immune sera and pooled sham sera were used throughout this study. All protocols involving animals were reviewed and approved by the Washington State University Institutional Animal Care and Use Committee.
Whole-cell C. burnetii protein extraction and preparation for 2D electrophoresis.
C. burnetii phase I or phase II cells (6 mg dry weight equivalent) in PBS with 0.25 M sucrose (0.25 M SP buffer) were centrifuged at 6000 g for 10 min at room temperature to pellet cells. The supernatant was discarded and pelleted cells were washed by gently resuspending them in cold Tris-buffered saline (TBS; 50 mM Tris/HCl, 150 mM NaCl, pH 7.4) and recentrifugation. Pelleted cells were resuspended in 250 μl Bio-Rad Ready Prep Sequential Extraction Buffer #1 (40 mM Tris base, pH 8.8) containing 60 U benzonase (Sigma-Aldrich), 1× Halt proteinase inhibitor (Thermo Scientific) and 2 mM MgCl2, and were sonicated with four 30 s pulses at 200 W. Sonicated cells were vortexed for 2 min, incubated for 3 min at room temperature, and this was repeated four times. Samples were then centrifuged at 20 800 g for 15 min to pellet insoluble material. Supernatant was collected and transferred to a fresh microfuge tube. Sequential Extraction Buffer #3 (150 μl) (Bio-Rad) containing 2 mM of the reducing agent tributyl phosphine (TBP) was added to the pelleted unsolubilized material. The tube was then vortexed for 2 min, incubated at room temperature for 3 min, and this was repeated four times. Tubes were spun at 20 800 g for 15 min at 20 °C. The supernatant was collected and pooled with the previous supernatant. The pelleted unsolubilized material was reextracted two more times using 125 μl aliquots of Sequential Extraction Buffer #3 with TBP following the same procedure and pooling all supernatants. The pooled supernatant extract was directly reduced and alkylated using the ReadyPrep Reduction-Alkylation kit (Bio-Rad) without prior 2D clean-up. The reduced and alkylated extract was aliquoted equally into six microfuge tubes and cleaned using the Bio-Rad ReadyPrep 2-D Cleanup kit according to the kit instructions. Cleaned pellets were resuspended in fresh rehydration buffer [5 M urea, 2 M thiourea, 2 mM TBP, 2 % (w/v) CHAPS and 2 % (w/v) 3-(decyldimethyl-ammonio) propanesulfonate inner salt (detergent SB 3-10, Sigma)] with vortexing and combined into one microfuge tube (total volume 750 μl). The extracted protein concentration was determined using the Bio-Rad RC/DC protein assay.
2D Electrophoresis.
pH 5–8 or pH 3–10 linear gradient ReadyStrip immobilized pH gradient (IPG) strips (11 cm, Bio-Rad) were passively rehydrated overnight with 250 μg whole-cell extracted protein in a volume of 200 μl rehydration buffer supplemented with 0.2 % (w/v) Bio-Lyte 3–10 ampholytes, 0.001 % (w/v) bromophenol blue, and 2 mM TBP, as per the IPG strip instruction manual. Electrophoresis of IPG strips was conducted on a Protean IEF Cell (Bio-Rad) for a total of 40 000 V h. Focused IPG strips were removed and equilibrated in SDS-PAGE buffer, utilizing the reduction/alkylation procedure described in the IPG strip manual. Second-dimension electrophoresis was done on Criterion 10.5–14 % Tris/HCl acrylamide gels (Bio-Rad). Gels for protein spot harvesting and liquid chromatography-tandem MS (LC-MS/MS) analysis were fixed in 10 % methanol, 7 % acetic acid for 45 min, washed briefly in distilled H2O, and stained overnight in Sypro Ruby gel stain (Bio-Rad) with orbital shaking. Stained gels were destained in 10 % methanol, 7 % acetic acid for 1 h, washed in distilled H2O and imaged with Quantity One software on a Pharos FX imager (Bio-Rad), and the gel images were printed. Gels were stored covered and in distilled H2O at 4 °C.
Immunoblotting and protein spot harvesting.
Parallel gels for immunoblotting were transferred to PVDF membranes (0.45 μm, Perkin Elmer) using cold Tris-glycine 20 % methanol transfer buffer. PVDF membranes were dried overnight, stained with Sypro Ruby blot stain (Bio-Rad) according to the manufacturer's instructions and imaged with Quantity One software on a Pharos FX imager (Bio-Rad), and the blot images were printed. Immunoblots were then blocked in blocking buffer [0.2 % I Block (Tropix) and 0.1 % Tween 20 in PBS] for 1 h. Blots were probed in blocking buffer with a 1 : 200 dilution of pooled immune sera or a 1 : 100 dilution of pooled sham sera, incubated for 1.5 h, and washed four times for 5 min each in blocking buffer. Blots were incubated for 1 h with peroxidase-conjugated rabbit anti-guinea pig IgG (whole molecule) (Sigma) diluted 1 : 15 000 in blocking buffer and washed five times for 5 min each followed by two quick washes in distilled H2O. Blots were developed using ECL Western Blotting Substrate (Pierce) and exposed to Biomax XAR films (Kodak).
2D Gel and immunoblot spot matching.
Exposed film was overlaid and aligned on the matching blot and immunoreactive spots were outlined by perforating with a 28 gauge needle. Perforated spots were excised using a scalpel. Excised blots were rescanned with Quantity One software on a Pharos FX imager and printed. Excised blots produce a white void where the excised blot pieces were removed that facilitates identifying immunoreactive spots on the stained blot by overlaying the stained blot image on the excised blot image on a standard light box. Immunoreactive spots on the stained blot image can then be precisely identified. PDQuest version 8.1 (Bio-Rad) 2D analysis software was used to align Sypro Ruby-stained blot images and stained gel images to match spots. Images were warped and spot centres were fitted by Gaussian modelling. Spots were normalized using the local regression model. Spots were matched using the PDQuest match tool. Neighbouring spot patterns and spot intensities were also examined using the PDQuest 3D analysis tool for additional confirmation of correct spot matching. Matched spots were identified and marked on corresponding printed gel images and used as a reference document when manually excising gel spots. Sypro Ruby-stained gels were made to fluoresce using a UV light box, and corresponding immunoreactive spots were removed using a number 11 scalpel and transferred to sterile microfuge tubes. Gel pieces were briefly centrifuged and liquid removed, and then stored at −20 °C until digested with trypsin.
In-gel trypsin digestion.
Gel pieces were washed in 100 μl 50 % acetonitrile (ACN), 50 % NH4HCO3 (pH 8.0) and centrifuged, and the wash solution was discarded. Gel pieces were dehydrated in 300 μl 100 % ACN and centrifuged, and ACN was removed, and gel pieces were air-dried for 5 min. Dehydrated gel pieces were rehydrated in a 3 μl solution of sequencing-grade modified trypsin (Promega) in 50 mM NH4HCO3 (50 μg ml−1) for 15 min. An additional 20 μl of 50 mM NH4HCO3 was then added to cover and fully swell gel pieces, which were incubated stationary for 16 h at 37 °C. Digestion reactions were vortexed for 3 min and centrifuged, and supernatants were transferred to sterile microfuge tubes and placed at 4 °C. Gel pieces were extracted with 0.1 % trifluoroacetic acid (TFA) for 45 min at 37 °C, vortexed for 4 min and centrifuged, and supernatants were pooled with previously collected supernatant. Gel pieces were similarly sequentially reextracted with 30 % ACN, 0.1 % TFA for 45 min and 60 % ACN for 30 min. All corresponding supernatants were pooled and dried down to approximately 20 μl in a SpeedVac concentrator.
MS and sequence analysis of tryptic peptide fragments.
LC-MS/MS analysis of tryptic peptides performed at the Environmental Biotechnology Institute, University of Idaho, was as described by Bansal et al. (2009). Briefly, solvent A was 0.1 % formic acid in water and solvent B was 0.1 % formic acid in ACN. Peptides were trapped on a 180 μm×20 mm Symmetry C18 5 μm trap column (Waters) prior to injection into a 75 μm×200 mm BEH 130 C18 nanoACQUITY Ultra Performance Liquid Chromatograph column (Waters). Flow rate was 0.4 μl min−1 and starting solvent concentrations were 95 % A and 5 % B with an increase in B concentration to 50 % by 44 min. Solvent B was then increased to 90 % over 5 min and held for an additional 5 min, then returned to 5 % over the next 5 min. A Micromass Q-TOF Premier tandem mass spectrometer (Waters) using electrospray ionization generated peptide sequence data. Raw peptide data files were converted to peak list files and uploaded to our local Mascot 2.1.0 (Matrix Science) server and searched against the National Center for Biotechnology Information (NCBI) non-redundant all entries database (20090217) and the annotated C. burnetii RSA 493 complete genome database (Seshadri et al., 2003).
Tryptic peptide analysis by LC-MS/MS at the Laboratory for Biotechnology and Bioanalysis 2 at Washington State University was as described by Lopez et al. (2005) with peptide separations as modified by Noh et al. (2008). Briefly, an Esquire HCT electrospray ion trap (Bruker Daltonics) with a C18 PepMap, 300 μm×1 mm, 5 μm trap column (LC Packings/Dionex) was used with an Ultimate Nano high pressure liquid chromatography system (LC Packings/Dionex). Peptides were separated on a polystyrene-divinylbenzene (PS-DVB) monolithic column (LC Packings/Dionex). Solvent A was 0.1 % formic acid, 3 % ACN. Solvent B was 0.1 % formic acid, 95 % ACN. The flow rate was 800 nl min−1 with a solvent A to B linear gradient change that reached 65 % solvent B at 95 min and then 100 % solvent B at 95.1 min, and held at 100 % solvent B until 115 min. Data were acquired in Bruker Auto (MS2) mode. Peptide sequence files were uploaded to our internal Mascot 2.1.0 (Matrix Science) server and searched against the NCBI non-redundant database (20090217) limited to lobed-finned fish and tetrapod clade, Mus and/or human, and the annotated C. burnetii RSA 493 complete genome database (Seshadri et al., 2003).
RESULTS
2D Electrophoresis and immunoblotting of phase I whole-cell extracts
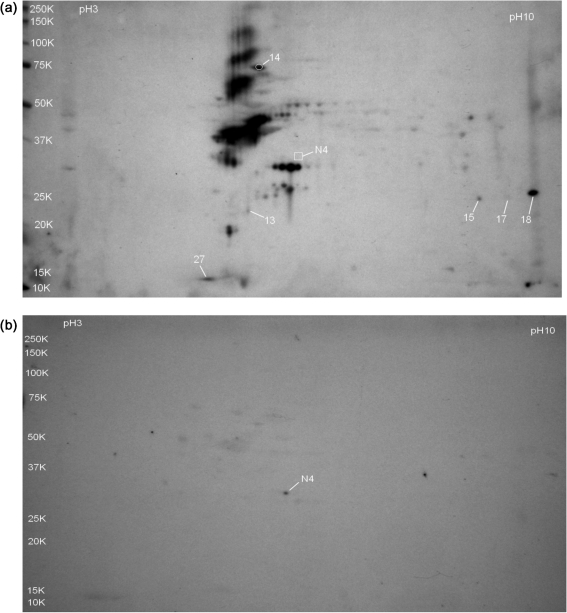
Initial attempts to identify immunogenic C. burnetii Nine Mile phase I proteins were conducted using whole-cell protein extracts of phase I organisms grown in embryonated chicken yolk sacs [phase I (egg)]. Four replicate pH 3–10 gradient blots were probed with immune sera and two replicate blots were probed with sham sera. Representative immune sera and sham sera probed blots are shown in Fig. 1(a and b), respectively. Spots identified as C. burnetii proteins are numbered. Sham-sera-reactive spot N4 had no detectable reactivity on any immune sera-probed blots and is included in Fig. 1(a) as a point of reference.
Fig. 1.
Immunoblots of whole-cell extracts of C. burnetii RSA 493 phase I grown in hen yolk sacs and separated by 2D gel electrophoresis on pH 3–10 IEF gradients and 10.5–14 % acrylamide SDS-PAGE gels. Labelled spots represent C. burnetii seroreactive proteins identified by LC-MS/MS and are listed in Table 1. Molecular mass standards (kDa) are indicated on the left. (a) Immunoblot probed with pooled immune guinea pig serum. Spot N4 was not reactive with immune sera and is included for reference. (b) Immunoblot probed with pooled serum from sham-immunized guinea pigs.
A total of 34 reactive spots were analysed by LC-MS/MS. Of the reactive spots, 24 were identified as chicken (Gallus) proteins (data not shown), seven spots were identified as C. burnetii proteins (spots 13–18 and 27, Fig. 1(a), Table 1), and three spots could not be identified. The antigenic C. burnetii proteins were CBU1706 (thioredoxin peroxidase), CBU1290 (DnaK), CBU1910 (com1) and CBU0229 (ribosomal protein L7/L12). CBU0495 (FabG) and CBU0780/CBU0955 (both response regulator GacA) were co-identified with CBU1910 in spots 15 and 18, respectively. The sham sera-reactive spot N4 was identified as CBU0263 (RpoA).
Table 1.
Phase I (egg) C. burnetii Nine Mile RSA 493 proteins identified by LC-MS/MS in seroreactive spots separated on pH 3–10 gradient gels
| Spot no.* | Locus tag† | Putative protein‡ | Protein score§ | Sequence coverage | No. of unique peptides with P value <0.05|| | Predicted Mr (kDa)/pI |
|---|---|---|---|---|---|---|
| 13 | CBU1706 | Thioredoxin peroxidase, AhpC/Tsa family | 71 | 20 % | 2 | 21.8/5.08 |
| 14 | CBU1290 | DnaK chaperone | 116 | 13 % | 1 | 70.7/5.14 |
| 15 | CBU0495 | 3-Oxoacyl-(acyl-carrier-protein) reductase FabG | 112 | 14 % | 2 | 26.3/7.63 |
| CBU1910 | com1 | 65 | 14 % | 1 | 27.6/9.08 | |
| 17 | CBU1910 | com1 | 55 | 14 % | 1 | 27.6/9.08 |
| 18 | CBU1910 | com1 | 165 | 23 % | 3 | 27.6/9.08 |
| CBU0780 | DNA-binding response regulators GacA | 54 | 6 % | 1 | 24.2/9.10 | |
| CBU0955 | 6 % | 1 | 24.1/9.52 | |||
| 27 | CBU0229 | Rp1L ribosomal protein L7/L12 | 84 | 38 % | 1 | 13.2/4.71 |
| N4 | CBU0263 | RpoA DNA-directed RNA polymerase, alpha | 205 | 12 % | 4 | 35.5/5.61 |
*Corresponds to spot shown in Fig. 1.
†TIGR annotation.
‡Described in Beare et al. (2009), Supplemental file 1, Supplemental Table S5.
§Mascot protein scores (standard scoring) are derived from combined peptide ion scores as a non-probabilistic basis for ranking protein hits.
||The number of unique peptides identified with ion scores >24 that indicate identity or extensive homology (P<0.05). Ion score is −10×log(P), where P is the probability that the match is a random event.
2D Electrophoresis, immunoblotting, and LC-MS/MS identification of phase II L929 proteins separated on pH 3–10 gradient gels
It is not uncommon to co-purify host cell proteins when isolating bacteria from an intracellular source (Ogawa et al., 2007). Since phase I organisms isolated from hen yolk sacs were used to immunize animals and generate immune sera, the detection of antibody-reactive chicken proteins in phase I extracts is not surprising. To reduce our immune sera reactivity to non-Coxiella antigens, whole-cell protein extracts of C. burnetii Nine Mile RSA 439 grown in murine L929 (phase II L929) cell cultures were used. RSA 439 is the phase II clone of RSA 493 phase I, with the only known difference in proteomes between these two strains resulting from the genomic deletion encompassing loci CBU0678 to CBU0698 [The Institute for Genomic Research (TIGR) annotation] associated with the truncated LPS of phase II organisms (Hoover et al., 2002). The only known difference in protein antigen seroreactivity between these two clones to phase-I-derived immune sera is due to a 21 kDa proteinase K-sensitive antigen that is uniquely reactive in extracts of phase I (Zhang et al., 2007).
A Sypro Ruby-stained pH 3–10 gradient 2D gel of C. burnetii Nine Mile RSA 439 phase II whole-cell protein extract is shown in Fig. 2(a) (representative of three gels). The numbered gel spots correspond to antibody-reactive spots on blots probed with immune sera (Fig. 2b, representative of three replicates) and sham sera (Fig. 2c). The C. burnetii proteins identified are listed in Table 2.
Fig. 2.
Whole-cell extracts of C. burnetii RSA 439 phase II grown in mouse L929 cell culture and separated by 2D gel electrophoresis on pH 3–10 IEF gradients and 10.5–14 % acrylamide SDS-PAGE gels. C. burnetii seroreactive proteins are listed in Table 2. Molecular mass standards (kDa) are indicated on the left. (a) Sypro Ruby-stained gel showing seroreactive spots. Immune sera-reactive spots are labelled with numbers alone. Sham sera-reactive spots are labelled with numbers with an N prefix. (b) Immunoblot probed with pooled immune guinea pig serum. Spot numbers with an N prefix were also sham sera-reactive. (c) Immunoblot probed with pooled serum from sham-immunized guinea pigs.
Table 2.
Phase II C. burnetii Nine Mile proteins identified by LC-MS/MS in seroreactive spots separated on pH 3–10 gradient gels
| Spot no.* | Locus tag† | Protein annotation‡ | Protein score§ | Sequence coverage | No. of unique peptides with P value <0.05|| | Predicted Mr (kDa)/pI |
|---|---|---|---|---|---|---|
| 9 | CBU0229 | Rp1L ribosomal protein L7/L12 | 391 | 63 % | 5 | 13.2/4.71 |
| 14 | CBU1290 | DnaK chaperone | 266 | 27 % | 4 | 70.7/5.14 |
| CBU0235 | FusA elongation factor EF-G | 247 | 26 % | 4 | 77.7/5.07 | |
| 15 | CBU0215 | Peptidase, C40/NplC-P60 family | 142 | 17 % | 2 | 58.0/6.06 |
| CBU0572 | Cytosol aminopeptidase | 228 | 36 % | 4 | 50.8/5.56 | |
| CBU1398 | SucB dihydrolipoamide succinyltransferase | 170 | 30 % | 2 | 45.9/5.54 | |
| 16 | CBU0236 | Elongation factor EF-Tu | 887 | 70 % | 14 | 43.5/5.32 |
| 17 | CBU0092 | Tol system component YbgF | 60 | 22 % | 1 | 32.0/5.49 |
| 18 | CBU1241 | Mdh malate dehydrogenase | 120 | 25 % | 2 | 35.4/5.07 |
| 19 | CBU0092 | Tol system component YbgF | 72 | 18 % | 1 | 32.0/5.49 |
| 20 | CBU1396 | SucD succinyl-CoA synthetase, alpha subunit | 187 | 36 % | 3 | 30.6/5.38 |
| CBU0092 | Tol system component YbgF | 89 | 26 % | 1 | 32.0/5.49 | |
| 32 | CBU1227 | Transcriptional regulatory protein QseB | 108 | 29 % | 1 | 25.2/7.88 |
| CBU1910 | com1 | 88 | 20 % | 2 | 27.6/9.08 | |
| 33 | CBU1227 | Transcriptional regulatory protein QseB | 129 | 22 % | 1 | 25.2/7.88 |
| CBU1910 | com1 | 101 | 13 % | 2 | 27.6/9.08 | |
| CBU0481 | Arginine transport ATP-binding protein ArtP | 66 | 34 % | 1 | 27.8/8.37 | |
| 34 | CBU0299 | Rph RNase PH | 136 | 36 % | 2 | 25.9/7.75 |
| 35 | CBU1227 | Transcriptional regulatory protein QseB | 163 | 22 % | 2 | 25.2/7.88 |
| CBU1910 | com1 | 218 | 29 % | 3 | 27.6/9.08 | |
| 36 | CBU1910 | com1 | 239 | 18 % | 3 | 27.6/9.08 |
| CBU0495 | 3-Oxoacyl-(acyl-carrier-protein) reductase FabG | 157 | 26 % | 2 | 26.3/7.63 | |
| CBU1227 | Transcriptional regulatory protein QseB | 74 | 17 % | 1 | 25.2/7.88 | |
| 37 | CBU1910 | com1 | 288 | 32 % | 4 | 27.6/9.08 |
| 37.5 | CBU1910 | com1 | 282 | 24 % | 4 | 27.6/9.08 |
| CBU0482 | Arginine-binding protein, periplasmic | 156 | 16 % | 2 | 29.8/9.40 | |
| 38 | CBU1910 | com1 | 331 | 31 % | 4 | 27.6/9.08 |
| CBU0481 | Arginine transport ATP-binding protein ArtP | 75 | 36 % | 1 | 27.8/8.37 | |
| 39 | CBU1910 | com1 | 145 | 24 % | 3 | 27.6/9.08 |
| 39.5 | CBU1910 | com1 | 167 | 28 % | 3 | 27.6/9.08 |
| 55 | CBU0937 | Conserved hypothetical protein | 513 | 33 % | 9 | 51.3/8.99 |
| N41 | CBU0263 | RpoA DNA-directed RNA polymerase, alpha | 283 | 47 % | 3 | 35.5/5.61 |
| N42 | CBU0236 | Elongation factor EF-Tu | 216 | 50 % | 2 | 43.5/5.32 |
| CBU0263 | RpoA RNA polymerase, alpha | 80 | 34 % | 0 | 35.5/5.61 | |
| N43 | CBU0263 | RpoA RNA polymerase, alpha | 156 | 37 % | 1 | 35.5/5.61 |
*Corresponds to spot shown in Fig. 2.
†TIGR annotation.
‡Described in Beare et al. (2009), Supplemental file 1, Supplemental Table S5.
§Mascot protein scores (standard scoring) are derived from combined peptide ion scores as a non-probabilistic basis for ranking protein hits.
||The number of unique peptides identified with ion scores >24 that indicate identity or extensive homology (P<0.05). Ion score is −10×log(P), where P is the probability that the match is a random event.
Examination of gel and blot images using the 3D viewer function of PDQuest showed distinct vertical troughs at the apparent boundary of the isoelectric gradient. Immunoreactive spots 1 and 49 were outside this boundary, migrated with the same apparent molecular mass as spot 14, and were similarly identified as CBU1290 (DnaK) and CBU0235 (EF-G). Reactive spots 3, 40 and 41 were also located outside the trough boundary and correlate with the molecular mass of reactive spots 37, 39 and 39.5, and were similarly identified as CBU1910 (com1). It appears that sample in the portion of the IPG strip beyond the electrodes is not focused in the first dimension but only in the second dimension. Reactive spot 2 was outside the trough, migrated with the same molecular mass as reactive spot 15, but was not identified as a protein in spot 15. This may be due to spot 2 being composed of all proteins with an approximate molecular mass of 46 kDa and the difficulties that this presents for detection by LC-MS/MS, particularly if the protein of interest is underrepresented or its peptides are more difficult to ionize. Spot 15 comprised multiple reactive doublets that were difficult to harvest individually and were harvested together to add some degree of confirmation to subsequent LC-MS/MS analysis. Spot 15 comprised CBU0215 (peptidase), CBU0572 (cytosol aminopeptidase) and CBU1398 (SucB). LC-MS/MS results for spots outside the trough boundary are not included in Table 2.
Several immune sera-reactive spots were identified as one protein: spot 9 (CBU0229, ribosomal protein L7/L12), spot 16 (CBU0236, EF-Tu), spot 17 (CBU0092, Tol system component YbgF), spot 18 (CBU1241, malate dehydrogenase), spot 19 (CBU0092, Tol–Pal system component YbgF), spot 34 (CBU0299, Rph RNase), spots 37, 39 and 39.5 (CBU1910, com1), and spot 55 (CBU0937, hypothetical exported protein). All other immune sera-reactive phase II spots analysed from pH 3–10 gradient gels identified more than one protein.
Spot 20 was identified as CBU1396 (SucD) and CBU0092 (Tol system component YbgF). CBU1910 (com1) was identified in all immune sera-reactive spots that focused on the cathode side of the gel with apparent molecular masses of approximately 25 kDa (spots 32, 33 and 35–39.5). The spots identified as only CBU1910 (com1) (spots 37, 39 and 39.5) all displayed the same apparent molecular mass that appeared to be slightly less than that of spots 32 and 33. Assuming the lack of post-translation modifications to com1, this indicates the primary protein represented by spots 32 and 33 is not com1. LC-MS/MS analysis of spots 32 and 33 did identify CBU1227 (transcriptional regulatory protein QseB) as the highest-scoring protein in both. Additionally, CBU0481 (arginine transport ATP-binding protein) was identified in spot 33. Spot 35 was identified as CBU1910 (com1) and CBU1227 (QseB). Spot 36 was a fairly abundant protein on stained gels and was identified as CBU1910 (com1), CBU0495 (FabG) and CBU1227 (QseB). The cathode side of spot 36 was designated spot 37, but more accurately should be described as the cathodic shoulder of spot 36 and was only identified as CBU1910 (com1). Similarly, spot 37.5 should be more accurately described as the anodic shoulder of spot 38. Analysis of both spot 37.5 and spot 38 identified CBU1910 (com1) as the highest Mascot-scoring protein. Spot 37.5 also contained CBU0482 (arginine-binding protein), while spot 38 co-identified CBU0481 (arginine transport ATP-binding protein). Spots 37, 37.5, 38 and 39 also identified CBU0578 (polyprenyl-phosphate β-d-glucosyltransferase) with Mascot scores that were significant (non-probalistic) by mudpit but not by standard scoring, and were omitted from Table 2. Interestingly, one peptide assigned to CBU0578 was detected in spots 37 and 37.5 with ion scores above the P<0.05 threshold, with intensities of 21 627 and 41 608, respectively, which were much higher than those of most peptides analysed in this study.
The blot probed with sera from sham-immunized guinea pigs was overexposed to assess cross-reacting antibody responses elicited by IFA in saline without antigen (Fig. 2c). Sham sera had very strong reactivity to three spots with the same molecular mass but slight differences in pI, spots N41, N42 and N43. The only protein identified in spots N41 and N43 was CBU0263 (RpoA). CBU0263 (RpoA) was also identified in spot N42. However, the highest-scoring protein identified in spot N42 was CBU0236 (EF-Tu). No immune-sera-reactive spots coincided with the location of spot N41 or spot N43, but the location of sham sera spot N42 did coincide with a weakly reactive spot on the immune sera-probed blot at a molecular mass less than the theoretical molecular mass of EF-Tu. Immune sera reactivity of this spot is most likely due to an unidentified protein and not EF-Tu. Strong sham sera reactivity was also detected on the horizontally diffuse spot N12, which was matched to immune sera-reactive spot 9, identified as CBU0229 (ribosomal protein L7/L12). Spots N50, N51 and N52 had weak sham sera reactivity and were also matched to immune sera-reactive spots, and were not analysed by LC-MS/MS. With the exception of spots N12, N42, N50, N51 and N52, no other numbered immune sera-reactive spots were matched to sham sera-reactive spots, indicating that the immune sera reactivity to these proteins is C. burnetii-specific.
To achieve greater spot separation and in an attempt to reduce the number of reactive spots with co-identified proteins, subsequent phase II (L929) extracts were run on narrower IEF gradients (pH 5–8) with longer second dimension runs.
2D Electrophoresis, immunoblotting, and LC-MS/MS identification of phase II L929 proteins separated on pH 5–8 gradient gels
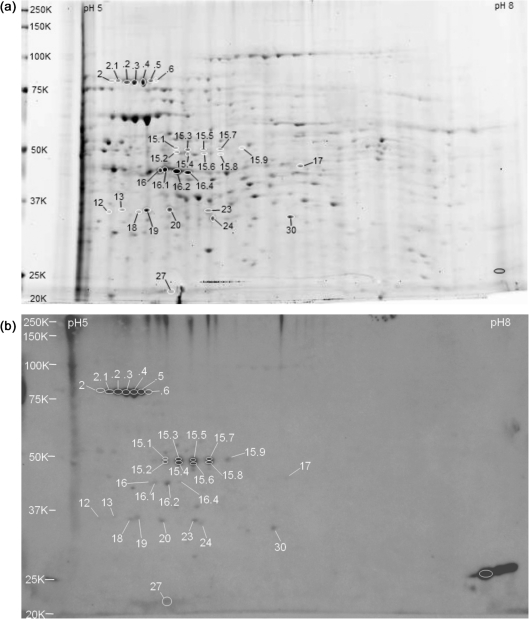
A Sypro Ruby-stained pH 5–8 gradient 2D gel of phase II proteins is shown in Fig. 3(a), representative of two replicates. The numbered spots on the gel correspond to matched antibody-reactive spots detected on the blots probed with immune sera shown in Fig. 3(b) (representative of two replicates) and sham sera shown in Fig. 3(c). Two immune sera-reactive spots were detected past the vertical trough boundaries. Based on our previous observations and identification of reactive proteins located outside the trough, these spots were not analysed by LC-MS/MS. Reactive proteins identified by LC-MS/MS are listed in Table 3.
Fig. 3.
Whole-cell extracts of C. burnetii RSA 439 phase II grown in mouse L929 cell culture and separated by 2D gel electrophoresis on pH 5–8 IEF gradients and 10.5–14 % acrylamide SDS-PAGE gels. C. burnetii seroreactive proteins are listed in Table 3. Molecular mass standards (kDa) are indicated on the left. (a) Sypro Ruby-stained gel showing seroreactive spots. Immune sera-reactive spots are labelled with numbers alone. Sham sera-reactive spots are labelled with numbers and an N prefix. (b) Immunoblot probed with pooled immune guinea pig serum. (c) Immunoblot probed with pooled serum from sham-immunized guinea pigs.
Table 3.
Phase II C. burnetii Nine Mile proteins identified by LC-MS/MS in seroreactive spots separated on pH 5–8 gradient gels
| Spot no.* | Locus tag† | Putative protein‡ | Protein score§ | Sequence coverage | No. of unique peptides with P value <0.05|| | Predicted Mr (kDa)/pI |
|---|---|---|---|---|---|---|
| 14.1 | CBU0235 | FusA elongation factor EF-G | 251 | 26 % | 4 | 77.7/5.07 |
| CBU1290 | DnaK chaperone | 94 | 26 % | 2 | 70.7/5.14 | |
| 14.2 | CBU0235 | FusA elongation factor EF-G | 316 | 12 % | 5 | 77.7/5.07 |
| CBU1290 | DnaK chaperone | 231 | 6 % | 4 | 70.7/5.14 | |
| 14.3 | CBU1290 | DnaK chaperone | 485 | 17 % | 7 | 70.7/5.14 |
| CBU0235 | FusA elongation factor EF-G | 167 | 8 % | 4 | 77.7/5.07 | |
| 14.4 | CBU1290 | DnaK chaperone | 644 | 34 % | 11 | 70.7/5.14 |
| 14.5 | CBU1290 | DnaK chaperone | 637 | 24 % | 11 | 70.7/5.14 |
| CBU0235 | FusA elongation factor EF-G | 129 | 3 % | 2 | 77.7/5.07 | |
| 15.1 | CBU1718 | GroEL chaperonin | 196 | 26 % | 3 | 58.2/5.14 |
| 15.3 | CBU0215 | Peptidase C40/NplC-P60 family | 105 | 13 % | 2 | 58.0/6.06 |
| 15.4 | CBU0215 | Peptidase C40/NplC-P60 family | 239 | 26 % | 4 | 58.0/6.06 |
| 15.5 | CBU0215 | Peptidase C40/NplC-P60 family | 231 | 17 % | 5 | 58.0/6.06 |
| CBU1398 | SucB dihydrolipoamide succinyltransferase | 105 | 10 % | 2 | 45.9/5.54 | |
| 15.6 | CBU0215 | Peptidase C40/NplC-P60 family | 183 | 25 % | 2 | 58.0/6.06 |
| CBU1398 | SucB dihydrolipoamide succinyltransferase | 61 | 24 % | 1 | 45.9/5.54 | |
| 15.7 | CBU1398 | SucB dihydrolipoamide succinyltransferase | 103 | 14 % | 2 | 45.9/5.54 |
| CBU0932 | GplK glycerol kinase | 93 | 10 % | 2 | 55.3/5.46 | |
| CBU0572 | Cytosol aminopeptidase | 86 | 18 % | 1 | 51.0/5.56 | |
| CBU0215 | Peptidase C40/NplC-P60 family | 65 | 11 % | 1 | 58.0/6.06 | |
| 15.8 | CBU0215 | Peptidase C40/NplC-P60 family | 114 | 13 % | 2 | 58.0/6.06 |
| CBU1398 | SucB dihydrolipoamide succinyltransferase | 87 | 12 % | 2 | 45.9/5.54 | |
| 16.2 | CBU0236 | Elongation factor EF-Tu | 877 | 65 % | 14 | 43.5/5.32 |
| 16.3 | CBU0236 | Elongation factor EF-Tu | 789 | 64 % | 12 | 43.5/5.32 |
| 16.4 | CBU0236 | Elongation factor EF-Tu | 1120 | 77 % | 18 | 43.5/5.32 |
| 17 | CBU0236 | Elongation factor EF-Tu | 75 | 26 % | 1 | 43.5/5.32 |
| 18 | CBU1241 | Mdh malate dehydrogenase | 160 | 11 % | 2 | 35.4/5.07 |
| 19 | CBU0092 | Tol system component YbgF | 250 | 39 % | 5 | 32.0/5.49 |
| 19.1 | CBU1396 | SucD succinyl-CoA synthetase, alpha subunit | 68 | 7 % | 2 | 30.6/5.38 |
| 20 | CBU0092 | Tol system component YbgF | 154 | 35 % | 4 | 32.0/5.49 |
| 20.1 | CBU1396 | SucD succinyl-CoA synthetase, alpha subunit | 399 | 35 % | 7 | 30.6/5.38 |
| 23 | CBU1385 | Elongation factor EF-Ts | 327 | 40 % | 8 | 31.8/5.85 |
| 24 | CBU0528 | RpsA ribosomal protein S1 | 280 | 13 % | 5 | 62.1/5.28 |
| 26 | CBU0858 | NadE NAD+ synthase glutamine-dependent | 632 | 26 % | 12 | 60.3/5.63 |
| 27 | CBU1943 | ATP synthase F1, alpha subunit | 153 | 9 % | 2 | 56.8/5.80 |
| 28 | CBU0103 | Peptidase, M20A family | 347 | 27 % | 6 | 52.8/5.35 |
| 29 | CBU0103 | Peptidase, M20A family | 562 | 37 % | 10 | 52.8/5.35 |
| 34 | CBU0750 | Arabinose-5-phosphate isomerase | 174 | 15 % | 2 | 34.5/6.79 |
| 38 | CBU1910 | com1 | 211 | 30 % | 4 | 27.6/9.08 |
| CBU1260 | OmpA-like protein | 99 | 16 % | 1 | 26.2/9.61 | |
| CBU0307 | OmpA-like protein | 84 | 15 % | 1 | 24.9/9.91 | |
| NS | CBU1718 | GroEL chaperonin | 511 | 45 % | 8 | 58.2/5.14 |
| N43 | CBU0263 | RpoA RNA polymerase, alpha subunit | 338 | 25 % | 5 | 35.5/5.61 |
*Corresponds to spot shown in Fig. 3.
†TIGR annotation.
‡Described in Beare et al. (2009), Supplemental file 1, Supplemental Table S5.
§Mascot protein scores (standard scoring) are derived from combined peptide ion scores as a non-probabilistic basis for ranking protein hits.
||The number of unique peptides identified with ion scores >24 that indicate identity or extensive homology (P<0.05). Ion score is −10×log(P), where P is the probability that the match is a random event.
Immune sera-reactive spot 14 seen on pH 3–10 gradient gels resolved into five reactive pI isoforms (spots 14.1–14.5) when separated on pH 5–8 gradient gels. These spots were all identified as comprising CBU1290 (DnaK) and CBU0235 (EF-G), with the exception of spot 14.4, in which only CBU1290 (DnaK) was identified. Reactive spot 16, identified as CBU0236 (EF-Tu) on the pH 3–10 gel, resolved into three reactive pI isoforms (spots 16.2, 16.3 and 16.4), all of which were identified as CBU236 (EF-Tu). Reactive spot 20 on the pH 3–10 gradient gel (identified as CBU1396 and CBU0092) resolved into two spots when separated on pH 5–8 gradient gels, designated spots 20 and 20.1. Spots 20 and 19 from the pH 5–8 gradient gel were identified as CBU0092 (Tol system component YbgF). Spots 20.1 and 19.1 were identified as CBU1396 (SucD). Spot 18 was identified as CBU1241 (malate dehydrogenase). CBU0236 EF-Tu was identified in spot 17. However, the apparent molecular mass of spot 17 was much lower than the theoretical molecular mass of EF-Tu and most likely is not responsible for the reactivity of this spot. Spot 23 was identified as CBU1385 (EF-Ts) and spot 34 identified as CBU0750 (arabinose-5-phosphate isomerase). Spots 28 and 29 were identified as CBU0103 (peptidase M20A). Weakly immune sera-reactive spots 24, 26 and 27 were identified as CBU0528 (ribosomal protein S1), CBU0858 (glutamine-dependent NAD+ synthase) and CBU1943 (ATP synthase F1, alpha subunit), respectively. Spots 15.1–15.8, analogous to spot 15 on the pH 3–10 gradient gel, appeared as reactive doublets. Interestingly, spot 15.1 was identified as CBU1718 (GroEL), and focused at a lower molecular mass and with a slightly more basic pI than its theoretical values. No proteins could be conclusively identified in spot 15.2. CBU0215 (peptidase) and CBU1398 (SucB) were identified in all other remaining 15 series spots with the exception of spots 15.3 and 15.4, which were identified as CBU0215 only. Spot 15.7 additionally contained CBU0932 (glycerol kinase) and CBU0572 (cytosol aminopeptidase).
The spot designated NS was very abundant on all gels but was not noticeably reactive above non-specific background levels on previous immune sera-probed blots. The reactivity of spot NS shown in Fig. 3(b) is greater than that previously detected. In view of the abundance of this protein on gels, the level of reactivity of this spot was considered weakly reactive. Spot NS was identified as CBU1718 (GroEL) with an apparent molecular mass and pI that matched the predicted values in contrast to its migration in spot 15.1. The amount of GroEL in spot NS and its weak seroreactivity make it unlikely that the reactivity of the much less abundant spot 15.1 would be due to GroEL.
The prominent immune sera-reactive spots identified as CBU1910 (com1) on pH 3–10 blots were not as evident on pH 5–8 immune sera-probed blots. Only one strongly reactive spot, spot 38, was identified as CBU1910 (com1). The expected pI of CBU1910 (com1) is 9.08 and therefore outside the pH range of pH 5–8 gradient strips, and the higher-pI isoforms of com1 may have migrated out of the IPG strip (Poznanovic et al., 2005).
Blots probed with sham sera reacted very strongly with three spots migrating with same apparent molecular mass but different pIs, identical to the pH 3–10 sham sera-probed blots, and were similarly designated spots N41, N42 and N43. The spot-matching function of PDQuest aligned these spots just to the cathode side of spot 20.1. However, only spot N43 could be unambiguously matched on the corresponding gel and was identified as CBU0263 (RpoA).
2D Electrophoresis, immunoblotting, and LC-MS/MS identification of phase I (L929) proteins separated on pH 5–8 gradient gels
Since a high percentage of reactive spots from phase I (egg) extracts were identified as Gallus proteins, whole-cell extracts of phase I grown in murine L929 cells were assayed. Due to the limited quantity of phase I (L929) cells, extracted protein was sufficient for only two pH 5–8 gradient gels. One gel was used for spot harvesting and the other was transferred and probed with immune sera. The stained pH 5–8 gradient gel is shown in Fig. 4(a). The numbered spots on the gel correspond to matched seroreactive spots detected on the immune sera-probed blot shown in Fig. 4(b). Reactive C. burnetii proteins identified by LC-MS/MS are listed in Table 4.
Fig. 4.
Whole-cell extracts of C. burnetii RSA 493 phase I grown in mouse L929 cell culture and separated by 2D gel electrophoresis on pH 5–8 IEF gradients and 10.5–14 % acrylamide SDS-PAGE gels. C. burnetii seroreactive proteins are listed in Table 4. Molecular mass standards (kDa) are indicated on the left. (a) Sypro Ruby-stained gel showing seroreactive spots. (b) Immunoblot probed with pooled immune guinea pig serum.
Table 4.
Phase I (L929) C. burnetii Nine Mile proteins identified by LC-MS/MS in seroreactive spots separated on pH 5–8 gradient gels
| Spot no.* | Locus tag† | Putative protein‡ | Protein score§ | Sequence coverage | No. of unique peptides with P value <0.05|| | Predicted Mr (kDa)/pI |
|---|---|---|---|---|---|---|
| 2 | CBU0235 | FusA elongation factor EF-G | 189 | 9 % | 3 | 77.7/5.07 |
| CBU1290 | DnaK chaperone | 59 | 7 % | 1 | 70.7/5.14 | |
| 2.1 | CBU0235 | FusA elongation factor EF-G | 223 | 18 % | 4 | 77.7/5.07 |
| CBU1290 | DnaK chaperone | 135 | 11 % | 2 | 70.7/5.14 | |
| 2.2 | CBU1290 | DnaK chaperone | 420 | 20 % | 8 | 70.7/5.14 |
| CBU0235 | FusA elongation factor EF-G | 270 | 12 % | 6 | 77.7/5.07 | |
| 2.3 | CBU1290 | DnaK chaperone | 660 | 32 % | 13 | 70.7/5.14 |
| CBU0235 | FusA elongation factor EF-G | 144 | 8 % | 2 | 77.7/5.07 | |
| 2.4 | CBU1290 | DnaK chaperone | 367 | 24 % | 5 | 70.7/5.14 |
| CBU0235 | FusA elongation factor EF-G | 57 | 2 % | 1 | 77.7/5.07 | |
| 2.5 | CBU1290 | DnaK chaperone | 183 | 11 % | 5 | 70.7/5.14 |
| 2.6 | CBU1290 | DnaK chaperone | 211 | 16 % | 3 | 70.7/5.14 |
| 12 | CBU0092 | Tol system component YbgF | 76 | 9 % | 1 | 32.0/5.49 |
| 13 | CBU1241 | Mdh malate dehydrogenase | 40 | 6 % | 1 | 35.4/5.07 |
| 15.1.2 | CBU1718 | GroEL chaperonin | 383 | 29 % | 5 | 58.2/5.14 |
| CBU1398 | SucB dihydrolipoamide succinyltransferase | 109 | 11 % | 3 | 45.9/5.54 | |
| CBU0215 | Peptidase, C40/NplC-P60 family | 40 | 1 % | 1 | 58.0/5.14 | |
| 15.3 | CBU1398 | SucB dihydrolipoamide succinyltransferase | 107 | 9 % | 2 | 45.9/5.54 |
| CBU1718 | GroEL chaperonin | 105 | 9 % | 2 | 58.2/5.14 | |
| CBU0215 | Peptidase, C40/NplC-P60 family | 42 | 5 % | 1 | 58.0/5.14 | |
| 15.4 | CBU1398 | SucB dihydrolipoamide succinyltransferase | 258 | 22 % | 5 | 45.9/5.54 |
| CBU1718 | GroEL chaperonin | 93 | 6 % | 1 | 58.2/5.14 | |
| CBU0215 | Peptidase, C40/NplC-P60 family | 85 | 6 % | 2 | 58.0/5.14 | |
| 15.5 | CBU1398 | SucB dihydrolipoamide succinyltransferase | 165 | 15 % | 2 | 45.9/5.54 |
| CBU0932 | GplK glycerol kinase | 64 | 2 % | 1 | 55.3/5.46 | |
| CBU0572 | Cytosol aminopeptidase | 59 | 5 % | 2 | 50.8/5.56 | |
| CBU0215 | Peptidase, C40/NplC-P60 family | 39 | 1 % | 1 | 58.0/6.06 | |
| 15.6 | CBU0215 | Peptidase, C40/NplC-P60 family | 84 | 4 % | 2 | 58.0/6.06 |
| 15.7 | CBU1398 | SucB dihydrolipoamide succinyltransferase | 150 | 14 % | 3 | 45.9/5.54 |
| CBU0932 | GplK glycerol kinase | 62 | 2 % | 1 | 55.3/5.46 | |
| CBU0572 | Cytosol aminopeptidase | 33 | 2 % | 1 | 50.8/5.56 | |
| 15.8 | CBU1398 | SucB dihydrolipoamide succinyltransferase | 63 | 11 % | 2 | 45.9/5.54 |
| 15.9 | CBU0932 | GplK glycerol kinase | 94 | 5 % | 1 | 55.3/5.46 |
| 16 | CBU0236 | Elongation factor EF-Tu | 192 | 27 % | 3 | 43.5/5.32 |
| 16.1 | CBU0236 | Elongation factor EF-Tu | 408 | 45 % | 8 | 43.5/5.32 |
| 16.2 | CBU0236 | Elongation factor EF-Tu | 613 | 53 % | 11 | 43.5/5.32 |
| 16.4 | CBU0236 | Elongation factor EF-Tu | 495 | 42 % | 10 | 43.5/5.32 |
| 17 | CBU0140 | Cell division protein FtsA | 91 | 8 % | 3 | 44.3/5.79 |
| 18 | CBU0092 | Tol system component YbgF | 84 | 16 % | 1 | 32.0/5.49 |
| 19 | CBU1241 | Mdh malate dehydrogenase | 200 | 17 % | 4 | 35.4/5.07 |
| 20 | CBU0092 | Tol system component YbgF | 73 | 9 % | 2 | 32.0/5.49 |
| 23 | CBU0092 | Tol system component YbgF | 80 | 7 % | 2 | 32.0/5.49 |
| 24 | CBU1396 | SucD succinyl-CoA synthetase, alpha | 206 | 31 % | 2 | 30.6/5.38 |
| 27 | CBU1706 | Thioredoxin peroxidase, AhpC/Tsa family | 46 | 5 % | 1 | 21.8/5.08 |
| 30 | CBU1385 | Elongation factor EF-Ts | 399 | 55 % | 6 | 31.8/5.85 |
*Corresponds to spot shown in Fig. 4.
†TIGR annotation.
‡Described in Beare et al. (2009), Supplemental file 1, Supplemental Table S5.
§Mascot protein scores (standard scoring) are derived from combined peptide ion scores as a non-probabilistic basis for ranking protein hits.
||The number of unique peptides identified with ion scores >24 that indicate identity or extensive homology (P<0.05). Ion score is −10×log(P), where P is the probability that the match is a random event.
The strongly reactive spots 2–2.6 were identified as CBU1290 (DnaK) and CBU0235 (EF-G), with only CBU1290 identified in spots 2.5 and 2.6. All spots in the 16 series were identified as CBU0236 (EF-Tu). A series of strongly reactive doublet spots corresponding to spots 15.1–15.8 were also evident. Manually excising spots 15.1 and 15.2 resulted in both spots being excised in the same gel piece at which point the individual spots could not be separated. Consequently, spots 15.1 and 15.2 were analysed by LC-MS/MS together, designated spot 15.1.2, and identified as CBU1718 (GroEL), CBU1398 (SucB) and CBU0215 (peptidase). Similar to previous LC-MS/MS analyses of the 15 series of spots, CBU1398 (SucB) and CBU0215 (peptidase) were identified across the series of spots and generally were identified together. Additionally, CBU0932 (glycerol kinase) and CBU0572 (cytosol aminopeptidase) were identified in spots 15.5 and 15.7. Also, spot 15.9 in this series displayed stronger immune sera reactivity on this blot and was identified as CBU0932 (glycerol kinase). Upon examination of previous immune sera-probed blots, a reactive spot analogous to spot 15.9 was distinguishable, although with less reactivity than on the present blot.
Immune sera-reactive spots 13 and 19 were identified as CBU1241 (malate dehydrogenase). Spots 12, 18, 20 and 23 all migrated with the same apparent molecular mass and all were identified as CBU0092 (Tol system component YbgF). Spot 24 was identified as CBU1396 (SucD). Spot 30, identified as CBU1385 (EF-Ts), had greater immune sera reactivity on this immunoblot than other immune sera-probed blots. The immune sera reactivity of spot 27 was diffuse but had a slightly defined focus, which was harvested as spot 27 and identified as CBU1706 (thioredoxin peroxidase). Weakly reactive spot 17 was identified as CBU0140 (cell division protein FtsA). Very strong reactivity was seen at the cathode edge (unlabelled circle), and had been consistently identified as CBU1910 on previous phase I and phase II pH 5–8 gradient gels, but was not harvested from this gel for LC-MS/MS analysis.
Summary of C. burnetii Nine Mile proteins identified in immunogenic 2D spots
Most reactive proteins were identified on multiple blots. However, CBU0140 (FtsA), CBU0299 (RNase PH), CBU0528 (ribosomal protein S1), CBU0750 (arabinose-5-phosphate isomerase), CBU0858 (glutamine-dependent NAD+ synthase), CBU1718 (GroEL) and CBU1943 (ATP synthase F1, alpha subunit) had sufficient intensity to be deemed immunoreactive on only one blot. CBU0103 (peptidase, M20A) was identified from one gel but was detectable on multiple pH 5–8 gradient immune sera-probed blots. A summary of the C. burnetii proteins identified in immune sera-reactive spots is shown in Table 5. CBU0229 (ribosomal protein L7/L12) was reactive with both immune sera and sham sera and is omitted from Table 5.
Table 5.
Summary of C. burnetii Nine Mile proteins specific to immune sera-reactive 2D spots
| Locus tag* | Putative protein† | Found in phase I, phase II or both | Human sera-reactive‡ |
|---|---|---|---|
| Immunoreactive proteins | |||
| CBU0092 | Tol system periplasmic component YbgF | I | 1,2 |
| CBU0103 | Peptidase, M20A family | II | |
| CBU0236 | Elongation factor EF-Tu | Both | 1,3 |
| CBU0299 | Rph RNase PH | II | |
| CBU0750 | Arabinose-5-phosphate isomerase | II | |
| CBU0932 | GplK glycerol kinase | Both | |
| CBU0937 | Hypothetical exported protein | II | 1 |
| CBU1241 | Mdh malate dehydrogenase | Both | |
| CBU1290 | DnaK chaperone | Both | 1,2 |
| CBU1385 | Elongation factor EF-Ts | Both | |
| CBU1396 | SucD, succinyl-CoA synthetase alpha chain | Both | |
| CBU1398 | SucB, dihydrolipoamide succinyltransferase E2 of 2-oxoglutarate dehydrogenase complex | Both | 2,4 |
| CBU1706 | Thioredoxin peroxidase, AhpC/Tsa antioxidant | I | 1 |
| CBU1910 | com1 outer-membrane protein | Both | 1,2 |
| Weakly immunoreactive proteins | |||
| CBU0140 | Cell division protein FtsA | I | |
| CBU0528 | Ribosomal protein S1P (RpsA) | II | 1 |
| CBU0858 | NAD+ synthase, glutamine-dependent | II | |
| CBU1718 | GroEL chaperonin | Both | 1,3,5 |
| CBU1943 | ATP synthase F1, alpha subunit | II | 3 |
| Proteins co-identified with a protein listed above | |||
| CBU0215 | Peptidase, C40/NplC-P60 family | Both | |
| CBU0235 | FusA elongation factor EF-G | Both | 1 |
| CBU0307 | OmpA-like outer-membrane protein | II | 1,4 |
| CBU0481 | Arginine transport ATP-binding protein ArtP | II | |
| CBU0482 | Arginine-binding protein, periplasmic | II | |
| CBU0495 | 3-Oxoacyl-ACP reductase FabG | I | |
| CBU0572 | Cytosol aminopeptidase | Both | 1 |
| CBU0780, CBU0955 | Response regulator GacA | I | |
| CBU1227 | Transcriptional regulatory protein QseB | II | |
| CBU1260 | OmpA-like transmembrane domain protein | II | 1 |
*TIGR annotation.
†Described in Beare et al. (2009), Supplemental file 1, Supplemental Table S5.
‡References: 1, Sekeyová et al. (2009); 2, Vigil et al. (2010); 3, Coleman et al. (2007); 4, Beare et al. (2008); 5, Williams et al. (1990).
Immunoreactive spots in the 15 series co-identified as CBU0215 (peptidase) and CBU1398 (SucB), and a definitive assignment of seroreactivity to either was not possible. Similarly, CBU0572 (cytosol aminopeptidase), CBU0235 (EF-G), CBU0482 (arginine-binding protein), CBU0495 (FabG), CBU1227 (transcriptional regulatory protein QseB), CBU0307 (OmpA-like), CBU0481 (arginine transport ATP-binding protein), CBU0780/CBU0955 (response regulator GacA) and CBU1260 (OmpA-like) were co-identified with seroreactive proteins, and their seroreactivity also cannot be conclusively determined in this study.
DISCUSSION
Nine novel seroreactive C. burnetii Nine Mile proteins were identified in the present study. Seven displayed good reactivity with immune sera, CBU0932 (glycerol kinase), CBU1241 (malate dehydrogenase), CBU1396 (SucD), CBU1385 (EF-Ts), CBU0299 (RNase PH), CBU0750 (arabinose-5-phosphate isomerase) and CBU0103 (peptidase, M20A family), while two were detected with fairly low reactivity, CBU0140 (cell division protein FtsA) and CBU0858 (glutamine-dependent NAD+ synthase). Eight other immune sera-reactive proteins identified in the present study were identified by Sekeyová et al. (2009) as reactive with sera from humans infected with C. burnetii using a 2D immunoproteomics platform: CBU0092 (Tol component YbgF), CBU0236 (EF-Tu), CBU0937 (hypothetical exported protein), CBU1290 (DnaK), CBU1706 (thioredoxin peroxidase), CBU1718 (GroEL) and CBU1910 (com1). Sekeyová et al. (2009) also identified CBU0235 (EF-G), CBU0572 (cytosol aminopeptidase), CBU0307 (OmpA-like) and CBU1260 (OmpA-like) as seroreactive. While we were able to identify these four additional proteins in immune sera-reactive spots, we were unable to conclusively assign reactivity due to their co-identification with proteins that were determined to be immune sera-reactive.
Several reactive proteins identified in this study were also reactive against convalescent human sera in other studies. Coleman et al. (2007) used a 2D platform to identify CBU0236 (EF-Tu), CBU1718 (GroEL), CBU1943 (ATP synthase F1, alpha subunit) and CBU0229 (ribosomal protein L7/L12) as seroreactive. Using a protein array platform, CBU1398 (SucB), CBU0307 (OmpA-like), CBU1910 (com1), CBU0092 (Tol system YbgF) and CBU0229 (L7/L12) were identified as immunoreactive with Q fever patient sera (Beare et al., 2008; Vigil et al., 2010). Vigil et al. (2010) also found CBU1290 (DnaK) to be strongly reactive with some patient sera but also strongly reactive against one naïve-group serum. The reactivity of CBU1398 (SucB) demonstrated in previous studies suggests that this protein is responsible for the reactivity seen in spots that comprised additional co-identified proteins in the present study. The similarity in antibody response between humans and guinea pigs against C. burnetii antigens supports the use of guinea pigs as an animal model for Q fever subunit vaccine studies.
The finding that sham sera reacted strongly with CBU0263 (RpoA) and CBU0229 (L7/L12) was somewhat of a surprise, and may be due to an antibody response to a microbial protein homologue originating from the normal guinea pig flora. Interestingly, Bartonella henselae ribosomal protein L7/L12 is reactive with serum from healthy human donors (Eberhardt et al., 2009).
CBU1718 (GroEL) has frequently been detected as reactive with immune sera. Despite being a very abundant protein on all gels in the present study, it was weakly reactive on only one immunoblot. Interestingly, Williams et al. (1990) found that convalescent and chronic-phase Q fever human sera both reacted with C. burnetii heat-shock protein HtpB (GroEL), although acute-phase sera did not.
Most immunoreactive proteins identified in this study have experimental molecular masses and pIs that are near their theoretical values, and their locations are in general agreement with those of other proteins identified in 2D proteomic reference maps of C. burnetii Nine Mile (Samoilis et al., 2007; Sekeyová et al., 2009). The most notable exception is the apparent molecular mass of CBU1718 (GroEL) identified in spot 15.1. This molecular mass is less than the expected molecular mass of GroEL which was identified in the spot labelled NS and focused where expected. Interestingly, a lower-molecular-mass GroEL isoform has also been identified in Brucella abortus and migrates with a molecular mass and pI similar to those of GroEL identified in spot 15.1 (Connolly et al., 2006). The location where CBU0215 (peptidase) was consistently identified is at a molecular mass approximately 8 kDa less than expected, which may reflect potential processing to the mature form, the use of a start or stop codon different from that used for the predictive protein calculations, or protein degradation. The immune sera-reactive spot identified as CBU0299 (ribonuclease PH) had a theoretical molecular mass of 26 kDa but migrated with an experimental molecular mass that appeared closer to 30 kDa. CBU1910 (com1) has a theoretical molecular mass of 27.6 kDa but migrates with an apparent molecular mass of 25.7 kDa due to cleavage of the leader sequence, in agreement with our results (Hendrix et al., 1993).
The proteins identified as immunoreactive have a variety of functions and most have been associated with the membrane or cell wall. Multiple proteins identified as immunoreactive are involved in carbon metabolism. CBU1398 (SucB), CBU1396 (SucD) and CBU1241 (malate dehydrogenase) catalyse reactions in the formation of oxaloacetate from succinyl-CoA, and CBU0932 (glycerol kinase) functions in the utilization of carbon from lipids. While generally considered cytosolic proteins, the association of these carbon-metabolizing proteins with cell membranes has been described for a number of intracellular pathogens (Connolly et al., 2006; Boonjakuakul et al., 2007; Janovská et al., 2007). CBU0092 is the periplasmic component YbgF of the Tol–Pal system involved in maintaining outer membrane integrity. CBU0937 is a hypothetical exported protein that has an unknown function. CBU0103 (peptidase) belongs to the M20A peptidase family. Members of the M20A peptidase family have diverse specificities, but protein blast analysis of CBU0103 shows the most identity (59 %) to succinyl-diaminopimelate desuccinylase. CBU0750 (arabinose-5-phosphate isomerase) catalyses the interconversion of ribulose 5-phosphate and arabinose 5-phosphate, and is required for the synthesis of the 3-deoxy-d-manno-octulosonic acid (Kdo) component of LPS (Tzeng et al., 2002). CBU0858 (glutamine-dependent NAD+ synthase) amidates nicotinic acid adenine dinucleotide (NaAD+) to NAD+, which is involved in a diverse range of redox reactions. CBU0299 (RpH RNase PH) is an exonuclease that generates mature tRNA by removal of an excess RNA extension on the 3′ end of tRNA. CBU1290 (DnaK) belongs to the heat-shock protein 70 family (Hsp70) and is highly conserved among prokaryotes and eukaryotes. Hsp70 functions as a molecular chaperone and in the transport of proteins across membranes. DnaK can be membrane-associated in pathogenic bacteria and participates in binding to the host cell surface, which may facilitate cellular invasion (Knaust et al., 2007; Xolalpa et al., 2007). CBU0236 (EF-Tu), CBU1385 (EF-Ts) and CBU0528 (ribosomal protein S1) are involved in protein translation. EF-Tu may function as a cytoskeleton component, maintaining the shape of prokaryotes (Mayer, 2003). EF-Tu can also be surface-exposed and bind to host cell surfaces, potentially facilitating intracellular invasion (Balasubramanian et al., 2008; Barel et al., 2008). CBU0528 (S1) is a component of the 30S ribosome and can be associated with membranes or free in the cytosol, depending on the destination of the nascent peptide.
The cross-reactivity of C. burnetii proteins is problematic for the serodiagnosis of Q fever. C. burnetii antisera cross-react with recombinant SucB from Bartonella vinsonii subsp. burkhoffii (Gilmore et al., 2003), and antisera against Escherichia coli EF-G and ribosomal protein L7/L12 are cross-reactive with C. burnetii homologues (Baca, 1978). Additionally, a monoclonal antibody directed against CBU0236 (EF-Tu) cross-reacts with recombinant EF-Tu from Chlamydia trachomatis (Seshadri et al., 1999). The identification of immunogenic C. burnetii proteins with low homology to other proteins is paramount for the unambiguous serodiagnosis of Q fever. CBU0092 (Tol–Pal component YbgF) and CBU0937 (hypothetical exported protein) have low identities to other proteins (34 % for both), and may provide additional stringency in the serodiagnosis of Q fever.
The number of Q fever subunit vaccine studies is fairly limited. A native purified 67 kDa C. burnetii protein is protective in both mice and guinea pigs (Zhang et al., 1994), and Williams et al. (1990) found that native purified CBU0311 (P1) induces a protective response in mice. However, recombinant C. burnetii CBU1910 (com1), CBU0311 (P1), CBU0630 (Cb-Mip) and P28 are unable to protect mice from subsequent challenge (Zhang & Samuel, 2003). Shannon & Heinzen (2009) have proposed that the protection induced by native but not recombinant C. burnetii antigens may be due to the requirement for native epitopes or other protective Coxiella antigens, possibly LPS, associated with purified native protein. Interestingly, contaminating Francisella tularensis LPS has been found to be required for protection when using Hsp60 (GroEL) purified from F. tularensis (Hartley et al., 2004). Additionally, a recombinant fusion protein of CBU0311 (P1) and CBU1718 (GroEL) is protective in mice, while individual recombinant P1 and GroEL are not (Li et al., 2005), indicating that an effective subunit vaccine against Q fever may require multiple antigenic subunits. Clearly there is a need to identify and characterize additional C. burnetii antigens in order to expedite the development of a safe and efficacious Q fever subunit vaccine. The C. burnetii immunoreactive antigens identified in the present study may represent potential subunit vaccine candidates. Induction of an IgG response suggests the involvement of T cells, which are required for the development of a protective immune response against C. burnetii (Zhang et al., 2007). T cell responses to these antigens need to be characterized to further validate their potential as subunit vaccine candidates.
Acknowledgments
We thank the Laboratory for Biotechnology and Bioanalysis 2 at Washington State University, Pullman, WA, and the Environmental Biotechnology Institute at the University of Idaho, Moscow, ID, for LC-MS/MS analyses. We also thank Adam Vigil at the University of California, Irvine, for his critical review of the manuscript. This work was supported by National Institutes of Health National Institutes of Allergy and Infectious Disease Grant R01-AI057768.
Abbreviations
ACN, acetonitrile
CMRV, chloroform, methanol residue vaccine
IFA, incomplete Freund's adjuvant
IPG, immobilized pH gradient
LC-MS/MS, liquid chromatography-tandem MS
WCKI, whole-cell killed vaccine
WCV, whole-cell vaccine
References
- Andoh, M., Naganawa, T., Hotta, A., Yamaguchi, T., Fukushi, H., Masegi, T. & Hirai, K. (2003). SCID mouse model for lethal Q fever. Infect Immun 71, 4717–4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andoh, M., Zhang, G., Russell-Lodrigue, K. E., Shive, H. R., Weeks, B. R. & Samuel, J. E. (2007). T cells are essential for bacterial clearance, and gamma interferon, tumor necrosis factor alpha, and B cells are critical for disease development in Coxiella burnetii infection in mice. Infect Immun 75, 3245–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arricau Bouvery, N., Souriau, A., Lechopier, P. & Rodolakis, A. (2003). Experimental Coxiella burnetii infection in pregnant goats: excretion routes. Vet Res 34, 423–433. [DOI] [PubMed] [Google Scholar]
- Ascher, M. S., Williams, J. C. & Berman, M. A. (1983). Dermal granulomatous hypersensitivity in Q fever: comparative studies of the granulomatous potential of whole cells of Coxiella burnetii phase I and subfractions. Infect Immun 42, 887–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baca, O. G. (1978). Comparison of ribosomes from Coxiella burnetii and Escherichia coli by gel electrophoresis, protein synthesis, and immunological techniques. J Bacteriol 136, 429–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian, S., Kannan, T. R. & Baseman, J. B. (2008). The surface-exposed carboxyl region of Mycoplasma pneumoniae elongation factor Tu interacts with fibronectin. Infect Immun 76, 3116–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal, R., Deobald, L. A., Crawford, R. L. & Paszczynski, A. J. (2009). Proteomic detection of proteins involved in perchlorate and chlorate metabolism. Biodegradation 20, 603–620. [DOI] [PubMed] [Google Scholar]
- Barel, M., Hovanessian, A. G., Meibom, K., Briand, J. P., Dupuis, M. & Charbit, A. (2008). A novel receptor–ligand pathway for entry of Francisella tularensis in monocyte-like THP-1 cells: interaction between surface nucleolin and bacterial elongation factor Tu. BMC Microbiol 8, 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beare, P. A., Chen, C., Bouman, T., Pablo, J., Unal, B., Cockrell, D. C., Brown, W. C., Barbian, K. D., Porcella, S. F. & other authors (2008). Candidate antigens for Q fever serodiagnosis revealed by immunoscreening of a Coxiella burnetii protein microarray. Clin Vaccine Immunol 15, 1771–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beare, P. A., Unsworth, N., Andoh, M., Voth, D. E., Omsland, A., Gilk, S. D., Williams, K. P., Sobral, B. W., Kupko, J. J. & other authors (2009). Comparative genomics reveal extensive transposon-mediated genomic plasticity and diversity among potential effector proteins within the genus Coxiella. Infect Immun 77, 642–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonjakuakul, J. K., Gerns, H. L., Chen, Y. T., Hicks, L. D., Minnick, M. F., Dixon, S. E., Hall, S. C. & Koehler, J. E. (2007). Proteomic and immunoblot analyses of Bartonella quintana total membrane proteins identify antigens recognized by sera from infected patients. Infect Immun 75, 2548–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman, S. A., Fisher, E. R., Cockrell, D. C., Voth, D. E., Howe, D., Mead, D. J., Samuel, J. E. & Heinzen, R. A. (2007). Proteome and antigen profiling of Coxiella burnetii developmental forms. Infect Immun 75, 290–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly, J. P., Comerci, D., Alefantis, T. G., Walz, A., Quan, M., Chafin, R., Grewal, P., Mujer, C. V., Ugalde, R. A. & Delvecchio, V. G. (2006). Proteomic analysis of Brucella abortus cell envelope and identification of immunogenic candidate proteins for vaccine development. Proteomics 6, 3767–3780. [DOI] [PubMed] [Google Scholar]
- Eberhardt, C., Engelmann, S., Kusch, H., Albrecht, D., Hecker, M., Autenrieth, I. B. & Kempf, V. A. J. (2009). Proteomic analysis of the bacterial pathogen Bartonella henselae and identification of immunogenic proteins for serodiagnosis. Proteomics 9, 1967–1981. [DOI] [PubMed] [Google Scholar]
- Gilmore, R. D., Jr, Carpio, A. M., Kosoy, M. Y. & Gage, K. L. (2003). Molecular characterization of the sucB gene encoding the immunogenic dihydrolipoamide succinyltransferase protein of Bartonella vinsonii subsp. berkhoffi and Bartonella quintana. Infect Immun 71, 4818–4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackstadt, T., Peacock, M. G., Hitchkock, P. J. & Cole, R. L. (1985). Lipopolysaccharide variation in Coxiella burnetii: intrastrain heterogeneity in structure and antigenicity. Infect Immun 48, 359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley, M. G., Green, M., Choules, G., Rogers, D., Rees, D. G. C., Newstead, S., Sjostedt, A. & Titball, R. W. (2004). Protection afforded by heat shock protein 60 from Francisella tularensis is due to copurified lipopolysaccharide. Infect Immun 72, 4109–4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix, L. R. & Mallavia, L. P. (1984). Active transport of proline by Coxiella burnetii. J Gen Microbiol 130, 2857–2863. [DOI] [PubMed] [Google Scholar]
- Hendrix, L. R., Mallavia, L. P. & Samuel, J. E. (1993). Cloning and sequencing of Coxiella burnetii outer membrane protein gene com1. Infect Immun 61, 470–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover, T. A., Culp, D. W., Vodkin, M. H., Williams, J. C. & Thompson, H. A. (2002). Chromosomal DNA deletions explain phenotypic characteristics of two antigenic variants, phase II and RSA 514 (Crazy), of the Coxiella burnetii Nine Mile strain. Infect Immun 70, 6726–6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzo, A. A., Marmion, B. P. & Hackstadt, T. (1991). Analysis of the cells involved in the lymphoproliferative response to Coxiella burnetii antigens. Clin Exp Immunol 85, 98–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janovská, S., Pávková, I., Hubálek, M., Lenčo, J., Macela, A. & Stulík, J. (2007). Identification of immunoreactive antigens in membrane proteins enriched fraction from Francisella tularensis LVS. Immunol Lett 108, 151–159. [DOI] [PubMed] [Google Scholar]
- Knaust, A., Weber, M. V. R., Hammerschmidt, S., Bergmann, S., Frosch, M. & Kurzai, O. (2007). Cytosolic proteins contribute to surface plasminogen recruitment of Neisseria meningitidis. J Bacteriol 189, 3246–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q., Niu, D., Wen, B., Chen, M., Qiu, L. & Zhang, J. (2005). Protective immunity against Q fever induced with recombinant P1 antigen fused with HspB of Coxiella burnetii. Ann N Y Acad Sci 1063, 130–142. [DOI] [PubMed] [Google Scholar]
- Lopez, J. E., Siems, W. F., Palmer, G. H., Brayton, K. A., McGuire, T. C., Norimine, J. & Brown, W. C. (2005). Identification of novel antigenic proteins in a complex Anaplasma marginale outer membrane immunogen by mass spectrometry and genome mapping. Infect Immun 73, 8109–8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurin, M. & Raoult, D. (1999). Q fever. Clin Microbiol Rev 12, 518–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, F. (2003). Cytoskeletons in prokaryotes. Cell Biol Int 27, 429–438. [DOI] [PubMed] [Google Scholar]
- Noh, S. M., Brayton, K. A., Brown, W. C., Norimine, J., Munske, G. R., Davitt, C. M. & Palmer, G. H. (2008). Composition of the surface proteome of Anaplasma marginale and its role in protective immunity induced by outer membrane immunization. Infect Immun 76, 2219–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa, M., Renesto, P., Azza, S., Moinier, D., Fourquet, P., Gorvel, J. P. & Raoult, D. (2007). Proteome analysis of Rickettsia felis highlights the expression profile of intracellular bacteria. Proteomics 7, 1232–1248. [DOI] [PubMed] [Google Scholar]
- Poznanovic, S., Schwall, G., Zengerling, H. & Cahill, M. A. (2005). Isoelectric focusing in serial immobilized pH gradient gels to improve protein separation in proteomic analysis. Electrophoresis 26, 3185–3190. [DOI] [PubMed] [Google Scholar]
- Russell-Lodrigue, K. E., Zhang, G. Q., McMurray, D. N. & Samuel, J. E. (2006). Clinical and pathologic changes in a guinea pig aerosol challenge model of acute Q fever. Infect Immun 74, 6085–6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell-Lodrigue, K. E., Andoh, M., Poels, W. J., Shive, H. R., Weeks, B. R., Zhang, G. Q., Tersteeg, C., Masegi, T., Hotta, A. & other authors (2009). Coxiella burnetii isolates cause genogroup-specific virulence in mouse and guinea pig models of acute Q fever. Infect Immun 77, 5640–5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samoilis, G., Psaroulaki, A., Vougas, K., Tselentis, Y. & Tsiotis, G. (2007). Analysis of whole cell lysate from the intracellular bacterium Coxiella burnetii using two gel-based protein separation techniques. J Proteome Res 6, 3032–3041. [DOI] [PubMed] [Google Scholar]
- Sekeyová, Z., Kowalczewska, M., Decloquement, P., Pelletier, N., Špitalská, E. & Raoult, D. (2009). Identification of protein candidates for the serodiagnosis of Q fever endocarditis by an immunoproteomic approach. Eur J Clin Microbiol Infect Dis 28, 287–295. [DOI] [PubMed] [Google Scholar]
- Seshadri, R., Hendrix, L. R. & Samuel, J. E. (1999). Differential expression of translational elements by life cycle variants of Coxiella burnetii. Infect Immun 67, 6026–6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshadri, R., Paulsen, I. T., Eisen, J. A., Read, T. D., Nelson, K. E., Nelson, W. C., Ward, N. L., Tettelin, H., Davidsen, T. M. & other authors (2003). Complete genome sequence of the Q-fever pathogen Coxiella burnetii. Proc Natl Acad Sci U S A 100, 5455–5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon, J. G. & Heinzen, R. A. (2009). Adaptive immunity to the obligate intracellular pathogen Coxiella burnetii. Immunol Res 43, 138–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon, J. G., Cockrell, D. C., Takahashi, K., Stahl, G. L. & Heinzen, R. A. (2009). Antibody-mediated immunity to the obligate intracellular bacterial pathogen Coxiella burnetii is Fc receptor- and complement-independent. BMC Immunol 10, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng, Y. L., Datta, A., Strole, C., Kumar Kollie, V. S., Birck, M. R., Taylor, W. P., Carlson, R. W., Woodard, R. W. & Stephens, D. S. (2002). KpsF is the arabinose-5-phosphate isomerase required for 3-deoxy-d-manno-octulosonic acid biosynthesis and for both lipooligosaccharide assembly and capsular polysaccharide expression in Neisseria meningiditis. J Biol Chem 277, 24103–24113. [DOI] [PubMed] [Google Scholar]
- Vigil, A., Ortega, R., Nakajima-Sasaki, R., Pablo, J., Molina, D. M., Chao, C. C., Chen, H. W., Ching, W. M. & Felgner, P. L. (2010). Genome-wide profiling of humoral immune response to Coxiella burnetii infection by protein array. Proteomics 10, 2259–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodkin, M. H. & Williams, J. C. (1986). Overlapping deletion in two spontaneous phase variants of Coxiella burnetii. J Gen Microbiol 132, 2587–2594. [DOI] [PubMed] [Google Scholar]
- Voth, D. E. & Heinzen, R. A. (2007). Lounging in a lysosome: the intracellular lifestyle of Coxiella burnetii. Cell Microbiol 9, 829–840. [DOI] [PubMed] [Google Scholar]
- Waag, D. M., England, M. J. & Pitt, M. L. M. (1997). Comparative efficacy of a Coxiella burnetii chloroform : methanol residue (CMR) vaccine and a licensed cellular vaccine (Q-Vax) in rodents challenged by aerosol. Vaccine 15, 1779–1783. [DOI] [PubMed] [Google Scholar]
- Waag, D. M., England, M. J., Tammariello, R. F., Byrne, W. R., Gibbs, P., Banfield, C. M. & Pitt, M. L. M. (2002). Comparative efficacy and immunogenicity of Q fever chloroform : methanol residue (CMR) and phase I cellular (Q-Vax) vaccines in cyanomolgus monkeys challenged by aerosol. Vaccine 20, 2623–2634. [DOI] [PubMed] [Google Scholar]
- Williams, J. C. & Cantrell, J. L. (1982). Biological and immunological properties of Coxiella burnetii vaccines in C57BL/10ScN endotoxin-nonresponder mice. Infect Immun 35, 1091–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, J. C., Peacock, M. G. & McCaul, T. F. (1981). Immunological and biological characterization of Coxiella burnetii, phases I and II, separated from host components. Infect Immun 32, 840–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, J. C., Damrow, T. A., Wagg, D. M. & Amano, K. I. (1986). Characterization of phase I Coxiella burnetii chloroform-methanol residue vaccine that induces active immunity against Q fever in C57BL/10 mice. Infect Immun 51, 851–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, J. C., Hoover, T. A., Waag, D. M., Banerjee-Bhatnagar, N., Bolt, C. R. & Scott, G. H. (1990). Antigenic structure of Coxiella burnetii. A comparison of lipopolysaccharide and protein antigens as vaccines against Q fever. Ann N Y Acad Sci 590, 370–380. [DOI] [PubMed] [Google Scholar]
- Xolalpa, W., Vallecillo, A. J., Lara, M., Mendoza-Hernandez, G., Comini, M., Spallek, R., Singh, M. & Espitia, C. (2007). Identification of novel bacterial plasminogen-binding proteins in the human pathogen Mycobacterium tuberculosis. Proteomics 7, 3332–3341. [DOI] [PubMed] [Google Scholar]
- Zhang, G. Q. & Samuel, J. E. (2003). Identification and cloning potentially protective antigens of Coxiella burnetii using sera from mice experimentally infected with Nine Mile phase I. Ann N Y Acad Sci 990, 510–520. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. X., Zhi, N., Yu, S. R., Li, Q. J., Yu, G. Q. & Zhang, X. (1994). Protective immunity induced by 67 K outer membrane protein of phase I Coxiella burnetii in mice and guinea pigs. Acta Virol 38, 327–332. [PubMed] [Google Scholar]
- Zhang, G., Russell-Lodrigue, K. E., Andoh, M., Zhang, Y., Hendrix, L. R. & Samuel, J. E. (2007). Mechanisms of vaccine-induced protective immunity against Coxiella burnetii infection in BALB/c mice. J Immunol 179, 8372–8380. [DOI] [PubMed] [Google Scholar]